Abstract
The HSP70 family of heat shock proteins consists of molecular chaperones of approximately 70kDa in size that serve critical roles in protein homeostasis. These adenosine triphosphatases unfold misfolded or denatured proteins and can keep these proteins in an unfolded, folding-competent state. They also protect nascently translating proteins, promote the cellular or organellar transport of proteins, reduce proteotoxic protein aggregates and serve general housekeeping roles in maintaining protein homeostasis. The HSP70 family is the most conserved in evolution, and all eukaryotes contain multiple members. Some members of this family serve specific organellar- or tissue-specific functions; however, in many cases, these members can function redundantly. Overall, the HSP70 family of proteins can be thought of as a potent buffering system for cellular stress, either from extrinsic (physiological, viral and environmental) or intrinsic (replicative or oncogenic) stimuli. As such, this family serves a critical survival function in the cell. Not surprisingly, cancer cells rely heavily on this buffering system for survival. The overwhelming majority of human tumors overexpress HSP70 family members, and expression of these proteins is typically a marker for poor prognosis. With the proof of principle that inhibitors of the HSP90 chaperone have emerged as important anticancer agents, intense focus has now been placed on the potential for HSP70 inhibitors to assume a role as a significant chemotherapeutic avenue. In this review, the history, regulation, mechanism of action and role in cancer of the HSP70 family are reviewed. Additionally, the promise of pharmacologically targeting this protein for cancer therapy is addressed.
Overview
The human HSP70 family consists of at least eight members; some of these have organelle-specific localization and tissue-specific expression, but in general, many members are believed to have overlapping function in the cell (reviewed in refs 1–3). HSP70 proteins all play a role in the mediation of correct protein folding, and consequently, in the maintenance of protein homeostasis. These proteins also enhance cell survival following a multitude of stresses, including elevated temperature, hypoxia, oxidative stress, altered pH, heavy metals and others. This survival role is reflected in the ability of HSP70 to buffer the toxicity of denatured and misfolded proteins that accumulate during stress. Unlike HSP90, there are no truly specific ‘client’ proteins of HSP70. Instead, this protein binds to stretches of exposed hydrophobic residues on unfolded and misfolded proteins, holding them in an intermediately folded state and preventing their aggregation. Unlike HSP90, HSP70 can directly unfold misfolded proteins, in an adenosine triphosphate (ATP)-dependent fashion. The eight members of this family are highly homologous, exhibiting between 52 and 99% amino acid identity (Figure 1). At least one HSP70 protein is found in most organisms, including archaebacteria, and most organisms have multiple members. This protein is widely regarded to be the most conserved protein in evolution; for example, the human HSP70 protein and the bacterial orthologue DnaK share 50% amino acid identity. Although HSP70 is generally considered to be a stress-induced survival protein, it should be noted that several family members also play key housekeeping roles. These roles include facilitating protein transport between organelles and subcellular compartments, folding of newly synthesized proteins, dissolution of protein complexes and uncoating of clathrin-coated vesicles.
Fig. 1.
Diagram of HSP70 functional domains, along with percent homology of each domain to HSP70-1, characteristics of messenger RNA expression and stress-induced regulation.
Though they share considerable homology and function, the eight human members of the HSP70 family can be loosely categorized by differences in subcellular localization, tissue-specific expression and stress-induced expression (Figure 1). Two members are organelle-specific and are required for promoting protein folding and maintaining proteostasis in these respective organelles: the endoplasmic reticulum-specific form is called HSP70-5 (also known as BiP or Grp78), which plays a role in sensing misfolded proteins in the endoplasmic reticulum and signaling endoplasmic reticulum stress in cells. The mitochondrion contains a protein with similar function, known as HSP70-9 (also called mortalin or Grp75). The remaining six members of the HSP70 family encode proteins that localize predominantly to the cytosol, but can also localize to the nucleus. At least three HSP70 family members show distinct patterns of tissue expression and clearly play predominant roles in those tissues. For example, HSP70-2 protein is constitutively expressed at extremely low levels in all cells but is abundantly expressed in the brain and the testes (4). Similarly, the HSP70-1t gene (also called HSPA1L) is predominantly expressed in the testes (1,5). Another member, HSP70-6, is undetectable in most tissues but is abundantly expressed in blood and immune cells (5) and can be induced after severe stress insults (1). The remaining three members can be distinguished by their abundance during stress. The gene encoding HSC70 (also called HSP70-8) is constitutively expressed as low levels in all cells and is not induced by heat shock or other stresses. This protein is believed to play the bulk of essential housekeeping functions of the HSP70 family and also plays a key role in chaperone-mediated autophagy; not surprisingly, knockout of this gene is lethal in mice (6). The remaining two genes are closely linked in the major histocompatibility class III gene cluster on chromosome 6p21.3. These are highly homologous tandem genes called HSP70-1a and -b (also called HSP70A1A and A1B); because these two proteins only differ by two amino acids, they are generally referred to together as HSP70-1. HSP70-1 can account for up to 2% of total protein in a stressed cell. In general, HSC70 is believed to carry out the bulk homeostatic functions of this chaperone, whereas the two stress-induced genes referred to as HSP70-1 are believed to carry out the major stress-induced survival role of this protein family.
Structure
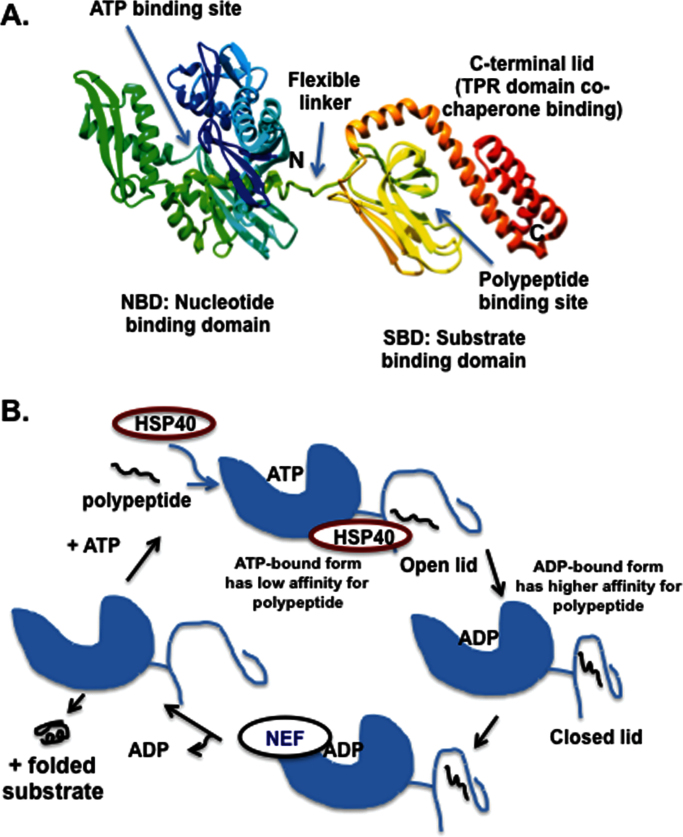
The HSP70 proteins have a highly conserved domain structure comprising the following major domains: an ~44kDa N-terminal nucleotide-binding domain (NBD) that exhibits ATPase activity and is highly conserved, a middle flexible linker region; an ~15kDa substrate-binding domain (SBD), which interacts with stretches of hydrophobic amino acids in peptides and an ~10kDa α-helical C-terminal domain that is believed to form a ‘lid’ that closes over the substrate and also mediates co-chaperone binding (7–10). The last four amino acids comprise an EEVD (amino acid designations) motif capable of interacting with tetratricopeptide repeat domain containing-proteins (see structure, Figure 2A). Interestingly, the C terminus of this protein is the least conserved; this probably allows different HSP70 family members to interact with distinct co-chaperones.
Fig. 2.
(A) The solution structure of Escherichia coli DnaK in the ADP and peptide-bound state (PDB 2KHO), with the NBD, SBD and C-terminal helical ‘lid’ denoted. (B) Functional cycles of HSP70 interaction with the J-domain protein HSP40, substrate polypeptide and NEF; nucleotide hydrolysis is facilitated by HSP40 and increases the affinity for substrate polypeptide by mediating the closure of the C-terminal lid. NEFs assist with ADP release, and this facilitates substrate release.
HSP70 is essentially a protein unfolding machine, which binds and releases stretches of hydrophobic amino acids in a regulated, ATP-hydrolysis-driven cycle (Figure 2B). As isolated molecules, the ATP hydrolysis rates are extremely slow (11), and co-chaperones are important for the function of HSP70. For example, members of the co-chaperone family HSP40 (also called J-domain proteins) bind to the NBD of HSP70 and stimulate the ATPase activity of this protein (12). Co-chaperones that bind to the NBD-like BAG-1 are nucleotide exchange factors (NEFs) that catalyze the release of adenosine diphosphate (ADP) (reviewed in ref. 13). Finally, a third class of co-chaperones bind to the C terminus of HSP70 and mediate the recruitment and fate of substrates; these proteins, like Chip and HOP, are extremely important in regulating HSP70 function (reviewed in ref. 14). HOP is a tetratricopeptide repeat domain protein that couples the interaction of HSP70 with HSP90. Chip, another tetratricopeptide repeat protein, inhibits the HSP40-stimulated ATPase activity of HSP70, and by virtue of its E3-ubiquitin ligase activity is believed to re-focus HSP70 to direct grossly misfolded proteins for ubiquitylation and degradation, instead of refolding. There is extensive communication and movement between the SBD and NBD (15,16). Specifically, peptide binding to the SBD induces a conformational change that is propagated back to the NBD, stimulating ATP hydrolysis. Hydrolysis of ATP is then signaled back to the SBD, resulting in closing of the α-helical ‘lid’ and enhancing substrate affinity. Finally, ADP–ATP exchange is mediated by NEFs like BAG-1, and this catalyzes substrate release. Allostery in the HSP70 protein is expertly reviewed in ref. 17.
Regulation of Hsp70 gene expression
Only three of the eight HSP70 family members show stress-inducible expression. The two closely linked genes, referred to as Hsp70-1, are the major stress-induced members of this family. The homolog HSP70-6 is also stress-induced, but only by severe stress (18). The human HSP70-1 promoter has been extensively analyzed; the basal expression of this gene is controlled by a TATA box and two CCAAT boxes, along with a serum response element, a metal response element and binding sites for the transcription factors Sp1, c-MYC and Foxa1 (19–21). In unstressed conditions, HSP70-1 is expressed at low levels but is cell cycle regulated and preferentially expressed in the G1 and S phases of the cell cycle (22,23). Rapid expression of HSP70-1 in response to cellular stresses, including heat shock, can occur in minutes and is mediated in large part by at least two heat shock elements (HSEs) in the promoter of this gene. The HSE is the binding site for the heat-shock inducible transcription factor heat shock factor 1 (HSF1; reviewed in ref. 24). In an unstressed cell, the HSP70-1 promoter is maintained with RNA polymerase II poised in a pre-initiated but paused state (25). Additionally, the transcription factor HSF1 is kept in an inactive complex in the cytosol and is complexed predominantly as inactive monomers with HSP90 and HSP70 (24). Following elevated temperature or other stresses, the accumulation of misfolded proteins in the cell is believed to redirect HSP90 to bind to misfolded proteins, freeing HSF1 and allowing it to translocate to the nucleus, trimerize and bind to HSEs in the promoters of HSP70-1 and HSP70-6 (reviewed in refs 24 and 26). The binding of HSF1 to HSEs in the HSP70 promoter triggers RNA polymerase II to escape from the paused pre-initiation complex and commence elongation (25). Interestingly, the HSP70-1a and -1b genes are intronless (27); therefore, they are not subject to the general repression of gene splicing that occurs during elevated temperatures or other stresses.
It has been acknowledged for many years that HSP70-1 is frequently overexpressed in transformed, relative to normal, cells. Although it seems likely that this gene is overexpressed in cancer because of the high levels of proteotoxic stress in tumors and subsequent activation of HSF1, it is also of note that there are at least three cancer-relevant regulators of HSF1 that have been identified, which likely contribute to the frequent overexpression of the Hsp70 gene in cancer cells. These are the mammalian target of rapamycin kinase mTORC1, the deacetylase and longevity factor SIRT1 and the p53 family member ΔNp63α. Each of these proteins is frequently activated in cancer cells, and all of them positively regulate HSF1 expression and/or function (28–30).
Knock-out mice are viable and fertile but show genomic instability and stress-induced phenotypes
Although the knockout of HSC70 is lethal, the knockout of both copies of HSP70.1 and HSP70.3 (equivalent to the linked Hsp70-1a and -1b genes in human) in the mouse resulted in progeny that were viable and fertile, though somewhat smaller than wild-type littermates (31). Not surprisingly, these knock-out mice were markedly more sensitive to multiple stresses, including ischemia, pancreatitis and inflammation (32–35). Interestingly, HSP70.1/70.3 homozygous knock-out mice also showed evidence for increased genomic instability; cells from the bone marrow of these mice showed evidence for higher rates of chromosomal aberrations compared with normal littermate mice, along with decreased telomerase activity and increased end-to-end chromosome fusions (31). These data implicate HSP70 in the control of genomic stability. Along these lines, researchers seeking to understand the process of mitosis have noted HSP70 protein decorating mitotic spindles and centrosomes for many years (36–39). These and other findings suggest that, like heat shock and other stresses, genomic instability and aneuploidy might be stresses that are signaled to the cell and are protected by the expression and function of HSP70.
HSP70 and cancer
The accumulated data on HSP70-1 strongly argue that this chaperone can play a causal role in cancer initiation. Specifically, overexpression of HSP70-1 was found to confer tumorigenicity to mouse fibrosarcoma cells, and to render these cells resistant to killing by cytotoxic T cells and macrophages in vitro (40). Overexpression of HSP70-1 in T cells of transgenic mice leads to an increased rate of T-cell lymphoma in these mice (41). Finally, in immortalized Rat-1 fibroblasts, overexpression of HSP70-1 was shown to confer loss of contact inhibition, along with growth in soft agar and tumorigenicity in vivo (42). Importantly, in cells expressing a tetracycline-regulated HSP70-1, the transformed phenotype required continuous expression of this gene, as reversal of tetracycline administration led to loss of transformed properties (42). More recently, HSP70-1 was found to be required for transformation mediated by the Her2/neu receptor (43), and overexpression of HSP70-2 has been seen in breast cancer cell lines (44). These combined findings strongly indicate that HSP70 can function as an oncogene.
Although some evidence implicates HSP70-2 in human cancer, the predominant form of HSP70 overexpressed in human cancer is the major, cytosolic, stress-induced gene HSP70-1, it is this form that will be discussed throughout the remainder of this review (unless otherwise noted); hereafter for simplicity, it will be referred to as HSP70 (gene) or HSP70 (protein). Evidence that HSP70 is overexpressed in cancer, and that high expression of this chaperone correlates with increased tumor grade and poor prognosis, is extensive (reviewed in refs 45 and 46). For example, HSP70 overexpression is a marker of early hepatocellular and prostate cancer (47,48). Overexpression of this protein is a marker for advanced disease and lymph node metastasis in colorectal carcinoma (49,50) and breast cancer (50). HSP70 overexpression correlates with Ki67 positivity in lung cancer (51), is a marker for undifferentiated ovarian cancer (52) and is correlated with increased proliferation and tumor size in uterine cervical cancer (53). Levels of HSP70 can be correlated with clinical stage in melanoma (54) and oral cancer (55), as well as increased grade and shorter overall survival in bladder cancer (56). Finally, high staining of HSP70 is associated with poor survival and worse prognosis in acute myeloid leukemia, and in cancers of the breast, endometrium and cervix/uterus (57–60). It is generally held that elevated HSP70 expression in transformed cells protects these cells from apoptosis, from the stress associated with aneuploidy and accumulated mutant proteins and from the proteotoxic stress associated with abnormally rapid proliferation.
In addition to correlations between HSP70 level with tumor grade and poorer survival, there are also correlations with drug resistance. High HSP70 was found to mediate cisplatin resistance in prostate cancer in vitro (61) and imatinib resistance in chronic myeloid leukemia, both in vitro and in vivo (62). Additionally, overexpression of HSP70 in a fibrosarcoma cell line conferred resistance to topotecan and gemcitabine (63). Interestingly, HSP70 also plays a role in the resistance of cancer cells to immune-mediated destruction. For example, overexpression of HSP70 was found to protect tumor cells from cytotoxicity by monocytes (64). Moreover, HSP70 in tumor-derived exosomes was found to stimulate STAT3 activity in myeloid-derived suppressor cells and to contribute to myeloid-derived suppressor cell expansion, thereby restraining tumor immune surveillance (65).
Antisense inhibition of Hsp70 is cytotoxic to tumor but not normal cells
Jäättelä was the first to show that silencing of HSP70 with antisense RNA-induced massive cell death in breast cancer cell lines, but was non-toxic to non-tumorigenic breast epithelial cells, as well as normal human fibroblasts. This cell death occurred in a manner that was not dependent on p53, or inhibited by Bcl2 (66). Similar findings, indicating that antisense-mediated HSP70 inhibition in lung, oral, colon, prostate, liver and brain cancer cell lines caused apoptosis, were reported by others (67–69). Notably, injection of xenograft tumors with adenoviral vectors expressing antisense RNA to HSP70 effectively eradicated tumors in immunocompromised mice (69). Although it seems clear that silencing HSP70 alone, in the absence of silencing HSC70, is toxic to some cancer cell lines, Workman reported that in Hct116 colorectal cancer cells and A2780 ovarian cancer cells, only ‘dual’ silencing of both HSP70 and HSC70 was accompanied by decreased cell viability (70). Therefore, there are likely to be cell-specific differences in the sensitivity to HSP70 inhibition.
Cancer-relevant pathways regulated by HSP70
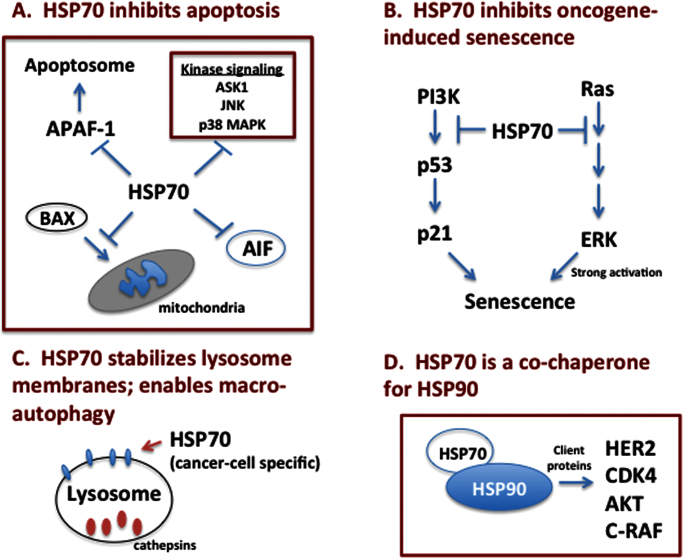
The currently available data indicate that cancer cells become ‘addicted’ to HSP70 through this chaperone’s activity on multiple cell signaling and survival pathways. Four of these cancer-relevant activities of HSP70 are described in this sudy and are summarized in Figure 3.
Fig. 3.
Cancer-relevant pathways affected by HSP70. (A) HSP70 inhibits the intrinsic and extrinsic apoptosis pathways, by inhibiting BAX translocation to mitochondria, the recruitment of APAF-1 to the apoptosome, the activity of stress-induced kinases and the function of AIF-1. (B) HSP70 inhibits both p53-dependent and -independent senescence. (C) HSP70 localizes to lysosome membranes specifically in cancer cells, stabilizes lysosome function and allows for autophagy, a key cancer survival pathway. (D) HSP70 is an obligate co-chaperone for HSP90 and is essential for the proper folding and function of HSP90 chaperone proteins like HER2, AKT, CDK4 and C-RAF.
Apoptosis
Overexpression of HSP70 can provide a selective survival advantage to tumor cells in part due to its ability to inhibit multiple pathways of cell death, including both intrinsic and extrinsic apoptosis. With regard to the intrinsic apoptosis pathway, HSP70 can bind directly to the pro-apoptotic BCL2 family member BAX and prevent it from translocating to mitochondria, where the latter disrupts mitochondrial membranes following an apoptotic stimulus (71,72). Additionally, interaction with HSP70 prevents the recruitment of APAF-1 and procaspase-9 to the apoptosome (73). With regard to the extrinsic pathway, in BCR-ABL-expressing cells, HSP70 binds to the death receptors DR4 and DR5, inhibiting the assembly of the death-inducing signaling complex, or DISC (74). HSP70 also inhibits BH3 interacting domain death agonist-mediated apoptosis, downstream of the tumor necrosis factor-α pathway (75). HSP70 binds and inhibits several stress-induced kinases, including apoptosis signal regulating kinase, c-jun N-terminal kinase and p38 mitogen-activated protein kinase; in each case, the interaction inhibits the ability of the kinase to function in apoptosis (76–78). Finally, HSP70 inhibits caspase-independent cell death, by binding directly to apoptosis-inducing factor (AIF) and inhibiting AIF-induced chromatin condensation (79).
Senescence
Sherman’s group showed that HSP70 also plays a role in the control of senescence in tumor cells. Silencing of this gene in tumor but not normal cells using small interfering RNA can induce senescence, mediated in part by reduced stability of MDM2 (the ubiquitin ligase for p53) along with concomitant stabilization of p53 and transactivation of the p53 target gene p21 (80). Silencing of HSP70 in cancer cells can also induce senescence in a p53-independent manner, by virtue of the ability of this protein to bind and inhibit extracellular-regulated kinases (ref. 81). In sum, HSP70 clearly plays a protective role against both p53-dependent and independent senescence. This role is likely to be highly cancer-relevant, as evident from findings that HSP70 is required for HER2/neu-mediated transformation of mammary epithelial cells; specifically, in the absence of HSP70, overexpression of HER2/neu induces senescence (43).
Autophagy
Jäättelä first showed that stress-inducible HSP70 localizes to the lysosomal and endosomal membranes of cancer but not normal cells. Following cancer-specific translocation to the lysosomal compartment, this group found that HSP70 promotes cell survival by inhibiting lysosomal membrane permeabilization, a hallmark of stress-induced death (82,83). This group went on to show that HSP70 stabilizes lysosomes by binding to the endo-lysosomal lipid bis(monoacylglycero)phosphate, an essential co-factor for lysosomal sphingolipid catabolism (84). Consistent with an essential role for lysosomes in autophagy, Leu et al. (85) were the first to show that silencing of HSP70 in cancer cells led to impaired autophagy, as evident by abnormal accumulation and oligomerization of the autophagy scaffold protein p62SQSTM1. Autophagy is a critical survival pathway for cancer cells, and autophagy inhibitors like hydroxychloroquine can inhibit tumor progression (86) as well as synergize with chemotherapeutic drugs (87). Notably, Leu et al. (85) identified the compound 2-phenylethynesulfonamide (PES) as a potent and effective HSP70 inhibitor; later, our group analyzed several derivatives of this compound and found a consistent correlation between autophagy inhibition and anticancer activity of PES derivatives (88). The combined data support a cancer-specific survival role for HSP70 in lysosome stabilization and autophagy.
HSP90
Despite great expectations and a much-anticipated value in the clinic, the development of HSP70 inhibitors has lagged far behind that of HSP90 inhibitors. Unfortunately, the clinical efficacy of HSP90 inhibitors has been generally disappointing. One possible reason for this is that treatment of cancer cell lines with HSP90 inhibitors generally leads to significant activation of HSF1 and upregulation of HSP70; indeed, upregulation of HSP70 is a key biomarker for the inhibition of HSP90 (89). Because of these findings, it has been generally hypothesized that combining HSP70 inhibitors with HSP90 inhibitors might lead to enhanced efficacy of the latter. Interestingly, however, it was discovered that HSP70 inhibition ‘alone’ effectively disrupts the HSP90 chaperone system. This is believed to be because HSP70 and the constitutive family member HSC70 are critical co-chaperones for HSP90 and are both involved in the delivery of client proteins to HSP90 (90). Workman first showed that targeting both HSC70 and HSP70 with small interfering RNA resulted in proteasome-dependent degradation of HSP90 client proteins (including C-RAF and CDK4), along with significant apoptosis in tumor but not normal cells (70). Similarly, Massey et al. (91) showed that the compound Ver-155008, which inhibits both HSC70 and HSP70 (as well as the mitochondrial HSP70 family member Grp78), induces degradation of the HSP90 client proteins HER2 and RAF-1. Leu et al. (92) showed that the HSP70 inhibitor PES, which interacts with both HSP70 and HSC70, causes sequestration of HSP90 client proteins, including epidermal growth factor receptor, HER2/ErbB2 and AKT, into an insoluble fraction in tumor cells. Interestingly, this group went on to show that PES also caused these HSP90 client proteins to be sequestered in a detergent-insoluble fraction in tumors from mice treated intraperitoneally with PES (92). These data point to inhibition of HSP90 client protein function as a potentially critical anticancer mechanism of action of HSP70 inhibitors. Although it is clear from several groups that HSP70 inhibitors can effectively target and inhibit the HSP90 chaperone machinery, it should be noted that multiple groups have reported that HSP70 inhibition markedly enhances the cytotoxicity of HSP90 inhibitors, for several different tumor types (70,91,93,94). Therefore, despite some similarities in action, there is merit to the idea of combining inhibitors of HSP70 and HSP90.
Pharmacological targeting of HSP70
Following the appreciation that HSP70 is an ideal anticancer target, several groups focused on the discovery of HSP70 inhibitors for cancer therapy. At present, over a dozen inhibitors of this chaperone have been reported. For the sake of brevity, only those inhibitors that have been tested as anticancer agents in pre-clinical or clinical trials are covered in this review. For a more comprehensive analysis of all HSP70 inhibitors, the reader is directed to several excellent reviews on this topic (95–98).
Deoxyspergualin/dihydropyrimidines
The first compound found to bind and modulate the function of HSP70 was the natural agent 15-deoxyspergualin. Although deoxyspergualin enhances, not inhibits, the ATPase activity of HSP70, this compound was found to have significant antitumor activity in a mouse leukemia model (99). The anticancer activity of this compound in clinical trials, however, failed to reveal any efficacy (100). Brodsky’s search for compounds related to deoxyspergualin revealed a class of dihydropyrimidines that block HSP70 ATPase activity (101); a second-generation version of these, called MAL3-101, blocks proliferation of cancer cells (93,102) and showed efficacy in a xenograft model of multiple myeloma (93). This promising compound, along with some related derivatives, awaits further pre-clinical and clinical testing.
MKT-077
The rhodocyanine MKT-077 was discovered to accumulate selectively in the mitochondria of tumor cells and to exhibit antiproliferative activity against cancer but not normal cell lines (103). In mouse xenograft studies, this agent showed promising antitumor activity (104). The selective toxicity of MKT-077 to tumor cells was discovered to be due to the ability of this compound to bind to the mitochondrial form of HSP70, HSP70-9 (also called Grp75) (105). Later studies supported an interaction between MKT-077 with HSC70 as well (106). Phase I clinical trials were conducted on this agent, and the dose-limiting toxicity was renal toxicity, thereby limiting further use of the compound (107). Interestingly, MKT-077 is an allosteric drug; it binds to a negatively charged pocket close to the ATP-binding site on HSP70, and selectively binds and inhibits the ADP-bound version of this protein (106). More recently, Gestwicki and Dickey developed a derivative of MKT-077 called YM-1; YM-1 was cytotoxic to multiple tumor cell lines, but not normal or immortalized cells, and restored tamoxifen sensitivity to a tamoxifen-resistant MCF7 cell line (108). YM-1 is, therefore, a promising prototype for the novel class of allosteric HSP70 inhibitors.
Peptide inhibitors
Garrido identified a domain of AIF that interacts with the SBD of HSP70 and reasoned that a mini-peptide comprising this domain, which they designate ADD70, might be a potent inhibitor of this chaperone. Transfection of tumor cells with this peptide sensitized human tumor cell lines to a variety of death-inducing stimuli and required the presence of HSP70 for cytotoxicity, confirming this chaperone as its target (109). This group found promising anticancer activity of this peptide when the peptide was transfected into tumor xenografts in mice; interestingly, this required the presence of an intact immune system and was caused by an increased number of CD8+ tumor-infiltrating T cells in ADD70-transfected tumor lines (110). Because of problems getting large peptides into cells, this group extended their findings by screening for peptide aptamers that bind to HSP70; they identified two aptamers, A8 and A17, which bind to HSP70. A 13 amino acid peptide from A17 was able to bind and inhibit HSP70, and induced the regression of subcutaneous tumors in vivo after local or systemic administration (111).
Ver-155008
In a rationalized approach, Massey used the crystal structure of HSC70 bound to BAG-1 to design analogs of ATP predicted to bind within the ATP-binding pocket and thereby inhibit this chaperone. The result was the identification of the ATP-analog VER-155008, which binds to HSP70 with a Kd of 0.3 µM and which inhibits the proliferation of a range of cancer cell lines (91,112). In pre-clinical analyses, the anticancer activity of this compound was somewhat limited (91). However, this compound serves as a useful scaffold, and it will be interesting to see whether derivatives can be fashioned with improved bioavailability.
PES and PES-Cl
PES was first discovered by Gudkov as a compound that inhibited the trafficking of p53 to mitochondria (113); however, the molecular target of this compound was not identified. Following their discovery that PES was selectively cytotoxic to multiple tumor but not normal cell lines, Leu et al. (85) biotintylated this compound and identified interacting proteins by mass spectrometry; this resulted in the identification of HSP70 as a PES-interacting protein. Notably, this group showed that silencing Hsp70 markedly reduced cytotoxicity by PES, supporting this protein as the relevant target of this compound. This group found that PES binds to the SBD of HSP70, likely to a deep hydrophobic pocket in the C-terminal α-helical lid (88). Consistent with that binding site, incubation of cells with PES prevents the ability of HSP70 to bind to critical chaperones, including Chip and HOP (85). Mechanistically, the groups of Leu et al. (85) have identified multiple mechanisms for cell death by this HSP70 inhibitor: it inhibits nuclear factor-κB activation (consistent with the identification of IkB-α as an HSP70 target) and inhibits autophagy (consistent with a role for HSP70 in lysosome stabilization). This compound inhibits HSP90 client protein function (92), induces G2/M arrest and inhibits the catalytic activity of the anaphase promoting complex/cyclosome in cell-free systems (88). Notably, in pre-clinical models of spontaneous B-cell lymphoma, treatment with PES (40mg/kg) or a modified version celled PES-Cl (20mg/kg) once per week for 20 weeks significantly protected mice from lymphoma, with no evidence of toxicity to the liver, kidney or other organs (85,88). Other groups have reported significant cytotoxic activity of PES against leukemia cell lines, but not normal hematopoietic cells (114,115), along with synergy with other anticancer compounds (115).
Two new functions have recently been ascribed to HSP70 and can be inhibited by PES. HSP70, along with HSP90, was recently found to co-localize with the purinosome, a dynamic multiprotein complex of enzymes involved in purine synthesis. Treatment of tumor cells with PES disrupts the purinosome, and this compound synergizes with the antimetabolite methotrexate to kill HeLa cells (116). In addition, both HSC70 and HSP70 can be found associated with the nascent polypeptides on actively translating ribosomes; in cells exposed to proteotoxic stress, these chaperones are believed to become sequestered by misfolded proteins, resulting in exposure of nascent-translating polypeptides and translational pausing of early elongating ribosomes (117). Consistent with this premise, treatment of cells with PES (as well as the compound described above, Ver-155008) causes ribosome pausing and decreased protein translation (117). PES and its derivatives thus exist as a promising class of HSP70 inhibitors for cancer therapy.
Other HSP70-relevant therapeutic tools (Cm70.1 antibody)
Multhoff first showed that HSP70 can be found localized to the plasma membrane of human tumor cells (118), where it is believed to help to maintain the stability of the tumor cell plasma membrane. Interestingly, only tumors but not the corresponding normal tissues were found to stain positively for the mouse monoclonal antibody cmHsp70.1, which specifically recognizes a C-terminal epitope of HSP70 that is exposed on the plasma membrane of tumor cells (119). Screening of nearly 1000 primary human tumor biopsies and the corresponding normal tissues indicated that human tumors but not normal tissues frequently express HSP70 on their cell surface (120). Notably, immunization of tumor-bearing mice with the peptide corresponding to the exposed C-terminal residues of HSP70 selectively induced antibody-dependent cellular cytotoxicity of human xenograft tumors (121), suggesting that this peptide, and possibly also the Cm70.1 monoclonal antibody, will be useful as therapeutic and/or imaging tools.
Conclusions and future directions
The field of HSP70 and cancer is now at a promising crossroads: many groups have confirmed activity of these inhibitors in cancer but not normal cells, and several new inhibitors have been identified. Therefore, we can expect exciting news from this field in the near future. In the meantime, several hurdles must be overcome. First and foremost, researchers in the field need to agree on a standard set of assays for HSP70 inhibition. For example, our group is the only group to use abrogation of autophagy as a measure of HSP70 inhibition, though we have recently found that other HSP70 inhibitors, including MKT-077 and Ver-155008, also inhibit autophagy (M.Murphy, unpublished results). Most groups have shown that HSP70 inhibitors cause sequestration of HSP90 client proteins to the insoluble compartment; yet, this is not a universally used assay. It is still unclear why HSP70 inhibitors exhibit such strong cytotoxicity for tumor but not normal cells. Although it is generally assumed that normal cells express little HSP70, this is not true for cultured primary cells; these express significant amounts of HSP70, yet are not affected by HSP70 inhibitors (M.Murphy, unpublished data). Along this line, Leu et al. (92) found that the HSP70 inhibitor PES interacts with HSP70 preferentially in tumor, but not normal cells, indicating that this protein may exhibit conformational or post-translational differences between tumor and normal cells; how HSP70 may be modified or structured differently in tumor cells remains to be elucidated. Techniques to assess binding affinity of inhibitors to HSP70, versus HSC70 and other HSP70 family members, need to be performed in order to understand which family members are bound and inhibited by each drug. Ideally, NMR or crystal structures of HSP70 with compounds should be sought whenever possible. Differences in reactivity and function between HSP70 and HSC70, such as the recently described sensitivity to redox status (122), need to be identified and exploited by researchers searching for inhibitors of these proteins. Although little is mentioned in this review, HSP70 clearly also has an extracellular role as an immunogenic molecule (reviewed in ref. 2). The role of HSP70, and of HSP70 inhibitors, in antitumor immunity needs to be further investigated; for example, can these inhibitors be used to block the ability of tumors to evade immune cell suppression, and should these inhibitors therefore be used alongside antibody-mediated therapy, such as herceptin and ipilumimab? We are only at the beginning of our pre-clinical analyses, and more effort should be made to determine whether there are particular tumor types, or particular tumor genotypes, that correlate with sensitivity to HSP70 inhibitors. Finally, the field is in dire need of improved communication: a conference on HSP70 in cancer, with emphasis on the development and use of inhibitors, would help researchers share ideas and reagents. With a push in this direction, we should be able to get these compounds into the clinic and take better advantage of this promising avenue for successful cancer therapy.
Funding
National Institutes of Health (CA102184 and 139319 to M.M.).
Acknowledgements
The author thanks Donna George and Julie Leu for critical reading of this manuscript, members of the Murphy lab for support and Mark Andrake (Fox Chase Cancer Center) for generating the ribbon model. This review is dedicated to K.L.A., H.B. and B.M.
Conflict of interest statement: None declared.
Glossary
Abbreviations:
- ADP
adenosine diphosphate
- AIF
apoptosis-inducing factor
- ATP
adenosine triphosphate
- HSE
heat shock element
- HSF1
heat shock factor 1
- HSP
heat shock protein
- NBD
nucleotide-binding domain
- NEF
nucleotide exchange factor
- PES
phenylethynesulfonamide
- SBD
substrate-binding domain.
References
- 1. Daugaard M., et al. (2007). The heat shock protein 70 family: highly homologous proteins with overlapping and distinct functions. FEBS Lett., 581, 3702–3710 [DOI] [PubMed] [Google Scholar]
- 2. Rérole A.L., et al. (2011). Hsp70: anti-apoptotic and tumorigenic protein. Methods Mol. Biol., 787, 205–230 [DOI] [PubMed] [Google Scholar]
- 3. Brodsky J.L., et al. (2006). Hsp70 molecular chaperones: emerging roles in human disease and identification of small molecule modulators. Curr. Top. Med. Chem., 6, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 4. Bonnycastle L.L., et al. (1994). Cloning, sequencing, and mapping of the human chromosome 14 heat shock protein gene (HSPA2). Genomics., 23, 85–93 [DOI] [PubMed] [Google Scholar]
- 5. Su A.I., et al. (2004). A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U. S. A., 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Florin L., et al. (2004). Nuclear translocation of papillomavirus minor capsid protein L2 requires Hsc70. J. Virol., 78, 5546–5553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bertelsen E.B., et al. (2009). Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc. Natl. Acad. Sci. U. S. A., 106, 8471–8476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flaherty K.M., et al. (1990). Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature., 346, 623–628 [DOI] [PubMed] [Google Scholar]
- 9. Bork P., et al. (1992). An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. U. S. A., 89, 7290–7294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhu X., et al. (1996). Structural analysis of substrate binding by the molecular chaperone DnaK. Science., 272, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell R., et al. (1998). Kinetic characterization of the ATPase cycle of the DnaK molecular chaperone. Biochemistry., 37, 596–607 [DOI] [PubMed] [Google Scholar]
- 12. Laufen T., et al. (1999). Mechanism of regulation of hsp70 chaperones by DnaJ cochaperones. Proc. Natl. Acad. Sci. U. S. A., 96, 5452–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bukau B., et al. (2006). Molecular chaperones and protein quality control. Cell., 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 14. Allan R.K., et al. (2011). Versatile TPR domains accommodate different modes of target protein recognition and function. Cell Stress Chaperones., 16, 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mayer M.P., et al. (2000). Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol., 7, 586–593 [DOI] [PubMed] [Google Scholar]
- 16. Zhuravleva A., et al. (2012). An interdomain energetic tug-of-war creates the allosterically active state in Hsp70 molecular chaperones. Cell., 151, 1296–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zuiderweg E.R., et al. (2013). Allostery in the Hsp70 chaperone proteins. Top. Curr. Chem., 328, 99–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parsian A.J., et al. (2000). The human Hsp70B gene at the HSPA7 locus of chromosome 1 is transcribed but non-functional. Biochim. Biophys. Acta., 1494, 201–205 [DOI] [PubMed] [Google Scholar]
- 19. Wu B.J., et al. (1986). Human HSP70 promoter contains at least two distinct regulatory domains. Proc. Natl. Acad. Sci. U. S. A., 83, 629–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Morgan W.D. (1989). Transcription factor Sp1 binds to and activates a human hsp70 gene promoter. Mol. Cell. Biol., 9, 4099–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Morgan W.D., et al. (1987). Two transcriptional activators, CCAAT-box-binding transcription factor and heat shock transcription factor, interact with a human hsp70 gene promoter. Mol. Cell. Biol., 7, 1129–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Milarski K.L., et al. (1986). Expression of human HSP70 during the synthetic phase of the cell cycle. Proc. Natl. Acad. Sci. U. S. A., 83, 9517–9521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Taira T., et al. (1997). A novel G1-specific enhancer identified in the human heat shock protein 70 gene. Nucleic Acids Res., 25, 1975–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akerfelt M., et al. (2010). Heat shock factors: integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol., 11, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mason P.B., Jr, et al. (1997). Cooperative and competitive protein interactions at the hsp70 promoter. J. Biol. Chem., 272, 33227–33233 [DOI] [PubMed] [Google Scholar]
- 26. Cotto J.J., et al. (1999). Stress-induced activation of the heat-shock response: cell and molecular biology of heat-shock factors. Biochem. Soc. Symp., 64, 105–118 [PubMed] [Google Scholar]
- 27. Hunt C., et al. (1985). Conserved features of eukaryotic hsp70 genes revealed by comparison with the nucleotide sequence of human hsp70. Proc. Natl. Acad. Sci. U. S. A., 82, 6455–6459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chou S.D., et al. (2012). mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS One., 7, e39679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Westerheide S.D., et al. (2009). Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science., 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu G., et al. (2005). DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res., 65, 758–766 [PubMed] [Google Scholar]
- 31. Hunt C.R., et al. (2004). Genomic instability and enhanced radiosensitivity in Hsp70.1- and Hsp70.3-deficient mice. Mol. Cell. Biol., 24, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee S.H., et al. (2001). Targeted hsp70.1 disruption increases infarction volume after focal cerebral ischemia in mice. Stroke., 32, 2905–2912 [DOI] [PubMed] [Google Scholar]
- 33. Van Molle W., et al. (2002). HSP70 protects against TNF-induced lethal inflammatory shock. Immunity., 16, 685–695 [DOI] [PubMed] [Google Scholar]
- 34. Shim E.H., et al. (2002). Targeted disruption of hsp70.1 sensitizes to osmotic stress. EMBO Rep., 3, 857–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hwang J.H., et al. (2005). Spontaneous activation of pancreas trypsinogen in heat shock protein 70.1 knock-out mice. Pancreas., 31, 332–336 [DOI] [PubMed] [Google Scholar]
- 36. Rattner J.B. (1991). hsp70 is localized to the centrosome of dividing HeLa cells. Exp. Cell Res., 195, 110–113 [DOI] [PubMed] [Google Scholar]
- 37. Oka M., et al. (1998). Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem., 273, 29727–29737 [DOI] [PubMed] [Google Scholar]
- 38. Sconzo G., et al. (1999). Constitutive hsp70 is essential to mitosis during early cleavage of Paracentrotus lividus embryos: the blockage of constitutive hsp70 impairs mitosis. Biochem. Biophys. Res. Commun., 260, 143–149 [DOI] [PubMed] [Google Scholar]
- 39. Agueli C., et al. (2001). A constitutive 70kDa heat-shock protein is localized on the fibres of spindles and asters at metaphase in an ATP-dependent manner: a new chaperone role is proposed. Biochem. J., 360, 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jäättelä M. (1995). Over-expression of hsp70 confers tumorigenicity to mouse fibrosarcoma cells. Int. J. Cancer., 60, 689–693 [DOI] [PubMed] [Google Scholar]
- 41. Seo J.S., et al. (1996). T cell lymphoma in transgenic mice expressing the human Hsp70 gene. Biochem. Biophys. Res. Commun., 218, 582–587 [DOI] [PubMed] [Google Scholar]
- 42. Volloch V.Z., et al. (1999). Oncogenic potential of Hsp72. Oncogene., 18, 3648–3651 [DOI] [PubMed] [Google Scholar]
- 43. Meng L., et al. (2011). Heat shock protein Hsp72 plays an essential role in Her2-induced mammary tumorigenesis. Oncogene., 30, 2836–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rohde M., et al. (2005). Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev., 19, 570–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ciocca D.R., et al. (2005). Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones., 10, 86–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Calderwood S.K., et al. (2006). Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem. Sci., 31, 164–172 [DOI] [PubMed] [Google Scholar]
- 47. Chuma M., et al. (2003). Expression profiling in multistage hepatocarcinogenesis: identification of HSP70 as a molecular marker of early hepatocellular carcinoma. Hepatology., 37, 198–207 [DOI] [PubMed] [Google Scholar]
- 48. Abe M., et al. (2004). Plasma levels of heat shock protein 70 in patients with prostate cancer: a potential biomarker for prostate cancer. Clin. Prostate Cancer., 3, 49–53 [DOI] [PubMed] [Google Scholar]
- 49. Hwang T.S., et al. (2003). Differential, stage-dependent expression of Hsp70, Hsp110 and Bcl-2 in colorectal cancer. J. Gastroenterol. Hepatol., 18, 690–700 [DOI] [PubMed] [Google Scholar]
- 50. Lazaris A.C.h., et al. (1997). Proliferating cell nuclear antigen and heat shock protein 70 immunolocalization in invasive ductal breast cancer not otherwise specified. Breast Cancer Res. Treat., 43, 43–51 [DOI] [PubMed] [Google Scholar]
- 51. Małusecka E., et al. (2001). Expression of heat shock proteins HSP70 and HSP27 in primary non-small cell lung carcinomas. An immunohistochemical study. Anticancer Res., 21, 1015–1021 [PubMed] [Google Scholar]
- 52. Athanassiadou P., et al. (1998). Expression of p53, bcl-2 and heat shock protein (hsp72) in malignant and benign ovarian tumours. Eur. J. Cancer Prev., 7, 225–231 [DOI] [PubMed] [Google Scholar]
- 53. Ralhan R., et al. (1995). Differential expression of Mr 70,000 heat shock protein in normal, premalignant, and malignant human uterine cervix. Clin. Cancer Res., 1, 1217–1222 [PubMed] [Google Scholar]
- 54. Lazaris A.C., et al. (1995). Immunohistochemical expression of C-myc oncogene, heat shock protein 70 and HLA-DR molecules in malignant cutaneous melanoma. Virchows Arch., 426, 461–467 [DOI] [PubMed] [Google Scholar]
- 55. Kaur J., et al. (1998). Expression of 70-kDa heat shock protein in oral lesions: marker of biological stress or pathogenicity. Oral Oncol., 34, 496–501 [DOI] [PubMed] [Google Scholar]
- 56. Syrigos K.N., et al. (2003). Clinical significance of heat shock protein-70 expression in bladder cancer. Urology., 61, 677–680 [DOI] [PubMed] [Google Scholar]
- 57. Ciocca D.R., et al. (1993). Heat shock protein hsp70 in patients with axillary lymph node-negative breast cancer: prognostic implications. J. Natl. Cancer Inst., 85, 570–574 [DOI] [PubMed] [Google Scholar]
- 58. Thanner F., et al. (2003). Heat-shock protein 70 as a prognostic marker in node-negative breast cancer. Anticancer Res., 23, 1057–1062 [PubMed] [Google Scholar]
- 59. Nanbu K., et al. (1998). Prognostic significance of heat shock proteins HSP70 and HSP90 in endometrial carcinomas. Cancer Detect. Prev., 22, 549–555 [DOI] [PubMed] [Google Scholar]
- 60. Thomas X., et al. (2005). Expression of heat-shock proteins is associated with major adverse prognostic factors in acute myeloid leukemia. Leuk. Res., 29, 1049–1058 [DOI] [PubMed] [Google Scholar]
- 61. Ren A., et al. (2008). Down-regulation of mammalian sterile 20-like kinase 1 by heat shock protein 70 mediates cisplatin resistance in prostate cancer cells. Cancer Res., 68, 2266–2274 [DOI] [PubMed] [Google Scholar]
- 62. Pocaly M., et al. (2007). Overexpression of the heat-shock protein 70 is associated to imatinib resistance in chronic myeloid leukemia. Leukemia., 21, 93–101 [DOI] [PubMed] [Google Scholar]
- 63. Sliutz G., et al. (1996). Drug resistance against gemcitabine and topotecan mediated by constitutive hsp70 overexpression in vitro: implication of quercetin as sensitiser in chemotherapy. Br. J. Cancer., 74, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jäättelä M., et al. (1993). Heat-shock proteins protect cells from monocyte cytotoxicity: possible mechanism of self-protection. J. Exp. Med., 177, 231–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chalmin F., et al. (2010). Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J. Clin. Invest., 120, 457–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nylandsted J., et al. (2000). Selective depletion of heat shock protein 70 (Hsp70) activates a tumor-specific death program that is independent of caspases and bypasses Bcl-2. Proc. Natl. Acad. Sci. U. S. A., 97, 7871–7876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaur J., et al. (2000). Induction of apoptosis by abrogation of HSP70 expression in human oral cancer cells. Int. J. Cancer., 85, 1–5 [DOI] [PubMed] [Google Scholar]
- 68. Frese S., et al. (2003). Cell death induced by down-regulation of heat shock protein 70 in lung cancer cell lines is p53-independent and does not require DNA cleavage. J. Thorac. Cardiovasc. Surg., 126, 748–754 [DOI] [PubMed] [Google Scholar]
- 69. Nylandsted J., et al. (2002). Eradication of glioblastoma, and breast and colon carcinoma xenografts by Hsp70 depletion. Cancer Res., 62, 7139–7142 [PubMed] [Google Scholar]
- 70. Powers M.V., et al. (2008). Dual targeting of HSC70 and HSP72 inhibits HSP90 function and induces tumor-specific apoptosis. Cancer Cell., 14, 250–262 [DOI] [PubMed] [Google Scholar]
- 71. Gotoh T., et al. (2004). hsp70-DnaJ chaperone pair prevents nitric oxide- and CHOP-induced apoptosis by inhibiting translocation of Bax to mitochondria. Cell Death Differ., 11, 390–402 [DOI] [PubMed] [Google Scholar]
- 72. Stankiewicz A.R., et al. (2005). Hsp70 inhibits heat-induced apoptosis upstream of mitochondria by preventing Bax translocation. J. Biol. Chem., 280, 38729–38739 [DOI] [PubMed] [Google Scholar]
- 73. Saleh A., et al. (2000). Negative regulation of the Apaf-1 apoptosome by Hsp70. Nat. Cell Biol., 2, 476–483 [DOI] [PubMed] [Google Scholar]
- 74. Guo F., et al. (2005). Mechanistic role of heat shock protein 70 in Bcr-Abl-mediated resistance to apoptosis in human acute leukemia cells. Blood., 105, 1246–1255 [DOI] [PubMed] [Google Scholar]
- 75. Gabai V.L., et al. (2002). Hsp72 and stress kinase c-jun N-terminal kinase regulate the bid-dependent pathway in tumor necrosis factor-induced apoptosis. Mol. Cell. Biol., 22, 3415–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Park H.S., et al. (2002). Heat shock protein hsp72 is a negative regulator of apoptosis signal-regulating kinase 1. Mol. Cell. Biol., 22, 7721–7730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Park H.S., et al. (2001). Hsp72 functions as a natural inhibitory protein of c-Jun N-terminal kinase. EMBO J., 20, 446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gabai V.L., et al. (1997). Hsp70 prevents activation of stress kinases. A novel pathway of cellular thermotolerance. J. Biol. Chem., 272, 18033–18037 [DOI] [PubMed] [Google Scholar]
- 79. Ravagnan L., et al. (2001). Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol., 3, 839–843 [DOI] [PubMed] [Google Scholar]
- 80. Yaglom J.A., et al. (2007). High levels of heat shock protein Hsp72 in cancer cells suppress default senescence pathways. Cancer Res., 67, 2373–2381 [DOI] [PubMed] [Google Scholar]
- 81. Gabai V.L., et al. (2009). Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol. Cell. Biol., 29, 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nylandsted J., et al. (2004). Heat shock protein 70 promotes cell survival by inhibiting lysosomal membrane permeabilization. J. Exp. Med., 200, 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Daugaard M., et al. (2007). Lens epithelium-derived growth factor is an Hsp70-2 regulated guardian of lysosomal stability in human cancer. Cancer Res., 67, 2559–2567 [DOI] [PubMed] [Google Scholar]
- 84. Kirkegaard T., et al. (2010). Hsp70 stabilizes lysosomes and reverts Niemann-Pick disease-associated lysosomal pathology. Nature., 463, 549–553 [DOI] [PubMed] [Google Scholar]
- 85. Leu J.I., et al. (2009). A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell., 36, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Amaravadi R.K., et al. (2007). Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J. Clin. Invest., 117, 326–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Carew J.S., et al. (2007). Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood., 110, 313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Balaburski G.M., et al. (2013). A modified HSP70 inhibitor shows broad activity as an anticancer agent. Mol. Cancer Res., 11, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Banerji U., et al. (2005). Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J. Clin. Oncol., 23, 4152–4161 [DOI] [PubMed] [Google Scholar]
- 90. Morishima Y., et al. (2000). Stepwise assembly of a glucocorticoid receptor.hsp90 heterocomplex resolves two sequential ATP-dependent events involving first hsp70 and then hsp90 in opening of the steroid binding pocket. J. Biol. Chem., 275, 18054–18060 [DOI] [PubMed] [Google Scholar]
- 91. Massey A.J., et al. (2010). A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol., 66, 535–545 [DOI] [PubMed] [Google Scholar]
- 92. Leu J.I., et al. (2011). HSP70 inhibition by the small-molecule 2-phenylethynesulfonamide impairs protein clearance pathways in tumor cells. Mol. Cancer Res., 9, 936–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Braunstein M.J., et al. (2011). Antimyeloma effects of the heat shock protein 70 molecular chaperone inhibitor MAL3-101. J. Oncol., 2011, 232037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Davenport E.L., et al. (2010). Targeting heat shock protein 72 enhances Hsp90 inhibitor-induced apoptosis in myeloma. Leukemia., 24, 1804–1807 [DOI] [PubMed] [Google Scholar]
- 95. Evans C.G., et al. (2010). Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem., 53, 4585–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Powers M.V., et al. (2010). Targeting HSP70: the second potentially druggable heat shock protein and molecular chaperone? Cell Cycle., 9, 1542–1550 [DOI] [PubMed] [Google Scholar]
- 97. Patury S., et al. (2009). Pharmacological targeting of the Hsp70 chaperone. Curr. Top. Med. Chem., 9, 1337–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Goloudina A.R., et al. (2012). Inhibition of HSP70: a challenging anti-cancer strategy. Cancer Lett., 325, 117–124 [DOI] [PubMed] [Google Scholar]
- 99. Plowman J., et al. (1987). Preclinical antitumor activity and pharmacological properties of deoxyspergualin. Cancer Res., 47, 685–689 [PubMed] [Google Scholar]
- 100. Dhingra K., et al. (1994). Phase II study of deoxyspergualin in metastatic breast cancer. Invest. New Drugs., 12, 235–241 [DOI] [PubMed] [Google Scholar]
- 101. Fewell S.W., et al. (2001). Identification of an inhibitor of hsc70-mediated protein translocation and ATP hydrolysis. J. Biol. Chem., 276, 910–914 [DOI] [PubMed] [Google Scholar]
- 102. Rodina A., et al. (2007). Selective compounds define Hsp90 as a major inhibitor of apoptosis in small-cell lung cancer. Nat. Chem. Biol., 3, 498–507 [DOI] [PubMed] [Google Scholar]
- 103. Koya K., et al. (1996). MKT-077, a novel rhodacyanine dye in clinical trials, exhibits anticarcinoma activity in preclinical studies based on selective mitochondrial accumulation. Cancer Res., 56, 538–543 [PubMed] [Google Scholar]
- 104. Chiba Y., et al. (1998). MKT-077, localized lipophilic cation: antitumor activity against human tumor xenografts serially transplanted into nude mice. Anticancer Res., 18, 1047–1052 [PubMed] [Google Scholar]
- 105. Wadhwa R., et al. (2000). Selective toxicity of MKT-077 to cancer cells is mediated by its binding to the hsp70 family protein mot-2 and reactivation of p53 function. Cancer Res., 60, 6818–6821 [PubMed] [Google Scholar]
- 106. Rousaki A., et al. (2011). Allosteric drugs: the interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol., 411, 614–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Propper D.J., et al. (1999). Phase I trial of the selective mitochondrial toxin MKT077 in chemo-resistant solid tumours. Ann. Oncol., 10, 923–927 [DOI] [PubMed] [Google Scholar]
- 108. Koren J., III, et al. (2012). Rhodacyanine derivative selectively targets cancer cells and overcomes tamoxifen resistance. PLoS One., 7, e35566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Schmitt E., et al. (2003). Chemosensitization by a non-apoptogenic heat shock protein 70-binding apoptosis-inducing factor mutant. Cancer Res., 63, 8233–8240 [PubMed] [Google Scholar]
- 110. Schmitt E., et al. (2006). Heat shock protein 70 neutralization exerts potent antitumor effects in animal models of colon cancer and melanoma. Cancer Res., 66, 4191–4197 [DOI] [PubMed] [Google Scholar]
- 111. Rérole A.L., et al. (2011). Peptides and aptamers targeting HSP70: a novel approach for anticancer chemotherapy. Cancer Res., 71, 484–495 [DOI] [PubMed] [Google Scholar]
- 112. Williamson D.S., et al. (2009). Novel adenosine-derived inhibitors of 70kDa heat shock protein, discovered through structure-based design. J. Med. Chem., 52, 1510–1513 [DOI] [PubMed] [Google Scholar]
- 113. Strom E., et al. (2006). Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat. Chem. Biol., 2, 474–479 [DOI] [PubMed] [Google Scholar]
- 114. Steele A.J., et al. (2009). 2-Phenylacetylenesulfonamide (PAS) induces p53-independent apoptotic killing of B-chronic lymphocytic leukemia (CLL) cells. Blood., 114, 1217–1225 [DOI] [PubMed] [Google Scholar]
- 115. Kaiser M., et al. (2011). Antileukemic activity of the HSP70 inhibitor pifithrin-μ in acute leukemia. Blood Cancer J., 1, e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. French J.B., et al. (2013). Hsp70/Hsp90 chaperone machinery is involved in the assembly of the purinosome. Proc. Natl. Acad. Sci. U. S. A., 110, 2528–2533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Liu B., et al. (2013). Cotranslational response to proteotoxic stress by elongation pausing of ribosomes. Mol. Cell., 49, 453–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Multhoff G., et al. (1996). Cell surface expression of heat shock proteins and the immune response. Cell Stress Chaperones., 1, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Multhoff G., et al. (2011). Distinguishing integral and receptor-bound heat shock protein 70 (Hsp70) on the cell surface by Hsp70-specific antibodies. Cell Stress Chaperones., 16, 251–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Pfister K., et al. (2007). Patient survival by Hsp70 membrane phenotype: association with different routes of metastasis. Cancer., 110, 926–935 [DOI] [PubMed] [Google Scholar]
- 121. Stangl S., et al. (2011). Targeting membrane heat-shock protein 70 (Hsp70) on tumors by cmHsp70.1 antibody. Proc. Natl. Acad. Sci. U. S. A., 108, 733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Miyata Y., et al. (2012). Cysteine reactivity distinguishes redox sensing by the heat-inducible and constitutive forms of heat shock protein 70. Chem. Biol., 19, 1391–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]