Abstract
Herein we review our progress on the development of phosphopeptide-based prodrugs targeting the SH2 domain of STAT3 to prevent recruitment to cytokine and growth factor receptors, activation, nuclear translocation and transcription of genes involved in cancer. We developed high affinity phosphopeptides (KI = 46–200 nM). Corresponding prodrugs inhibited constitutive and IL-6 induced Tyr705 phosphorylation at 0.5–1 μM in a variety of human cancer cell lines. They were not cytotoxic at 5 μM in vitro but they inhibited tumor growth in a human xenograft breast cancer model in mice, accompanied by reduced VEGF expression and angiogenesis.
Keywords: phosphopeptide, prodrug, SH2 domain, STAT3, peptidomimetic
Signal Transducer and Activator of Transcription 3 (STAT3) is a Target for Anticancer Drug Design
Signal transducer and activator of transcription 3 (STAT3) is likely the most studied member of the STAT family of proteins.1-7 STAT3 participates in the transcription of numerous proteins and is hypothesized to play indispensable roles in the development of a large number of human cancers, metastasis, angiogenesis and immune evasion.8-10 Thus targeting STAT3 is regarded as a potential modality for the treatment of cancer.4,5,11-14 STAT3 signals by being recruited to phosphotyrosine residues on growth factor and cytokine receptors. On binding via its Src homology 2 (SH2) domain, Tyr705 becomes phosphorylated by associated JAK kinases, Src or the phosphotransferase activity of the receptor. Phosphorylated STAT3 (pSTAT3) dimerizes by reciprocal pTyr-SH2 domain interactions, is translocated to the nucleus, then acts as a transcription factor participating in the expression of acute phase response genes, vascular endothelial growth factor (VEGF), matrix metalloproteinase 9, Bcl proteins and others. Recently, other functions of STAT3 have been discovered. Unphosphorylated STAT3 was found to act as a co-transctription factor in complex with NFκB.15 In non-transcriptional roles, STAT3, phosphorylated on Ser727, was found to be located in electron-transport complexes in the mitochondria16 and in this state supported RAS transformation of cells.17 Removing STAT3 from cells using siRNA, antisense or like techniques, or overexpressing STAT3 or dominant negative versions, likely will impact multiple STAT3 functions. Precise determination of the role of Tyr705 phosphorylation can be accomplished by highly selective inhibitors targeted to the SH2 domain that block association with receptors and subsequent phosphorylation, dimerization and transcriptional activities. In this review we highlight our progress in the development of high affinity phosphopeptide ligands of the SH2 domain of STAT3, their conversion to cell-permeable, phosphatase-stable prodrugs and the evaluation of these in cellular and human cancer xenograft models of human cancer. We found that although selective inhibition of Tyr705 phosphorylation is not cytotoxic to cancer cells in vitro, in vivo tumor growth inhibition can be achieved which may be driven by reduced angiogenesis.
Targeting SH2 Domains
SH2 domains are 100 amino acid domains that recognize phosphotyrosine and two to four residues to its C-terminus.18,19 These domains are involved in the recruitment of signal transduction proteins to activated receptors of growth factors and cytokines and aberrant signaling by these pathways contributes to a variety of diseases such as cancer and asthma. The SH2 domains of Src, Lck, Grb2 and p85 were easily expressed and structures of complexes with phosphopeptides obtained by X-ray crystallography or NMR guided industrial, government and academic laboratories that developed several high affinity, elegant peptidomimetics.20-22 In spite of this great effort, there is a paucity of literature describing the biological activity of these materials and no SH2 domain-targeted phosphopeptide mimetics have advanced to clinical trials.
Two major challenges that have impeded the development of phosphopeptide-based SH2 domain inhibitors are the negative charge of the phosphate group that prevents passive diffusion across cell membranes and the lability of the phosphate group to phosphatase activity, which renders phosphopeptides unrecognizable. To overcome phosphatase-lability, researchers have replaced phosphate groups with carboxyl, phosphonate, malonate, phosphonomethyl, phosphonodifluoromethyl and heterocyclic groups that are negatively charged.23,24 Bioreversible esters have been employed to block the negative charge of the phosphate or phosphonate oxygens in a variety of compounds including SH2-domain-targeted peptides and mimetics.25 The Garbay group employed S-acylthioethyl groups on a series of phosphopeptides targeting the SH2 domain of Grb2.26,27 Cytotoxicity to cancer cell lines was observed at 1 μM concentration. Gay et al. employed the phenyl phosphoramidite approach to deliver phosphopeptides targeting Grb2 to tumor cells28,29 with concentrations ~25 μM required to inhibit the target. Stankovic reported pivaloyloxymethyl (POM) protection of phosphatase-stable phosphodifluoromethylphenylalanine in a Src SH2 domain inhibitor.30 They were only able to append one POM group to the phosphonate. Although the mono-POM prodrug entered cells, no biological evaluation was reported. McKinney et al. reported a bis-POM protected phosphonodifluoromethyl analog of a phosphopeptide mimetic targeting STAT4 and STAT6 but no biological data were presented.31
Development of High Affinity Phosphopeptide Mimics Targeting the SH2 Domain of STAT3
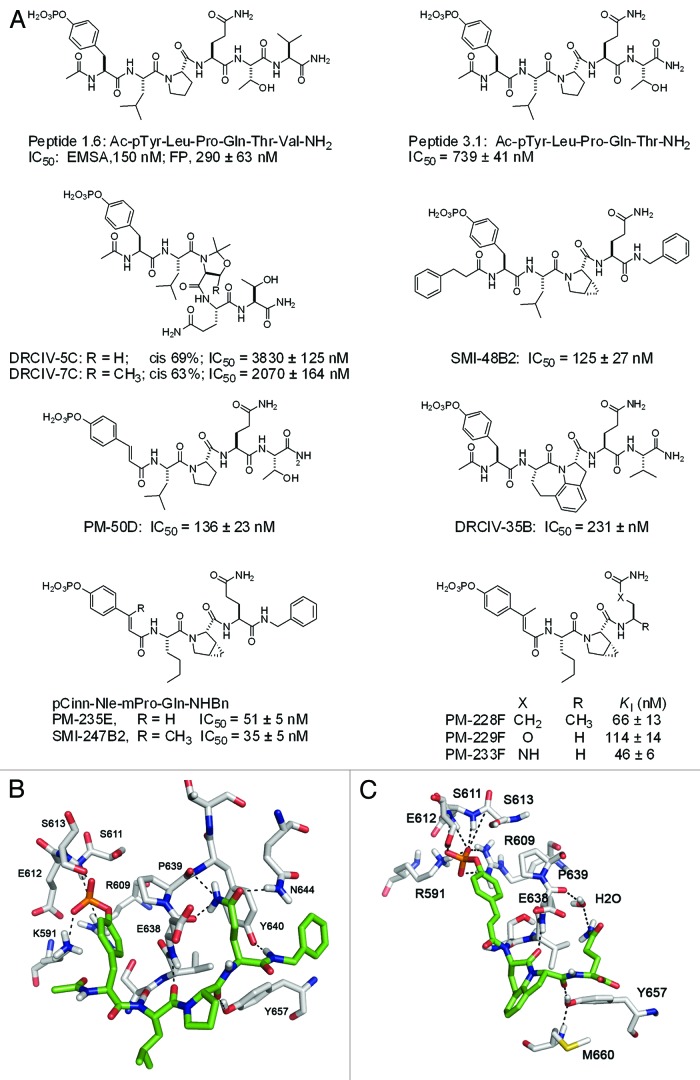
Several groups have engaged in the development of peptidomimetic inhibitors targeting the SH2 domain of STAT3. The team of James Turkson and Patrick Gunning has developed inhibitors derived from the Tyr705 sequence32,33 and from screening and medicinal chemistry approaches.34-39 Others have used the Tyr705 sequence40 as well as our lead, Peptide 1.6.41 To develop high affinity and selective phosphopeptides targeting the SH2 domain, our laboratory screened a set of candidates derived from putative STAT3 binding sites on receptors for IL-6, EGF, IL-10 and G-CSF and found that Ac-pTyr-Leu-Pro-Gln-Thr-Val-NH2 (termed Peptide 1.6) from the sequence surrounding Tyr904 of the IL-6 co-receptor gp130, was a high affinity ligand (Fig. 1). Peptide 1.6 inhibited STAT3-DNA complex formation with an IC50 = 150 nM, as judged by electrophoretic mobility shift assays.42 Wiederkehr-Adam et al. found similar peptides using a combinatorial phosphopeptide approach.43 Peptide 1.6 possesses the pTyr-Xaa-Yaa-Gln motif reported to be the recognition determinant of STAT3.44,45 Substitution of the glutamine of Peptide 1.6 with alanine, glutamic acid and asparagine reduced affinity thereby supporting the requirement for glutamine at pY+3 for high affinity binding to the SH2 domain of STAT3.42
Figure 1 (See previous page). Structures of phosphopeptides and phosphopeptide mimics targeting the SH2 domain of STAT3. (A) Highlights of structure-affinity relationship studies. (B) Model of Ac-pTyr-Leu-Pro-Gln-NHBn bound to the SH2 domain of STAT3 (see reference 57). (C) Model of pCinn-Haic-Gln-OH bound to the SH2 domain of STAT3 (see reference 55). In (B and C) hydrogen bonds are depicted as dotted lines. The inhibitor is depicted with the green coloring scheme and STAT3 in white.
To probe the molecular surface of the SH2 domain of STAT3 and to search for high-affinity modifications, we substituted natural and unnatural amino acids at each position of our lead peptide. We independently developed a fluorescence polarization assay to monitor the ability of phosphopeptides to compete with the N-terminally fluorescein-tagged version of Peptide 1.6 (FAM-Ala-pTyr-Leu-Pro-Thr-Val-NH2, FAM = 4 carboxyfluorescein) for binding to full length STAT3.46 Haan et al. expressed just the SH2 domain of STAT3 but it only bound a phosphopeptide at pH 5.5.47 Due to potential conformational variation at the lower pH that might not exist at physiological pH, we expressed full-length protein for our assays. Peptide 1.6 exhibited an IC50 of 290 nM. We utilized the truncated peptide Ac-pTyr-Leu-Pro-Thr-NH2 (termed Peptide 3.1, IC50 = 739 nM, Fig. 1) as the template for our studies. In spite of the reduced affinity of peptide 3.1, its smaller size meant significantly less synthetic steps in the mostly manual syntheses that produced the phosphopeptides we assayed. Affinity was recaptured with the modifications we incorporated. We showed that hydrophobic groups could be appended to the N-terminal nitrogen of pTyr (position pY−1) suggesting a hydrophobic patch exists on the protein surface adjacent to the phosphotyrosine binding pocket.46 Leucine at pY+1 could be substituted with a variety of hydrophobic residues. Aliphatic amino acids such as norleucine and cyclohexylalanine provided higher affinity than aromatic phenylalanine.46 Methylation of the nitrogen of Leu at pY+1 abrogated binding,46 which supports experimentally the hydrogen bond between the NH of Leu706 and the C = O of Ser636 observed in the crystal structure of the STAT3 dimer published by Becker, et al.48
Alanine scanning showed that proline at position pY+2 contributed significantly to binding42 and throughout our studies 19 proline analogs were substituted at this position to probe this site.46,49-51 Of this group, cis-3,4-methanoproline (mPro) provided a two-fold increase in affinity46 and this amino acid was utilized in later structure-affinity relationship studies. Peptide bonds containing proline (Xaa-Pro) can exist either in the cis or trans conformation. Pseudoproline derivatives, 2,2-dimethyl-1,3 oxazole-4-carboxylate and 2,2-dimethyl-5-methyl-1,3 oxazole-4-carboxylate, result in predominantly cis-peptide bonds, as opposed to native proline, predominantly trans.52 Incorporation of these pseudoproline residues resulted in 63–69% cis conformation (proline, 2%) and decreased affinity 3- to 5-fold suggesting that the when bound to the SH2 domain of STAT3, Leu-Pro is in the trans conformation (Fig. 1, peptides DRCIV-5C and DRCIV-7C).49
Overall, we substituted 45 Gln surrogates at pY+3 to probe the binding site.46,53-55 Methyl substitutions on the side chain amide nitrogen were not tolerated and isosteric methionine sulfoxide resulted in a > 10-fold loss in affinity.46 These results indicated the importance of hydrogen bond donation by the side chain amide group of Gln. Various cyclic and aliphatic glutamine surrogates were tolerated with slight losses in affinity.49,53-55
Threonine at pY+4 was replaced with a variety of groups: organic, heterocyclic and peptidic.46,55 The most effective substitution was a simple benzylamide.46 Taking these lessons into account, we incorporated a hydrophobic N-terminus, mPro, and a C-terminal benzyl amide to create SMI-48B2 (Fig. 1), which had an IC50 of 125 nM, a five-fold increase in affinity over Peptide 3.1.
Conformationally Constrained Phosphopeptides
Properly constrained peptide inhibitors can lead to increased affinity by presenting the contact groups in the proper orientation for binding to the target protein. By constraining the molecule to the bioactive conformation, the system does not lose the entropy of rotation of all of the peptides’ bonds on binding, leading to a favorable entropic term in the free energy equation. The dihedral angle of C-Cα-Cβ-Cγ of the phosphotyrosine residue in the STAT3 crystal structure is 174 degrees.48 The phosphotyrosine mimic, 4-phosphoryloxycinnamate (pCinn), constrains this angle to 180° and resulted in a 5-fold increase in affinity of peptide 3.1 (PM-50D, IC50 = 136 nM, Fig. 1).50 Interestingly, pCinn resulted in a 11-fold loss in affinity for a phosphopeptide inhibitor of the Src SH2 domain.56 Examination of the crystal structure of STAT3 bound to DNA48 as well as models generated by us57 led to the hypothesis that addition of a methyl group on the β-carbon of pTyr or pCinn would lead to greater hydrophobic interaction with the side chain methylene groups of Glu638, which lines the phosphotyrosine binding pocket. We developed synthetic methodology for β-methylcinnamate and found that this substitution increased affinity 1.5–3 fold in a series of peptides (e.g., PM-235E vs. SMI-247B2, Fig. 1).53
To constrain the central dipeptide, Leu-Pro was substituted with a series of azabicyclo[4.3.0]-nonane-9-carboxylates (ABN), in which the side chain of leucine was incorporated in a 6-membered ring fused to the 5-membered ring of proline.58 All stereoisomers of this bicylic lactam reduced activity.50 However, substitution with the tricyclic heterocycle, Haic, increased affinity of our peptides > three-fold (DRCIV-35B, IC50 = 231 nM, Fig. 1).50 Chen et al. incorporated azabicyclo[6.3.0]undecane (ABU) and found that this substitution increased affinity 20-fold.59 All of these dipeptide replacements constrain the ψ dihedral angle of the pY+1 residue. The size of the ring fused to the five-membered ring of proline is important. The eight-membered ring in ABU appears to allow the most optimal orientation of the Gln with respect to the phosphotyrosine, as compared with the seven-membered ring of Haic and the six-membered ring of ABN.
Constrained peptidomimetics exhibited high affinity. Among the more notable candidates were a series containing glutamine surrogates (R)-4-aminopentamide, in which the α-carboxyl group of Gln was reduced to a methyl group (PM-228F, KI = 66 ± 13 nM), straight chain ethylcarbamate (PM-229F, KI = 114 ± 14 nM) and a straight chain ethylurea (PM-233F, KI = 46 ± 6 nM) (Fig. 1).
Structure of Phosphopeptides Bound to the SH2 Domain of STAT3
Structures of protein-ligand complexes are extremely useful in drug development programs. Unfortunately, STAT3 was difficult to crystallize and in the one structure we obtained, the electron density for the peptide (PM-50D) was too weak to determine its structure.60 However, molecular modeling approaches provided some insights of phosphopeptide-SH2 domain interactions. In the first model we examined potential interactions between the phosphopeptide, Ac-pTyr-Leu-Pro-Gln-NHBn, and STAT3 using the structure of a phosphopeptide complexed with STAT161 as a template.57,62 This model showed three hydrogen bonds between the Gln CONH2 of the inhibitor and the protein, highlighting the importance of this residue for recognition and affinity (Fig. 1B).46,54 In the second, docking and molecular dynamics simulations of the peptidomimetic inhibitor, pCinn-Haic-Gln-OH, showed that the glutamine binds in a slightly different pocket (Fig. 1C). A loop of STAT3 (residues 659–668) moves so that Met660 forms a hydrophobic interaction with the five- and six-membered rings of Haic. The main chain NH of Met660 hydrogen bonds with the OH of Tyr657 which is involved with a hydrogen bond with the C = O of Haic.50
Inhibition of STAT3 Phosphorylation in Intact Cells and Development of Phosphopeptide Mimic Prodrugs
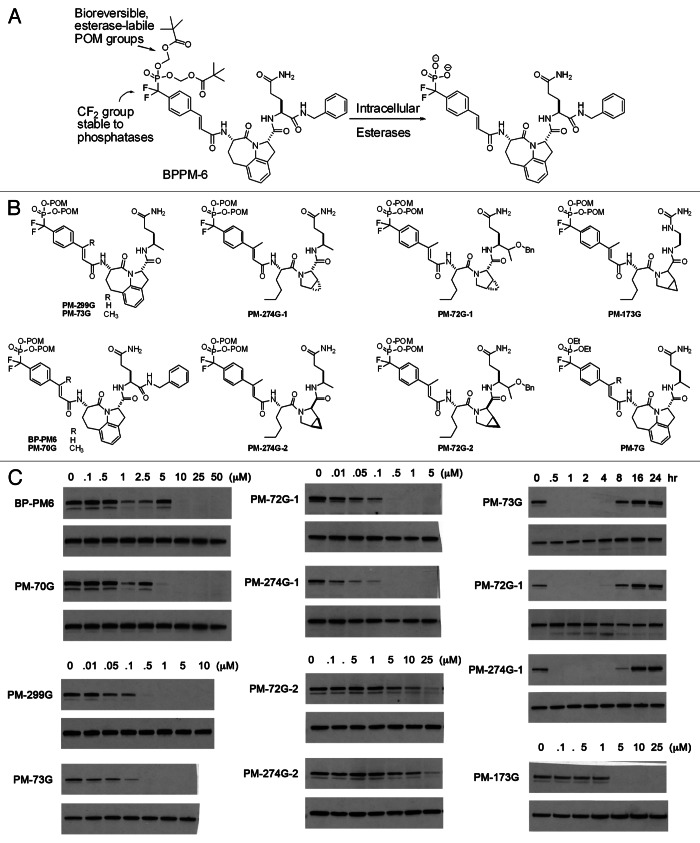
To inhibit STAT3 phosphorylation in intact cells we employed a prodrug approach.53,63 The phosphate was replaced with the phosphatase-stable phosphonodifluoromethyl group.64 The negative charge of the phosphonate oxygens was blocked with the pivaloyloxymethyl group (POM), which is cleaved by carboxyl esterases (Fig. 2A).65 Our first prodrug, BP-PM6 (Fig. 2A), inhibited constitutive phosphorylation of STAT3 in human MDA-MB-468 breast tumor cells at a concentration of 10 μM, supporting the hypothesis that the compound entered the cells, was stripped of its POM groups by esterases and bound to the SH2 domain of STAT3, preventing receptor recruitment and phosphorylation of Tyr705.63 The reduction of pSTAT3 also suggests that STAT3 is phosphorylated and dephosphorylated in a dynamic equilibrium, which we can perturb with phosphopeptide mimics.
Figure 2 (See previous page). Inhibition of the phosphorylation of STAT3 Tyr705 in intact cells. (A) Prodrug strategy showing the phosphatase-stabilizing CF2 substitution and the POM blocking groups. Cleavage of the POM groups liberates the negatively charged phosphonate for binding to the SH2 domain of STAT3. (B) Structures of prodrugs used to study STAT3 phosphorylation. (C) Inhibition of constitutive Tyr705 phosphosphorylation in MDA-MB-468 breast cancer cells. The left column shows the effect of addition of a methyl group on the β-position of the cinnamate (BP-PM6 vs. PM-70G and PM-299G vs. PM-73G) and the increase in potency on substituting the CONBn group with a simple methyl group (BP-PM6 vs. PM 299G and PM-70G vs. PM-73G). The center column shows inhibition by the Nle-mPro-containing prodrugs PM-72G-1 and PM-274G-1 and the reduced potency of the stereoieomers possessing “D” mPro. The right column shows the time course of inhibition of Tyr705 phosphorylation (5 μM prodrug) and the dose response of the prodrug of one of the highest affinity phosphopepides, PM-173G. Gels are presented in pairs in which the upper is pTyr705 STAT3 and the lower is total STAT3. With the exception of the time course study, cells were exposed to prodrugs for 2 h before lysis and protein determination by western blots.
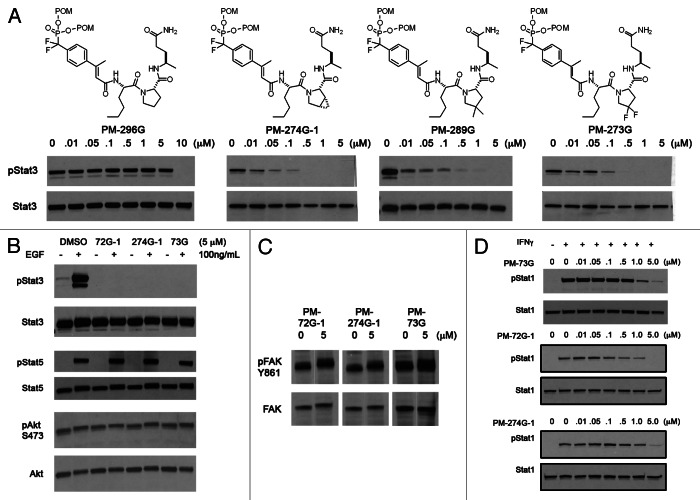
We converted several phosphopeptide mimetics into cell-permeable prodrugs (Fig. 2B). Although the range in affinity of the phosphate bearing root structures was within a factor of 3, the structures of these prodrugs had a striking effect on potency of inhibition of constitutive pSTAT3 in intact MDA-MB-468 cells in culture.51,53,63 Addition of a methyl group to the β-position of the cinnamoyl moiety produced a slight increase in potency (BP-PM6 vs. PM-70G and PM-299G vs. PM-73G, Fig. 2C), which reflected the increase in affinity of the corresponding phosphopeptides for STAT3.53 Interestingly, the potency of prodrugs in which the C-terminal benzylamide group (CONHCH2C6H5) was replaced by a simple methyl group was enhanced > 10-fold (BP-PM6 vs. PM-299G and PM-70G vs. PM-73G, Fig. 2C).53 This was not reflective of the intrinsic affinity of the corresponding phosphopeptides in which the benzylamide-containing peptides were 2-fold more avid than the corresponding methyl substituted peptides.53 Replacing the CONHBn with an isosteric ether CH(CH3)OBn resulted in highly potent inhibition of constitutive pSTAT3 (PM-72G-1).55 Replacement with a methyl group retained the potency in cells (Fig. 2C, PM-72G1 vs. PM-274G-1) whereas in the corresponding phosphopeptides the CH(CH3)OBn resulted in slightly more affinity for isolated STAT3 (2.5-fold) than the methyl group.55 Prodrugs containing mPro were very highly potent inhibitors of STAT3 phosphorylation (PM-72G-1 and PM-274G-1, Fig. 2C).53 As mPro is no longer commercially available and its synthesis by any of the reported methods is expensive and low yield,66,67 we sought less expensive proline derivatives.51 In a prodrug containing native proline (PM-296G, Fig. 3A), complete inhibition occurred at 10 μM. However, prodrugs containing the substituted prolines, mPro, 4,4-dimethylproline and 4,4-difluoroproline were all significantly more potent. Complete inhibition occurred at 500 nM.51 It is unclear at this time why this difference occurs. However, one could speculate that there may be proteolysis of the proline peptide and that the substituted prolines may not fit in the active site of the putative protease.
Figure 3. (A) Effect of proline on the inhibition of STAT3. (B) Prodrugs do not inhibit EGF induced STAT5 phosphorylation or Ser473Akt phosphorylation. (C) Prodrugs do not inhibit Src phosphorylation of Tyr861 of FAK. (D) Prodrugs inhibit IFNγ-stimulated phosphorylation of STAT1 but at 10-fold higher concentration than constitutive STAT3. In (A) cells were treated with prodrug for 2 h before lysis and protein determination by western blots. In (B–D) cells were treated with prodrug for 1.5 h at which time EGF or IFNγ was added. After 30 min cells were lysed and protein levels determined by western blots.
Three prodrugs, PM-73G, PM-274G and PM-72G, were studied in detail (Fig. 2B). These compounds had β-methyl cinnamate and were varied at both the central dipeptide and the glutamine surrogate. After a two-hour exposure of MDA-MB-468 cells, significant inhibition of constitutive phosphorylation of STAT3 was observed at 100 nM and complete inhibition occurred at 500 nM (Fig. 2C). Commercially available mPro is a mixture of “L” and “D” enantiomers, and in the cases of PM-72G and PM-274 two prodrugs were isolated in the synthesis, designated with either a -1 or -2 to reflect the order of elution from the preparative HPLC. The second stereoisomers were much less potent, requiring 25 μM for complete inhibition, which reflects the relative affinities of the phosphopeptides.46 Time course experiments showed significant inhibition at 30 min with recovery of pSTAT3 at about 8 h.53 These materials inhibited Tyr705 phosphorylation in a variety of human cancer cell lines including U266 (multiple myeloma), MDA-MB-231, SUM190, SUM 149 (breast), HCC-827 (lung) and SKOV3-ip (ovarian). They also inhibited IL-6 stimulated phosphorylation in MeWo and A375 (melanoma) and HeyA8 (ovarian) cells.53
Based on similarity of binding free energies of phosphopeptides to a set of SH2 domains, Ladbury and colleagues postulated that selective inhibition of SH2 domains within cells is unlikely.68,69 To test this hypothesis, we assayed for the effect of our STAT3 inhibitors on the activities of SH2 domain-driven pathways. Administration of epidermal growth factor (EGF) to MDA-MB-468 cells resulted in phosphorylation of STAT5. Our prodrugs did not inhibit that process, suggesting that they do not bind appreciably to STAT5 (Fig. 3B). Phosphatidylinositol-3-kinase is recruited to receptors via the SH2 domains of the p85 regulatory domain, which activates the kinase domain leading to the phosphorylation of Akt. Our prodrugs did not inhibit constitutive Ser473 phosphorylation, suggesting that they did not bind to p85 (Fig. 3B). Via its SH2 domain, Src kinase binds to the focal adhesion kinase and selectively phosphorylates Tyr861.70 Our prodrugs did not impact this process indicating selectivity for STAT3 over Src (Fig. 3C). Significant inhibition of interferon-γ stimulated phosphorylation of STAT1 was observed at 1 μM but 5 μM was required for complete inhibition (Fig. 3D). These are 10-fold higher concentrations that observed for the inhibition of pSTAT3. The amino acid sequences and the three dimensional structures of the phosphopeptide binding regions of STAT1 and STAT3 are nearly identical,57 so cross reactivity is not surprising. However, at high concentration (25 μM) selectivity for STAT3 over these processes was abolished.
These results suggest that it is indeed possible to dial in specificity for specific SH2 domains in intact cells, but concentrations must be carefully regulated. Two features of our peptides contribute to the selectivity for the SH2 domain of STAT3: (1) the cinnamic acid-derived pTyr mimic, which reduced affinity for the Src SH2 domain56 and (2) the glutamine surrogate. Most SH2 domains, e.g., Src, p8522 and STAT5,71 recognize hydrophobic residues at pY+3 and the hydrophilic side chain amide of the Gln mimic would not be accommodated in these binding pockets. The Ladbury analysis focused on the SH2 domain of Src.
Effect of STAT3 Inhibition on the Growth and Survival of Cancer Cells In Vitro
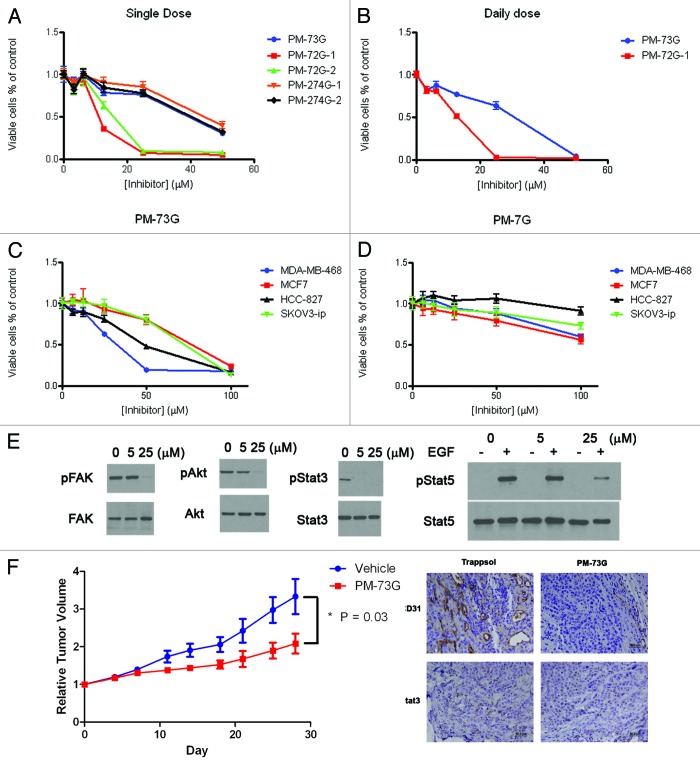
The possible linkage between STAT3 phosphorylation inhibition and cell survival has been a point of marked controversy based on many studies. Our efforts have for the first time provided effective tools for dissecting these responses and reveal them as distinct. We examined the effect prodrugs on proliferation of MDA-MB-468 breast cancer cells using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium (MTT) or sulforhodamine B (SRB) assays. PM-73G, PM-274G-1 and PM-274G-2 showed little or no ability to inhibit growth up to 50 μM (Fig. 4A), which is a concentration 100 times that at which constitutive pSTAT3 was completely inhibited. However, both stereoisomers of PM-72G inhibited growth at with an IC50 of 10–15 μM. Since PM-72G-2, which contains the “D” stereoisomer of mPro and required 25 μM to inhibit pSTAT3, also inhibits cell growth, it is likely that inhibition by both of the PM-72G versions is due to off-target effects due to the C-terminal Gln-benzyl ether group, which is missing in structural analog PM-274G and in PM-73G. Realizing that levels of pSTAT3 recover after about eight hours following exposure to prodrug, assays were repeated with daily administration of PM-73G and PM-72G-1 (Fig. 4B). The IC50 for PM-73G was between 25 μM and 50 μM, which is 50 times higher than the 0.5 μM concentration that completely inhibited pSTAT3. Growth and survival assays were also performed on multiple tumor cell lines including melanoma (A375 and MeWo), lung (HCC-827, H1299, H1819, H520 H528 and A549), breast (MCF7, MDA-MB-231, SUM190 and SUM149), ovarian (SKOV3-ip) and multiple myeloma (U266) cells, again with no appreciable toxicity detected at 5 μM. A direct comparison of MCF7 cells (no constitutive pSTAT3) and SKOV3-ip, HCC-827 and MDA-MB-468 cells (constitutive pSTAT3) with daily administration of PM-73G revealed no correlation between STAT3 phosphorylation and cytotoxocity (Fig. 4C).53 No inhibition of cyclin D1 or Bcl protein expression at 5 μM of PM-73G, PM-72G-1 or PM-274G-1 was observed. PM-7G does not have cleavable phosphonate blocking groups and it displays minimal effect on survival of MCF7, SKOV3-ip, HCC-827 and MDA-MB-468 cells (Fig. 4D). Thus the cinnamoyl-Haic-Apa sub-structure is not cytoxic on its own and the phosphonate contributes to the observed cell death.
Figure 4. (A–D) Prodrugs are minimally cytotoxic to cultured cancer cell lines. (E) At high concentration (25 μM), PM-73G displays off-target effects by inhibition of the phosphorylation of STAT5, Ser473Akt and Tyr861FAK. (F) Intraperitoneal administration of PM-73G inhibits breast tumor growth and angiogenesis.
At the relatively high concentration of 25 μM in MDA-MB-468 cells, PM-73G inhibited phosphorylation of FAK pTyr861 and Akt Ser473 after two hour treatment, as well as EGF-stimulated STAT5 phosphorylation (Fig. 4E). Thus, at high concentrations, selectivity for individual SH2 domains is compromised and cytotoxicity correlates with off-target effects.
Collectively, these results challenge the hypothesis that Tyr705 phosphorylation of STAT3 is required for cell growth and proliferation in vitro. Further challenges come from inhibition of pSTAT3 by JAK kinase inhibitors. The first example came from Kreis et al., who noted that treatment of several melanoma lines with Pyridone 6 completely inhibited STAT3 phosphorylation but had no impact on cell growth.72 Hedvat et al. reported that the JAK2 inhibitor, AZD1480, at concentrations that completely inhibited pSTAT3, had no effect on the proliferation of MDA-MB-468 (breast), DU145 (prostate) and MDAH2774 (ovarian) cancer cells in vitro.73 This was reported in subsequent publications from this group74,75 and others.76 Looyenga et al. found that the JAK1/2 inhibitor ruxolitinib did not affect growth of lung cancer cell lines in vitro.77 Treatment of ovarian cancer cells with the anti-IL-6 monoclonal antibody siltuximab inhibited STAT3 phosphorylation but did not affect proliferation of cells grown on plastic as adherent cultures.78 Thus it would appear that selective inhibition of Tyr705 phosphorylation is not cytotoxic to cancer cells of epithelial origin in vitro. Furthermore, if an agent or compound is killing these cells, it is acting by off-target effects. Controversy still exists as Zhang et al. recently reported that an apparently selective small molecule (not a phosphopeptide mimic) targeting the SH2 domain of STAT3 displays cytotoxicity in vitro.39
Effect of Selective STAT3 Inhibition In Vivo
In spite of the lack of cytotoxicity, we evaluated the ability of PM-73G to inhibit tumor growth in vivo using the MDA-MB-468 breast tumor model.79 In an initial intratumoral (IT) administration trial, we found that tumor growth was inhibited which was accompanied by a reduction in tumor microvessel density and VEGF protein. IT administration of concentrations as low as 8 μM resulted in inhibition of pSTAT3 in tumor sections as determined by immunohistochemical staining of tumor sections using anti-pSTAT3 antibodies.
To determine the utility of systemic administration, mice bearing MDA-MB-468 tumors were treated with 170 mg/kg of PM-73G administered intraperitoneally (i.p.) daily for 5 d followed by two days rest over four weeks.79 (Rodents have circulating carboxyesterases which prematurely deprotect the POM group, thus necessitating such a high dose.) Tumors from the treated animals grew at much lower rates than those given the vehicle (20% Trappsol/PBS) (Fig. 4F). Immunohistochemical analysis revealed nearly complete inhibition of vascularization. Two hours after administration of PM-73G pSTAT3 levels were significantly reduced, compared with vehicle (Fig. 4F). Thus selectively inhibiting STAT3 phosphorylation impedes communication with the microenvironment, i.e., VEGF signaling and angiogenesis. Necroscopic examination revealed no organ toxicity and no changes in complete blood counts (cbcs). To the best of our knowledge, this is the first example of a phosphopeptide-based prodrug targeting an SH2 domain showing the ability to inhibit its target by systemic administration. This contrasts with the compounds of Zhang et al., which utilize carboxyphenyl groups to target the phosphotyrosine binding pocket.38,39
Of the four cell lines examined in Figure 4C, MDA-MB-468 was the most sensitive to growth inhibition in vitro. The others were not evaluated in vivo so it is unclear if the observed reduction in tumor growth and microvessel density is cell line-dependent. However, our results are similar to those recently reported for JAK2 kinase inhibitors AZD148075 and ruxolotinib77 that employed other cell lines. Whereas inhibition of STAT3 phosphorylation is not intrinsically cytotoxic, anti-tumor activity is the result of impaired communication between tumor cells and the microenvironment, e.g., VEGF production and activity. AZD1480 impacts immune cell recruitment to the tumor, supporting the proposed role(s) of STAT3 in immune surveillance and tumor immunity.75
Synthesis of Amino Acid Surrogates and Peptidomimetics
The structure-affinity and structure-activity studies described in this communication utilized phosphotyrosine, Leu-Pro dipeptide and glutamine surrogates that were not available commercially. Synthetic strategies had to be developed for these materials which was a major part of the program. Readers are referred to references 46, 54 and 55 for the synthesis of glutamine mimics; 49, 50, 51 and 58 for the synthesis of pseudoproline peptides, Leu-Pro mimics and proline analogs; 50, 53 and 63 for constrained tyrosine mimics; and 53 and 63 for the synthesis of the prodrugs.
Summary
At the outset of the program our laboratory had two goals: (1) to develop cell-permeable phosphopeptide mimics targeting an SH2 domain and (2) to use this technology to inhibit an important cancer target, STAT3. Our chemistry effort developed very high affinity phosphopeptides and were able to convert these into prodrugs which hit their target in vivo with systemic (ip) administration. Our data and that of Zhang et al.38,39 suggest that the SH2 domain is indeed druggable, despite the failed attempts of the industrial, government and academic labs mentioned above. As mentioned, the bis-POM prodrug strategy is useful for proof of principle studies, but it suffers from premature loss of one of the POM groups due to both esterase activity and chemical hydrolysis. Improvements to the bio-reversible ester strategy are ongoing. Although we do not have a clinical candidate as of yet, our selective phosphopeptide mimics have been useful tools for the study of Tyr705 phosphorylation. From our work and the studies of the JAK inhibitors, the dogma of STAT3 signaling is shifting. For epithelial tumors, it appears that phosphorylated STAT3 is not necessary for tumor cell survival. However, the original studies on the effects of STAT3 on VEGF expression and signaling80,81 have borne true. Inhibition of STAT3 Tyr705 phosphorylation appears to be new antiangiogenesis strategy.
Acknowledgments
We are especially grateful to Dr Timothy Schaefer who got us started on the STAT3 program. His departure from the MD Anderson Cancer Center was a setback for JAK-STAT science. We are grateful to the MD Anderson Cancer Center that provided us the seed money, which led to funding by the NCI, RO1 CA096652. We acknowledge the MDACC SPORE in ovarian Cancer P50 CA083639, the UT SPORE in lung cancer P50 CA070907 and the MDACC SPORE in Melanoma, P50 CA093459 and the CTT/TI-3D Chemistry and Molecularly-Targeted Therapeutic Development Grant Program for support of this work and the NCI Cancer Center Support Grant CA016672 for the support of our NMR facility and the Translational Chemistry Core Facility for mass spectrometry and X-ray crystallography. We are grateful to Dr Xiaomin Chen and Dr Zhiyong Ren who expressed copious amounts of STAT3 and who crystallized the protein.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/22682
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–88. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- 3.Bromberg J. Stat proteins and oncogenesis. J Clin Invest. 2002;109:1139–42. doi: 10.1172/JCI15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costantino L, Barlocco D. STAT 3 as a target for cancer drug discovery. Curr Med Chem. 2008;15:834–43. doi: 10.2174/092986708783955464. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher S, Turkson J, Gunning PT. Molecular approaches towards the inhibition of the signal transducer and activator of transcription 3 (Stat3) protein. ChemMedChem. 2008;3:1159–68. doi: 10.1002/cmdc.200800123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy DE, Darnell JE., Jr. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–62. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 8.Lee H, Pal SK, Reckamp K, Figlin RA, Yu H. STAT3: a target to enhance antitumor immune response. Curr Top Microbiol Immunol. 2011;344:41–59. doi: 10.1007/82_2010_51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kortylewski M, Yu H. Role of Stat3 in suppressing anti-tumor immunity. Curr Opin Immunol. 2008;20:228–33. doi: 10.1016/j.coi.2008.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haftchenary S, Avadisian M, Gunning PT. Inhibiting aberrant Stat3 function with molecular therapeutics: a progress report. Anticancer Drugs. 2011;22:115–27. doi: 10.1097/CAD.0b013e328341185b. [DOI] [PubMed] [Google Scholar]
- 12.Lavecchia A, Di Giovanni C, Novellino E. STAT-3 inhibitors: state of the art and new horizons for cancer treatment. Curr Med Chem. 2011;18:2359–75. doi: 10.2174/092986711795843218. [DOI] [PubMed] [Google Scholar]
- 13.Mankan AK, Greten FR. Inhibiting signal transducer and activator of transcription 3: rationality and rationale design of inhibitors. Expert Opin Investig Drugs. 2011;20:1263–75. doi: 10.1517/13543784.2011.601739. [DOI] [PubMed] [Google Scholar]
- 14.Berg T. Signal transducers and activators of transcription as targets for small organic molecules. Chembiochem. 2008;9:2039–44. doi: 10.1002/cbic.200800274. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, Stark GR. Roles of unphosphorylated STATs in signaling. Cell Res. 2008;18:443–51. doi: 10.1038/cr.2008.41. [DOI] [PubMed] [Google Scholar]
- 16.Wegrzyn J, Potla R, Chwae YJ, Sepuri NB, Zhang Q, Koeck T, et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–7. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gough DJ, Corlett A, Schlessinger K, Wegrzyn J, Larner AC, Levy DE. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–6. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–80. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 19.Waksman G, Kumaran S, Lubman O. SH2 domains: role, structure and implications for molecular medicine. Expert Rev Mol Med. 2004;6:1–18. doi: 10.1017/S1462399404007331. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Echeverria C. Inhibitors of signaling interfaces: targeting Src homology 2 domains in drug discovery. Protein Tyrosine Kinases 2006:31-52. [Google Scholar]
- 21.Metcalf CA III, Sawyer T. Src homology-2 domains and structure-based, small-molecule library approaches to drug discovery. In: Makriyannis A, Biegel D, eds. Drug Discovery Strategies and Methods. New York, NY: Marcel Dekker, Inc., 2004:23-59. [Google Scholar]
- 22.Sawyer TK. Src homology-2 domains: structure, mechanisms, and drug discovery. Biopolymers. 1998;47:243–61. doi: 10.1002/(SICI)1097-0282(1998)47:3<243::AID-BIP4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 23.Burke TR, Jr., Lee K. Phosphotyrosyl mimetics in the development of signal transduction inhibitors. Acc Chem Res. 2003;36:426–33. doi: 10.1021/ar020127o. [DOI] [PubMed] [Google Scholar]
- 24.Burke TR, Jr., Yao ZJ, Liu DG, Voigt J, Gao Y. Phosphoryltyrosyl mimetics in the design of peptide-based signal transduction inhibitors. Biopolymers. 2001;60:32–44. doi: 10.1002/1097-0282(2001)60:1<32::AID-BIP1002>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 25.Hecker SJ, Erion MD. Prodrugs of phosphates and phosphonates. J Med Chem. 2008;51:2328–45. doi: 10.1021/jm701260b. [DOI] [PubMed] [Google Scholar]
- 26.Liu WQ, Vidal M, Mathé C, Périgaud C, Garbay C. Inhibition of the ras-dependent mitogenic pathway by phosphopeptide prodrugs with antiproliferative properties. Bioorg Med Chem Lett. 2000;10:669–72. doi: 10.1016/S0960-894X(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu WQ, Vidal M, Olszowy C, Million E, Lenoir C, Dhôtel H, et al. Structure-activity relationships of small phosphopeptides, inhibitors of Grb2 SH2 domain, and their prodrugs. J Med Chem. 2004;47:1223–33. doi: 10.1021/jm031005k. [DOI] [PubMed] [Google Scholar]
- 28.Gay B, Suarez S, Caravatti G, Furet P, Meyer T, Schoepfer J. Selective GRB2 SH2 inhibitors as anti-Ras therapy. Int J Cancer. 1999;83:235–41. doi: 10.1002/(SICI)1097-0215(19991008)83:2<235::AID-IJC15>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 29.Gay B, Suarez S, Weber C, Rahuel J, Fabbro D, Furet P, et al. Effect of potent and selective inhibitors of the Grb2 SH2 domain on cell motility. J Biol Chem. 1999;274:23311–5. doi: 10.1074/jbc.274.33.23311. [DOI] [PubMed] [Google Scholar]
- 30.Stankovic CJ, Surendran N, Lunney EA, Plummer MS, Para KS, Shahripour A, et al. The role of 4-phosphonodifluoromethyl- and 4-phosphono-phenylalanine in the selectivity and cellular uptake of SH2 domain ligands. Bioorg Med Chem Lett. 1997;7:1909–14. doi: 10.1016/S0960-894X(97)00334-X. [DOI] [Google Scholar]
- 31.McKinney J, Raimundo BC, Cushing TD, Yoshimura H, Ohuchi Y, Hiratate A, et al. STAT4 and STAT6 binding dipeptide derivatives. Application: WO WO: (Tularik Inc., USA; Taisho Pharmaceutical Co., Ltd.). 2001:85 pp. [Google Scholar]
- 32.Turkson J, Kim JS, Zhang S, Yuan J, Huang M, Glenn M, et al. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261–9. [PubMed] [Google Scholar]
- 33.Turkson J, Ryan D, Kim JS, Zhang Y, Chen Z, Haura E, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276:45443–55. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 34.Gunning PT, Katt WP, Glenn M, Siddiquee K, Kim JS, Jove R, et al. Isoform selective inhibition of STAT1 or STAT3 homo-dimerization via peptidomimetic probes: structural recognition of STAT SH2 domains. Bioorg Med Chem Lett. 2007;17:1875–8. doi: 10.1016/j.bmcl.2007.01.077. [DOI] [PubMed] [Google Scholar]
- 35.Shahani VM, Yue P, Fletcher S, Sharmeen S, Sukhai MA, Luu DP, et al. Design, synthesis, and in vitro characterization of novel hybrid peptidomimetic inhibitors of STAT3 protein. Bioorg Med Chem. 2011;19:1823–38. doi: 10.1016/j.bmc.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahani VM, Yue P, Haftchenary S, Zhao W, Lukkarila JL, Zhang X, et al. Identification of Purine-Scaffold Small-Molecule Inhibitors of Stat3 Activation by QSAR Studies. ACS Med Chem Lett. 2011;2:79–84. doi: 10.1021/ml100224d. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Siddiquee KA, Gunning PT, Glenn M, Katt WP, Zhang S, Schrock C, et al. An oxazole-based small-molecule Stat3 inhibitor modulates Stat3 stability and processing and induces antitumor cell effects. ACS Chem Biol. 2007;2:787–98. doi: 10.1021/cb7001973. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Yue P, Fletcher S, Zhao W, Gunning PT, Turkson J. A novel small-molecule disrupts Stat3 SH2 domain-phosphotyrosine interactions and Stat3-dependent tumor processes. Biochem Pharmacol. 2010;79:1398–409. doi: 10.1016/j.bcp.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Yue P, Page BD, Li T, Zhao W, Namanja AT, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc Natl Acad Sci U S A. 2012;109:9623–8. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Nikolovska-Coleska Z, Yang CY, Gomez C, Gao W, Krajewski K, et al. Design and synthesis of a new, conformationally constrained, macrocyclic small-molecule inhibitor of STAT3 via ‘click chemistry’. Bioorg Med Chem Lett. 2007;17:3939–42. doi: 10.1016/j.bmcl.2007.04.096. [DOI] [PubMed] [Google Scholar]
- 41.Dourlat J, Valentin B, Liu WQ, Garbay C. New syntheses of tetrazolylmethylphenylalanine and O-malonyltyrosine as pTyr mimetics for the design of STAT3 dimerization inhibitors. Bioorg Med Chem Lett. 2007;17:3943–6. doi: 10.1016/j.bmcl.2007.04.107. [DOI] [PubMed] [Google Scholar]
- 42.Ren Z, Cabell LA, Schaefer TS, McMurray JS. Identification of a high-affinity phosphopeptide inhibitor of Stat3. Bioorg Med Chem Lett. 2003;13:633–6. doi: 10.1016/S0960-894X(02)01050-8. [DOI] [PubMed] [Google Scholar]
- 43.Wiederkehr-Adam M, Ernst P, Müller K, Bieck E, Gombert FO, Ottl J, et al. Characterization of phosphopeptide motifs specific for the Src homology 2 domains of signal transducer and activator of transcription 1 (STAT1) and STAT3. J Biol Chem. 2003;278:16117–28. doi: 10.1074/jbc.M300261200. [DOI] [PubMed] [Google Scholar]
- 44.Gerhartz C, Heesel B, Sasse J, Hemmann U, Landgraf C, Schneider-Mergener J, et al. Differential activation of acute phase response factor/STAT3 and STAT1 via the cytoplasmic domain of the interleukin 6 signal transducer gp130. I. Definition of a novel phosphotyrosine motif mediating STAT1 activation. J Biol Chem. 1996;271:12991–8. doi: 10.1074/jbc.271.22.12991. [DOI] [PubMed] [Google Scholar]
- 45.Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE, Jr., Yancopoulos GD. Choice of STATs and other substrates specified by modular tyrosine-based motifs in cytokine receptors. Science. 1995;267:1349–53. doi: 10.1126/science.7871433. [DOI] [PubMed] [Google Scholar]
- 46.Coleman DRI, 4th, Ren Z, Mandal PK, Cameron AG, Dyer GA, Muranjan S, et al. Investigation of the binding determinants of phosphopeptides targeted to the SRC homology 2 domain of the signal transducer and activator of transcription 3. Development of a high-affinity peptide inhibitor. J Med Chem. 2005;48:6661–70. doi: 10.1021/jm050513m. [DOI] [PubMed] [Google Scholar]
- 47.Haan S, Hemmann U, Hassiepen U, Schaper F, Schneider-Mergener J, Wollmer A, et al. Characterization and binding specificity of the monomeric STAT3-SH2 domain. J Biol Chem. 1999;274:1342–8. doi: 10.1074/jbc.274.3.1342. [DOI] [PubMed] [Google Scholar]
- 48.Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–51. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 49.Coleman DRI, Kaluarachchi K, Ren Z, Chen X, McMurray JS. Solid phase synthesis of phosphopeptides incorporating 2,2-dimethyloxazolidine pseudoproline analogs: evidence for trans Leu-Pro peptide bonds in Stat3 inhibitors. Int J Pept Res Ther. 2008;14:1–9. doi: 10.1007/s10989-007-9099-7. [DOI] [Google Scholar]
- 50.Mandal PK, Limbrick D, Coleman DR, Dyer GA, Ren Z, Birtwistle JS, et al. Conformationally constrained peptidomimetic inhibitors of signal transducer and activator of transcription. 3: Evaluation and molecular modeling. J Med Chem. 2009;52:2429–42. doi: 10.1021/jm801491w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mandal PK, Ren Z, Chen X, Kaluarachchi K, Liao WSL, McMurray JS. Structure-Activity Studies of Phosphopeptidomimetic Prodrugs Targeting the Src Homology 2 (SH2) Domain of Signal Transducer and Activator of Transcription 3 (Stat3) Int J Pept Res Ther. 2012 doi: 10.1007/s10989-012-9313-0. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dumy P, Keller M, Ryan DE, Rohwedder B, Woehr T, Mutter M. Pseudo-Prolines as a Molecular Hinge: Reversible Induction of cis Amide Bonds into Peptide Backbones. J Am Chem Soc. 1997;119:918–25. doi: 10.1021/ja962780a. [DOI] [Google Scholar]
- 53.Mandal PK, Gao F, Lu Z, Ren Z, Ramesh R, Birtwistle JS, et al. Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3. J Med Chem. 2011;54:3549–63. doi: 10.1021/jm2000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mandal PK, Heard PA, Ren Z, Chen X, McMurray JS. Solid-phase synthesis of Stat3 inhibitors incorporating O-carbamoylserine and O-carbamoylthreonine as glutamine mimics. Bioorg Med Chem Lett. 2007;17:654–6. doi: 10.1016/j.bmcl.2006.10.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mandal PK, Ren Z, Chen X, Xiong C, McMurray JS. Structure-affinity relationships of glutamine mimics incorporated into phosphopeptides targeted to the SH2 domain of signal transducer and activator of transcription 3. J Med Chem. 2009;52:6126–41. doi: 10.1021/jm901105k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shahripour A, Plummer MS, Lunney E, Para KS, Stankovic CJ, Rubin JR, Humblet C, Fergus JH, Marks JS, et al. Novel phosphotyrosine mimetics in the design of peptide ligands for pp60src SH2 domain. Bioorg Med Chem Lett. 1996;6:1209–14. doi: 10.1016/0960-894X(96)00208-9. [DOI] [Google Scholar]
- 57.McMurray JS. Structural basis for the binding of high affinity phosphopeptides to Stat3. Biopolymers. 2008;90:69–79. doi: 10.1002/bip.20901. [DOI] [PubMed] [Google Scholar]
- 58.Mandal PK, Kaluarachchi KK, Ogrin D, Bott SG, McMurray JS. An efficient synthesis of the constrained peptidomimetic 2-oxo-3-(N-9-fluorenyloxycarbonylamino)-1-azabicyclo[4.3.0]nonane-9-carboxylic acid from pyroglutamic acid. J Org Chem. 2005;70:10128–31. doi: 10.1021/jo0515935. [DOI] [PubMed] [Google Scholar]
- 59.Chen J, Bai L, Bernard D, Nikolovska-Coleska Z, Gomez C, Zhang J, et al. Structure-Based Design of Conformationally Constrained, Cell-Permeable STAT3 Inhibitors. ACS Med Chem Lett. 2010;1:85–9. doi: 10.1021/ml100010j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ren Z, Mao X, Mertens C, Krishnaraj R, Qin J, Mandal PK, et al. Crystal structure of unphosphorylated STAT3 core fragment. Biochem Biophys Res Commun. 2008;374:1–5. doi: 10.1016/j.bbrc.2008.04.049. [DOI] [PubMed] [Google Scholar]
- 61.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–71. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 62.McMurray JS, Ren Z, Mandal PK, Chen X. Model of intermolecular interactions between high affinity phosphopeptides and Stat3. Adv Exp Med Biol. 2009;611:543–4. doi: 10.1007/978-0-387-73657-0_238. [DOI] [PubMed] [Google Scholar]
- 63.Mandal PK, Liao WS, McMurray JS. Synthesis of phosphatase-stable, cell-permeable peptidomimetic prodrugs that target the SH2 domain of Stat3. Org Lett. 2009;11:3394–7. doi: 10.1021/ol9012662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burke TR, Jr., Smyth MS, Otaka A, Nomizu M, Roller PP, Wolf G, et al. Nonhydrolyzable phosphotyrosyl mimetics for the preparation of phosphatase-resistant SH2 domain inhibitors. Biochemistry. 1994;33:6490–4. doi: 10.1021/bi00187a015. [DOI] [PubMed] [Google Scholar]
- 65.Farquhar D, Khan S, Srivastva DN, Saunders PP. Synthesis and antitumor evaluation of bis[(pivaloyloxy)methyl] 2′-deoxy-5-fluorouridine 5′-monophosphate (FdUMP): a strategy to introduce nucleotides into cells. J Med Chem. 1994;37:3902–9. doi: 10.1021/jm00049a009. [DOI] [PubMed] [Google Scholar]
- 66.Fujimoto Y, Irreverre F, Karle JM, Karle IL, Witkop B. Synthesis and x-ray analysis of cis-3,4-methylene-L-proline, the new natural amino acid from horse chestnuts, and of its trans isomer. J Am Chem Soc. 1971;93:3471–7. doi: 10.1021/ja00743a030. [DOI] [PubMed] [Google Scholar]
- 67.Sagnard I, Sasaki NA, Chiaroni A, Riche C, Potier P. Enantioselective Synthesis of Cyclopropane a-Amino Acids: Synthesis of N-Boc-cis.(2S,3R,4S).3,4-Methanoproline and N-Boc-(2S,3R,4S)-3,4-Methanoglutamic Acid. Tetrahedron Lett. 1995;36:3149–52. doi: 10.1016/0040-4039(95)00482-R. [DOI] [Google Scholar]
- 68.Ladbury JE. Protein-protein recognition in phosphotyrosine-mediated intracellular signaling. Protein Reviews. 2005;3:165–84. doi: 10.1007/0-387-24532-4_8. [DOI] [Google Scholar]
- 69.Ladbury JE, Arold S. Searching for specificity in SH domains. Chem Biol. 2000;7:R3–8. doi: 10.1016/S1074-5521(00)00067-3. [DOI] [PubMed] [Google Scholar]
- 70.Calalb MB, Zhang X, Polte TR, Hanks SK. Focal adhesion kinase tyrosine-861 is a major site of phosphorylation by Src. Biochem Biophys Res Commun. 1996;228:662–8. doi: 10.1006/bbrc.1996.1714. [DOI] [PubMed] [Google Scholar]
- 71.Müller J, Schust J, Berg T. A high-throughput assay for signal transducer and activator of transcription 5b based on fluorescence polarization. Anal Biochem. 2008;375:249–54. doi: 10.1016/j.ab.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 72.Kreis S, Munz GA, Haan S, Heinrich PC, Behrmann I. Cell density dependent increase of constitutive signal transducers and activators of transcription 3 activity in melanoma cells is mediated by Janus kinases. Mol Cancer Res. 2007;5:1331–41. doi: 10.1158/1541-7786.MCR-07-0317. [DOI] [PubMed] [Google Scholar]
- 73.Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell. 2009;16:487–97. doi: 10.1016/j.ccr.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Scuto A, Krejci P, Popplewell L, Wu J, Wang Y, Kujawski M, et al. The novel JAK inhibitor AZD1480 blocks STAT3 and FGFR3 signaling, resulting in suppression of human myeloma cell growth and survival. Leukemia. 2011;25:538–50. doi: 10.1038/leu.2010.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xin H, Herrmann A, Reckamp K, Zhang W, Pal S, Hedvat M, et al. Antiangiogenic and antimetastatic activity of JAK inhibitor AZD1480. Cancer Res. 2011;71:6601–10. doi: 10.1158/0008-5472.CAN-11-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loveless ME, Lawson D, Collins M, Nadella MV, Reimer C, Huszar D, et al. Comparisons of the efficacy of a Jak1/2 inhibitor (AZD1480) with a VEGF signaling inhibitor (cediranib) and sham treatments in mouse tumors using DCE-MRI, DW-MRI, and histology. Neoplasia. 2012;14:54–64. doi: 10.1593/neo.111478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Looyenga BD, Hutchings D, Cherni I, Kingsley C, Weiss GJ, Mackeigan JP. STAT3 is activated by JAK2 independent of key oncogenic driver mutations in non-small cell lung carcinoma. PLoS One. 2012;7:e30820. doi: 10.1371/journal.pone.0030820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Coward J, Kulbe H, Chakravarty P, Leader D, Vassileva V, Leinster DA, et al. Interleukin-6 as a therapeutic target in human ovarian cancer. Clin Cancer Res. 2011;17:6083–96. doi: 10.1158/1078-0432.CCR-11-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Auzenne EJ, Klostergaard J, Mandal PK, Liao WS, Lu Z, Gao F, et al. A phosphopeptide mimetic prodrug targeting the SH2 domain of STAT3 inhibits tumor growth and angiogenesis. J Exp Ther Oncol. 2012 In press. [PMC free article] [PubMed] [Google Scholar]
- 80.Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21:2000–8. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 81.Gray MJ, Zhang J, Ellis LM, Semenza GL, Evans DB, Watowich SS, et al. HIF-1alpha, STAT3, CBP/p300 and Ref-1/APE are components of a transcriptional complex that regulates Src-dependent hypoxia-induced expression of VEGF in pancreatic and prostate carcinomas. Oncogene. 2005;24:3110–20. doi: 10.1038/sj.onc.1208513. [DOI] [PubMed] [Google Scholar]