Abstract
Silencing of abnormally activated genes can be accomplished in a highly specific manner using nucleic acid based approaches. The focus of this review includes the different nucleic acid based inhibition strategies such as antisense oligodeoxynucleotides, small interfering RNA (siRNA), dominant-negative constructs, G-quartet oligonucleotides and decoy oligonucleotides, their mechanism of action and the effectiveness of these approaches to targeting the STAT (signal transducer and activator of transcription) proteins in cancer. Among the STAT proteins, especially STAT3, followed by STAT5, are the most frequently activated oncogenic STATs, which have emerged as plausible therapeutic cancer targets. Both STAT3 and STAT5 have been shown to regulate numerous oncogenic signaling pathways including proliferation, survival, angiogenesis and migration/invasion.
Keywords: signal transducer and activator of transcription, nucleic acid based inhibitors, antisense oligonucleotide, siRNA, dominant-negative constructs, G-quartet oligonucleotides, decoy oligonucleotides
Introduction
Of the many approaches that have been developed, nucleic acid based strategies to target the STAT proteins involved in cancer progression have emerged as a rational tool to block STAT activation. Several nucleic acid based strategies have been developed including antisense oligonucleotide, small interfering RNA (siRNA), dominant-negative constructs, G-quartet oligonucleotides and decoy oligonucleotides to target STAT pathways.1 STAT pathways are activated by diverse external signals initiated by a plethora of cytokines and growth factors from cell surface receptors, eliciting rapid changes culminating in the transcriptional induction of target genes in the nucleus.2 Among the seven mammalian STAT proteins, constitutive activation of STAT3 and STAT5 has been reported in several different cancer lines and tumor tissues and hence are considered potential molecular therapeutic targets.3,4 Several lines of evidence suggest that a constitutively active form of STAT3 alone is sufficient to induce neoplastic transformation.2 This was demonstrated in mouse fibroblast cells where STAT3 activation was associated with oncogenic transformation by v-Src and STAT3 inhibition blocks the transformation of mouse fibroblasts.5 In addition, transformed mouse and rat fibroblasts by a constitutively activated mutant form of STAT3 (STAT3C), generated tumors in mice suggesting that STAT3 activation may contribute to tumor formation in human cancers.6 Further, inhibition of STAT5 through the use of dominant-negative inhibitory mutants blocks proliferation of transformed lymphoma cells both in vitro and in vivo models,7 indicating that STAT3 and STAT5 may represent promising molecular targets for cancer therapy.
This review will focus on targeting STAT3 and STAT5 using different nucleic acid based approaches. Nucleic acid based strategies have emerged as a powerful tool to successfully target molecules linked to cancer. These approaches are unique due to their high specificity and selectivity and minimal adverse effects or toxicity. Both STAT3 and STAT5 have structural similarities as they are clustered together on the long (q) arm of chromosome 17 and exist as two isoforms—α and β. The two isoforms of STAT3 are both derived from a single gene by alternative mRNA splicing. The α isoform is the full length STAT protein and the β isoform is the shorter truncated protein and lacks the C-terminal transactivation domain, and has often been used as a dominant negative version of STAT3.8,9 On the contrary, the STAT5 α and β forms are encoded by 2 closely-related genes. Each gene gives rise to a long and short isoform.10 Although STAT5 α and STAT5 β have 94% sequence identity, however they differ in their COOH-terminal transactivation domain and have distinct functional roles.11 STAT proteins consist of an N-terminal domain which is important in STAT dimer-dimer interactions, a coiled-coil domain, a DNA binding domain that forms complexes between STAT proteins and DNA, a linker domain, a Src homology-2 (SH-2) domain engages in dimerization between two activated STAT monomers through reciprocal phospho-tyrosine (pTyr)-SH2 domain interactions, and a C-terminal transactivation domain.8 Most inhibitors target disruption of the SH2 domain which engages in dimerization between two activated STAT monomers through reciprocal phospho-tyrosine (pTyr)-SH2 domain interactions. The dominant-negative strategy to target STAT3 have been designed to interrupt formation of STAT3 dimers by introducing mutation in the Tyr705 residue in the STAT3F mutant and in STAT3D mutant, the residues Glu434 and Glu435 have been mutated to block the DNA-binding activity.12 In the decoy oligonucleotide strategy, double stranded short oligonucleotides bind to dimerized phosphorylated STAT3 or STAT5, so that the genome docking site on STAT3 is occupied and inhibits STAT3 or STAT5 from binding to its DNA-binding site.13,14 Similarly, G-quartet oligonucleotides interact with the SH2 domains of STAT3 homodimers to destabilize dimer formation and disrupt DNA-binding activity.15 Although specificity of the nucleic acid based approaches for their targets have made them increasingly important as therapeutic molecules, off- target effects are also seen.16
Several nucleic acid based therapeutic approaches being developed appear to be promising in pre-clinical models.17 siRNA approaches can silence critical STAT3/5 target genes associated with tumor cell viability, proliferation and metastasis; however, clinical applications of siRNA are still under development.16,18 Preclinical results with G-quartet oligonucleotide indicated efficacy inhibiting STAT3 signaling in cancer models, however its clinical utility remains unknown.19 The STAT3 decoy demonstrated antitumor efficacy in several preclinical cancer models2 and recently completed a phase 0 clinical trial in head and neck cancer patients where pharmacodynamic effects were observed following a single intratumoral inoculation.20 While the parent formulation of the STAT3 decoy required local delivery, a cyclic formulation demonstrated increased thermal and enzymatic stability and may be amenable to systemic administration in humans.20
Nucleic Acid Based Therapies Targeting STATs and Their Mechanism of Action
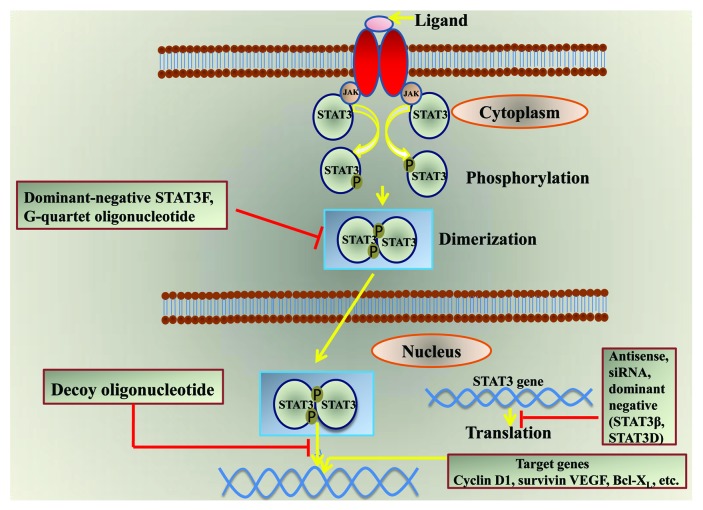
Nucleic acid based strategies developed to block STAT3 and STAT5 gene expression include antisense approaches [antisense RNA and small interfering RNA (siRNA)], dominant-negative constructs, G-quartet oligonucleotides and decoy oligonucleotides. The four different nucleic acid based approaches targeting STAT3 are shown in Figure 1.
Figure 1. Nucleic acid-based approaches targeting STAT3.
Antisense approaches
Several antisense strategies including antisense RNA and siRNA have successfully been used to blocked STAT-mediated gene expression in several preclinical cancer models.21 The advantage of an antisense approach is the potential affinity and specificity for targeting gene expression. An antisense RNA approach is based on short sequences of RNA that target a complementary coding sequence of mRNA and mediate gene suppression at the transcriptional level.22 High affinity binding either occurs in the cytoplasm or in the nucleus. Gene inactivation is initiated by formation of the DNA-RNA heteroduplex causing stearic blockade of the ribosome complex or by mRNA cleavage by RNaseH.23 Antisense oligonucleotides have been used to interrupt constitutively active STATs and block cells from undergoing malignant transformation.2 However, the major obstacle to the successful clinical use of antisense oligonucleotides include several off target effects such as elevation of liver enzymes and/or liver failure, splenomegaly, immune stimulation, thrombocytopenia and prolongation of the activated partial thromboplastin time have been reported in animal models.23 RNA interference (RNAi) is a sequence-specific post transcriptional gene silencing approach that has evolved into a powerful research tool for analyzing gene function in the treatment of cancer, and in the development of highly specific therapeutics.24 Chemically synthesized small interfering RNA (siRNA) represent a class of double stranded RNA molecules that can induce RNA interference (RNAi). siRNAs are 20–25 nucleotides in length metabolized from a large RNA molecule by an endogenous nuclease. The siRNA molecules in turn, bind to a protein complex, termed RNA-induced silencing complex (RISC), which unwinds the two strands of RNA molecules, allowing the antisense strand to bind to the targeted RNA molecule causing effective suppression of gene expression. Both in vitro and in vivo work have demonstrated the therapeutic potential of the RNAi method.25,26 siRNA targeting STAT3 and STAT5 have been used to silence gene expression in several cancers.16,18 Interference with STAT3 signaling using siRNA inhibited cell proliferation, induced apoptosis, downmodulated expression of STAT3 target genes and suppressed tumor growth in several different cancer models24,27-31 (see Table 1). Silencing STAT5 using STAT5 siRNA demonstrated decreased proliferation, invasion and metastasis in various cancers32,33 (see Table 1). Although an siRNA approach is being successfully used in the knockdown of gene expression, cationic lipids are needed for efficient uptake. Thus, the clinical utility of this class of therapeutics is limited due to challenges in drug delivery to the target organs.29 Initial clinical trials used systemically administered naked siRNA showed rapid clearance of the siRNA within minutes.30 Several approaches are being developed to enhance efficient uptake of siRNA. Hence cell-specific siRNA delivery systems are being considered to improve stability and uptake by the target cells. The first targeted delivery of siRNA in humans employed cyclodextrin polymer-based nanoparticles coupled to transferrin which were able to bind to transferrin receptors that are typically upregulated on cancer cells.30 To facilitate targeted delivery of STAT3-siRNA, an anti-Lewis-Y (Ley) monoclonal antibody (hu3S193) was used.29 Ley antigen is expressed in over 70% of epithelial cancers including breast, colon, ovary, prostate and lung cancers and selectively targets Ley-expressing tumors, with minimal uptake by the normal tissues. The STAT3siRNA-hu3S193 construct induced STAT3 knockdown by approximately 70% in association with inhibition of cellular proliferation by approximately 50% suggesting hu3S193 antibody may represent an effective vehicle for the targeted delivery of siRNAs.29 siRNA targeting STAT5 have also shown growth inhibition in cancer models32,33 (see Table 1). Further, suppression of STAT5 signaling using STAT5-siRNA in colorectal cancer cells, provided evidence that STAT5 is embedded in a complex signaling network and may engage in crosstalk with members of other pathways, such as MAPK.34 It is also becoming increasingly critical that cancers that rely on complicated crosstalk between a number of signaling pathways would require multi-targeted therapies.35 Although efforts are being made to improve the delivery of the siRNA to the target site, however, their efficient uptake is still the major obstacle for clinical use.
Table 1. siRNA targeting STAT3 and STAT5 in cancer.
| Preclinical cancer models | Target protein | Key findings |
|---|---|---|
| Laryngeal cancer23,30 |
STAT3 |
Decreased STAT3 expression, reduced tumor volume, suppressed growth and induced apoptosis |
| Pancreatic cancer26 |
STAT3 |
Inhibited cell proliferation, induced apoptosis, downmodulated STAT3 target genes and suppressed tumor growth |
| Breast cancer27 |
STAT3 |
Reduced STAT3 target gene expression and caused significant reduction in tumor volume |
| Esophageal carcinoma31 |
STAT5 |
Decreased proliferation, invasion and metastasis in association with an induction in apoptosis and an increase in the G0/G1 phase |
| Acute myeloid leukemia32 | STAT5 | Inhibited cell proliferation and survival |
Dominant-negative approaches
Dominant-negative approaches involve mutating the functional domain to generate a gene product that can interfere with the function of the normal gene leading to reduced levels of gene activation. Several dominant-negative constructs targeting STAT3 or STAT5 have been designed and developed. Gene therapy with STAT3β, a dominant-negative variant of STAT3 has been shown to interfere with STAT3-mediated gene regulation and block cell transformation. STAT3β is a naturally occurring splice variant of STAT3 that lacks the Ser727 phosphorylation site at the c-terminal transcriptional activation domain where a unique 7-amino acid sequence functions as a dominant-negative form of STAT3 in many cellular contexts.36,37 Evidence suggests that STAT3β functions as a dominant- negative form of STAT3 in melanoma model where overexpression of STAT3β resulted in the cell death of B16 melanoma cells in vitro and suppression of tumor growth in vivo.36 Inhibition of STAT3 activity by dominant-negative STAT3β in a head and neck squamous cell carcinoma (HNSCC) cell line provided evidence that STAT3 signaling mediates cell growth and apoptosis in HNSCC.38 Dominant-negative mutants of STAT3 such as STAT3D and STAT3F have been reported to inhibit the DNA binding activity of STAT339 and have shown effectiveness in both in vitro and in vivo tumor models.37 STAT3F contains a phenylalanine substitution for the c-terminal tyrosine phosphorylation site, which prevents it from undergoing dimerization and nuclear translocation, functioning in a dominant-negative manner.40 The second mutant, STAT3D, has alanine substitutions for Glu 434 and Glu 435 in the DNA binding region and is unable to bind DNA.40 STAT3D forms inactive heterodimers with endogenous STAT3 and inhibits signaling. In vitro and in vivo models have shown that dominant-negative forms of STAT3 can modulate the function of STAT3 and perturb cellular proliferation and transformation. STAT3F has been used to investigate the role of STAT3 in cytokine-dependent induction of target genes in HepG2, a human hepatocellular liver carcinoma cell line.41 The mutant STAT3 can bind to IL-6 induced activated gp130 receptor, but phenylalanine at residue 705 can no longer be phosphorylated, preventing activation of STAT3. In cervical cancer, blocking STAT3 by the dominant negative STAT3D inhibited VEGF production, which has been reported to contribute to tumor angiogenesis.42 Abrogation of STAT3 by dominant-negative STAT3D increased apoptosis in a human HNSCC xenograft model.43
Carboxyl-truncated variants of STAT5a and STAT5b function as dominant- negative forms of STAT5 isoforms.44 In breast cancer, STAT5aΔ740, which corresponds to a naturally occurring alternative splice variant and STAT5aΔ713, derived by truncation after amino acid residue Ala-713, was analogous to an 80 kDa STAT5a product of a nuclear protease and demonstrated comparable dominant-negative properties and suppressed transcriptional activity of wild-type STAT5a and STAT5b.45 The dominant-negative STAT5aΔ740 inhibited growth and induced apoptosis in estrogen responsive breast cancer cells and inhibited tumor growth.46 In prostate cancer cells, blocking STAT5 using the dominant-negative STAT5aΔ713 induced apoptosis.47 In HNSCC, the dominant-negative mutant Stat5b754 inhibited in vitro cell proliferation.48 Although the dominant negative approach has wide application to the study of a number of different kinds of proteins however it tends to be highly effective for proteins that need to assemble into multimers to be functional.
G-quartet oligonucleotides
G-quartet oligonucleotides (GQ-ODN) are a unique class of anticancer agents that interact directly with a target protein and interfere with its function. GQ-ODN consist of G-rich oligonucleotides, which form intramolecular four stranded G-quartet structures.49 G-quartets arise from the association of four G-bases and each G-base makes two H-bonds with its neighboring G-base to form a macrocycle and stack on top of each other to stabilize the polyguanylate assemblies and give rise to tetrad-helical structures.50 G-quartet oligonucleotides have been developed to modulate several biological processes including inhibition of the oncogene STAT3.51 Computer-based docking analysis revealed that GQ-ODN specifically blocked DNA-binding activity of STAT3 by interacting with the SH2 domains of STAT3 homodimers.15 Selective targeting of STAT3 over STAT1 by G-quartet oligonucleotides was based on a few critical amino acids as determined using computational analyses.15 However, in vitro and in vivo antitumor efficacy of G-quartet oligonucleotides targeting STAT3 required polyethyleneimine for effective delivery of G- quartet oligonucleotides to hepatocellular carcinoma cells. The antiproliferative activity of G-quartet oligonucleotides has also shown effectiveness in other cancer models such as prostate cancer,19 HNSCC15 NSCLC52 (see Table 2). A single stranded DNA expression vector has been used for efficient delivery of G-quartet oligonucleotides.53 A G-quartet oligonucleotide approach to target STAT5 has not been reported. However, the effectiveness of G-quartet oligonucleotides as a therapeutic modality may require further optimization of the physicochemical properties to increase selectivity and specificity in inhibiting the target protein to facilitate clinical development.
Table 2. G-quartet oligonucleotides targeting STAT3 and STAT5 in cancer.
| Preclinical cancer models | Target protein | Key findings |
|---|---|---|
| Head and neck squamous cell carcinoma14 |
STAT3 |
Reduced expression of STAT3 target genes, induced apoptosis and inhibited growth in vitro and in vivo |
| Prostate cancer18 |
STAT3 |
Induced apoptosis and reduced expression of target genes in vitro |
| Non-small cell lung cancer51 | STAT3 | Downmodulated expression of STAT3, p-STAT3 and Bcl-XL and induced apoptosis in vitro |
Decoy oligonucleotides
Of the several nucleic acid based strategies that have been introduced in the inhibition of gene expression, synthetic double stranded oligonucleotides (called “decoy oligonucleotide”) that mimic the consensus binding site within the cis-acting elements of its target genes and attenuate the binding of the transcription factor to promoter regions of its target genes to block their expression, have been tested successfully in a clinical trial.54 Decoy oligonucleotides are highly selective and have shown to be effective in both in vitro and animal models. The first decoy oligonucleotide was developed for tissue specific regulation of renin gene expression.55 Since then, several transcription factor decoys have been developed such as E2F-1, CREB and NFκB for several disease states.56 Treatment with an NFκB decoy in a prostate cancer cell line overexpressing the NFκB protein resulted in suppression of cell proliferation, induced apoptosis and reduced several downstream target gene expression.57 In human osteosarcoma and human cervical carcinoma cells, the E2F decoy oligonucleotide inhibited proliferation and target gene expression.58 The CRE decoy targeting against the cAMP response element (CRE) transcription factor, inhibited of growth of breast cancer models in association with inhibition of CRE-directed gene transcription.59
Our laboratory developed a highly specific double-stranded decoy oligonucleotide targeting STAT3. The STAT3 decoy is a 15-mer double-stranded oligonucleotide, with phosphorothioate modifications of three nucleotides at the 5′ and 3′ end, which corresponds to the DNA binding region within the c-fos promoter.60 Phosphorothioate-modified DNA was first synthesized by Eckstein and colleagues in the early 1960s,61 differs from natural DNA in that one of the nonbridging oxygen atoms in phosphodiester linkage is substituted with sulfur to protect the decoy oligonucleotide from nuclease degradation.22 The STAT3 decoy enters cells, competes for binding with the endogenous transcription factor, and has the potential to attenuate the binding of the transcription factor to promoter regions of target genes thereby inhibiting target gene expression.62 The STAT3 decoy demonstrated selective binding for STAT3 protein and inhibited the proliferation and survival of head and neck squamous cell carcinoma (HNSCC) cells in vitro60 and the growth of HNSCC xenograft tumors in vivo.63 Subsequent investigations by others demonstrated that the STAT3 decoy exhibited anti-tumor activity in a variety of preclinical models including cancers of the lung, breast, skin, brain, colorectal and ovary64-69 (see Table 3). Preclinical studies of the STAT3 decoy in animal models demonstrated that it was well tolerated and lacked toxicity.70 A phase 0 clinical trial was recently reported showing downregulation of STAT3 target gene expression in the post-treatment HNSCC tumor.20 STAT3 decoy was injected intratumorally in escalating dose ranging from 250 µg to 1 mg per injection into HNSCC tumors in patients undergoing surgical resection (5–6 patients per dose). Tumors were biopsied prior to treatment and after completion of surgery. There was no evidence of toxicity and decrease in STAT3 target gene expression was observed in the post-treatment STAT3 decoy group compared with the pre-treatment levels. Investigation of the effect of STAT3 decoy on STAT1 mediated signaling due to the high sequence homology between STAT3 and STAT1 (72% protein sequence) suggested that the therapeutic efficacy of STAT3 decoy are independent of STAT1 activation.71
Table 3. Decoy oligonucleotides targeting STAT3 and STAT5 in cancer.
| Preclinical cancer models | Target protein | Key findings |
|---|---|---|
| Head and neck squamous cell carcinoma59,62 |
STAT3 |
In vitro and in vivo antitumor efficacy associated with downmodulation of STAT3 target gene expression |
| Lung cancer63 |
STAT3 |
Induced apoptosis and downregulated STAT3 target genes both in vitro and in vivo and inhibited tumor growth |
| Breast cancer64 |
STAT3 |
Retarded tumor growth accompanied by immune activation |
| Skin cancer65 |
STAT3 |
Inhibited growth both in vitro and in vivo |
| Brain cancer66 |
STAT3 |
Suppressed in vivo tumor growth by inhibiting proliferation and promoting apoptosis |
| Colorectal cancer67 |
STAT3 |
Inhibition of phospho-STAT3 nuclear localization and in vitro cell death |
| Ovarian cancer68 |
STAT3 |
Inhibited cancer cell invasion and enhanced sensitivity to paclitaxel |
| Leukemia13 | STAT5 | Inhibited growth and proliferation |
However, the utility of the parent decoy oligonucleotide formulation is limited due to rapid degradation in the presence of nucleases.4 Hence, chemical modification to increase biostability of the transcription factor decoys is under active investigation. Locked nucleic acid (LNA) can replace nucleotides in the decoy backbone resulting in enhanced stability.72 LNA are nucleic acid analogs containing a methylene linkage between the 2′ oxygen and the 4′ carbon of the ribose ring.72 Positioning of LNA in the decoy oligonucleotide backbone is critical, to prevent any reduction of affinity of transcription factor decoy for its target sequence. Crinelli et al. reported that intra‐ and inter‐strand positioning of LNA is important for NFκB binding affinity, their inter‐strand positioning is critical for stability.72 Presently, data on the antitumor efficacy of LNA- modified transcription factor decoy is lacking. We modified our parent STAT3 decoy by creating a cyclic structure that demonstrated enhanced stability in serum and was found to be efficacious in a HNSCC xenograft model upon intravenous administration.20
Targeting STAT5 using a decoy oligonucleotide has been shown to inhibit the growth and proliferation of leukemia cells in vitro.14 There are no other reports to date of the STAT5 decoy in other cancer types.
Overall, the decoy oligonucleotide approach represents a potential approach for deriving novel anti-STAT therapeutic agents. While the phase 0 trial of the STAT3 decoy suggests activity in human tumors, the optimum physicochemical properties that will result in a suitable bioavailability profile, low toxicity, and good pharmacological properties remain incompletely understood.
Conclusion
Cumulative evidence has identified STAT3 and STAT5 as potential targets for cancer therapy. Disrupting STAT3 and STAT5 signaling in tumor cells by various nucleic acid based approaches such as antisense RNA, small interfering RNA (siRNA), dominant-negative constructs, G-quartet oligonucleotide and transcription factor decoys has been shown to induce apoptosis, inhibit cell proliferation, suppress angiogenesis and inhibit tumor growth in preclinical cancer models. Although nucleic acid based gene suppression technologies are making significant contribution as anticancer agents primarily because of their selective recognition of molecular targets and pathways and have the potential advantage of precisely targeting a gene, however the drawback of each approach such as poor cellular uptake and rapid in vivo degradation potentiates the development of novel delivery systems to facilitate cellular internalization with retained activity. Nuclease degradation of oligonucleotides can be circumvented by chemical derivatization of the backbone and/or by the protection and stability offered by DNA delivery systems. For efficient delivery of nucleic acid based therapeutics, both viral and non-viral delivery systems are being used. Despite the appreciable success of cationic lipids in gene transfer, safety is a major concern in human studies. Lipid based or magnetic nanoparticles are also being extensively studied to improve nucleic acid delivery to the target. Several nanoparticles are under development and some have shown effective for gene or small interfering RNA (siRNA) delivery. Although rapid developments have been made to facilitate uptake and delivery of nucleic acid based therapeutics using several novel delivery tools, however further research is needed to make these tools effective for clinical use.
Footnotes
Previously published online: www.landesbioscience.com/journals/jak-stat/article/22312
References
- 1.Durfort T, Tkach M, Meschaninova MI, Rivas MA, Elizalde PV, Venyaminova AG, et al. Small interfering RNA targeted to IGF-IR delays tumor growth and induces proinflammatory cytokines in a mouse breast cancer model. PLoS One. 2012;7:e29213. doi: 10.1371/journal.pone.0029213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leeman RJ, Lui VW, Grandis JR. STAT3 as a therapeutic target in head and neck cancer. Expert Opin Biol Ther. 2006;6:231–41. doi: 10.1517/14712598.6.3.231. [DOI] [PubMed] [Google Scholar]
- 3.Levy DE, Lee CK. What does Stat3 do? J Clin Invest. 2002;109:1143–8. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 5.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr. Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–8. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bromberg JFWM, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, et al. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/S0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 7.Frank D. Stat Signaling in Cancer: Insights into pathogenesis and treatment strategies. Signal transduction in cancer. 2003;115:p277. doi: 10.1007/0-306-48158-8_11. [DOI] [PubMed] [Google Scholar]
- 8.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2:536–50. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y, Qiu J, Dong S, Redell MS, Poli V, Mancini MA, et al. Stat3 isoforms, alpha and beta, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3beta mapping to its unique C-terminal end. J Biol Chem. 2007;282:34958–67. doi: 10.1074/jbc.M704548200. [DOI] [PubMed] [Google Scholar]
- 10.Azam M, Lee C, Strehlow I, Schindler C. Functionally distinct isoforms of STAT5 are generated by protein processing. Immunity. 1997;6:691–701. doi: 10.1016/S1074-7613(00)80445-8. [DOI] [PubMed] [Google Scholar]
- 11.Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, et al. Stat5b is essential for natural killer cell-mediated proliferation and cytolytic activity. J Exp Med. 1998;188:2067–74. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakajima K, Yamanaka Y, Nakae K, Kojima H, Ichiba M, Kiuchi N, et al. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 1996;15:3651–8. [PMC free article] [PubMed] [Google Scholar]
- 13.Barton BE, Murphy TF, Shu P, Huang HF, Meyenhofer M, Barton A. Novel single-stranded oligonucleotides that inhibit signal transducer and activator of transcription 3 induce apoptosis in vitro and in vivo in prostate cancer cell lines. Mol Cancer Ther. 2004;3:1183–91. [PubMed] [Google Scholar]
- 14.Wang X, Zeng J, Shi M, Zhao S, Bai W, Cao W, et al. Targeted blockage of signal transducer and activator of transcription 5 signaling pathway with decoy oligodeoxynucleotides suppresses leukemic K562 cell growth. DNA Cell Biol. 2011;30:71–8. doi: 10.1089/dna.2010.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jing N, Zhu Q, Yuan P, Li Y, Mao L, Tweardy DJ. Targeting signal transducer and activator of transcription 3 with G-quartet oligonucleotides: a potential novel therapy for head and neck cancer. Mol Cancer Ther. 2006;5:279–86. doi: 10.1158/1535-7163.MCT-05-0302. [DOI] [PubMed] [Google Scholar]
- 16.Cumaraswamy AT. A; Resetca, D; Minden, MD; and Gunning, PT Inhibitors of Stat5 protein signalling. Med Chem Commun. 2012;2:22–7. doi: 10.1039/c1md00175b. [DOI] [Google Scholar]
- 17.Figueiredo ML, Kao C, Wu L. Advances in preclinical investigation of prostate cancer gene therapy. 2007;Molecular therapy: the journal of the American Society of Gene Therapy15:1053–64. doi: 10.1038/sj.mt.6300181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang G, Huang C, Cao J, Huang KJ, Jiang T, Qiu ZJ. Lentivirus-mediated shRNA interference targeting STAT3 inhibits human pancreatic cancer cell invasion. World J Gastroenterol. 2009;15:3757–66. doi: 10.3748/wjg.15.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weerasinghe P, Li Y, Guan Y, Zhang R, Tweardy DJ, Jing N. T40214/PEI complex: a potent therapeutics for prostate cancer that targets STAT3 signaling. Prostate. 2008;68:1430–42. doi: 10.1002/pros.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen M, Thomas SM, Kim S, Yeh JI, Ferris RL, Johnson JT, et al. First-in-human trial of a STAT3 decoy oligonucleotide in head and neck tumors: implications for cancer therapy. Cancer Discov. 2012;2:694–705. doi: 10.1158/2159-8290.CD-12-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yue P, Turkson J. Targeting STAT3 in cancer: how successful are we? Expert Opin Investig Drugs. 2009;18:45–56. doi: 10.1517/13543780802565791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dean NM, Bennett CF. Antisense oligonucleotide-based therapeutics for cancer. Oncogene. 2003;22:9087–96. doi: 10.1038/sj.onc.1207231. [DOI] [PubMed] [Google Scholar]
- 23.Engelhard HH. Antisense Oligodeoxynucleotide Technology: Potential Use for the Treatment of Malignant Brain Tumors. Cancer Control. 1998;5:163–70. doi: 10.1177/107327489800500207. [DOI] [PubMed] [Google Scholar]
- 24.Gao LF, Wen LJ, Yu H, Zhang L, Meng Y, Shao YT, et al. Knockdown of Stat3 expression using RNAi inhibits growth of laryngeal tumors in vivo. Acta Pharmacol Sin. 2006;27:347–52. doi: 10.1111/j.1745-7254.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- 25.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 26.Aigner A. Delivery systems for the direct application of siRNAs to induce RNA interference (RNAi) in vivo. J Biomed Biotechnol. 2006;2006:71659. doi: 10.1155/JBB/2006/71659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Yang G, Jiang T, Cao J, Huang KJ, Qiu ZJ. Down-regulation of STAT3 expression by vector-based small interfering RNA inhibits pancreatic cancer growth. World J Gastroenterol. 2011;17:2992–3001. doi: 10.3748/wjg.v17.i25.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunigal S, Lakka SS, Sodadasu PK, Estes N, Rao JS. Stat3-siRNA induces Fas-mediated apoptosis in vitro and in vivo in breast cancer. Int J Oncol. 2009;34:1209–20. [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Kowolik CM, Swiderski PM, Kortylewski M, Yu H, Horne DA, et al. Humanized Lewis-Y specific antibody based delivery of STAT3 siRNA. ACS Chem Biol. 2011;6:962–70. doi: 10.1021/cb200176v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis ME. The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharm. 2009;6:659–68. doi: 10.1021/mp900015y. [DOI] [PubMed] [Google Scholar]
- 31.Gao LFXD, Xu DQ, Wen LJ, Zhang XY, Shao YT, Zhao XJ. Inhibition of STAT3 expression by siRNA suppresses growth and induces apoptosis in laryngeal cancer cells. Acta Pharmacol Sin. 2005;26:377–83. doi: 10.1111/j.1745-7254.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- 32.Jin D, Zang W, Wang T, Li M, Wan J, Zhao G. The effect of STAT5 silenced by siRNA on proliferation, apoptosis and invasion of esophageal carcinoma cell line EC9706. Chinese-German J Clin Oncol. 2010;9:692–6. doi: 10.1007/s10330-010-0717-z. [DOI] [Google Scholar]
- 33.Wang Z, Yang XW, Carroll M. Constitutive Phosphorylation of STAT5A and STAT5B in Acute Myeloid Leukemia. PennScience. 2009;7:13–6. [Google Scholar]
- 34.Xiong H, Su WY, Liang QC, Zhang ZG, Chen HM, Du W, et al. Inhibition of STAT5 induces G1 cell cycle arrest and reduces tumor cell invasion in human colorectal cancer cells. Lab Invest. 2009;89:717–25. doi: 10.1038/labinvest.2009.11. [DOI] [PubMed] [Google Scholar]
- 35.Silva CM. Role of STATs as downstream signal transducers in Src family kinase-mediated tumorigenesis. Oncogene. 2004;23:8017–23. doi: 10.1038/sj.onc.1208159. [DOI] [PubMed] [Google Scholar]
- 36.Niu G, Heller R, Catlett-Falcone R, Coppola D, Jaroszeski M, Dalton W, et al. Gene therapy with dominant-negative Stat3 suppresses growth of the murine melanoma B16 tumor in vivo. Cancer Res. 1999;59:5059–63. [PubMed] [Google Scholar]
- 37.Benekli M, Baer MR, Baumann H, Wetzler M. Signal transducer and activator of transcription proteins in leukemias. Blood. 2003;101:2940–54. doi: 10.1182/blood-2002-04-1204. [DOI] [PubMed] [Google Scholar]
- 38.Rubin Grandis J, Zeng Q, Drenning SD. Epidermal growth factor receptor--mediated stat3 signaling blocks apoptosis in head and neck cancer. Laryngoscope. 2000;110:868–74. doi: 10.1097/00005537-200005000-00016. [DOI] [PubMed] [Google Scholar]
- 39.de Koning JP, Soede-Bobok AA, Ward AC, Schelen AM, Antonissen C, van Leeuwen D, et al. STAT3-mediated differentiation and survival and of myeloid cells in response to granulocyte colony-stimulating factor: role for the cyclin-dependent kinase inhibitor p27(Kip1) Oncogene. 2000;19:3290–8. doi: 10.1038/sj.onc.1203627. [DOI] [PubMed] [Google Scholar]
- 40.Jost M, Huggett TM, Kari C, Boise LH, Rodeck U. Epidermal growth factor receptor-dependent control of keratinocyte survival and Bcl-xL expression through a MEK-dependent pathway. J Biol Chem. 2001;276:6320–6. doi: 10.1074/jbc.M008210200. [DOI] [PubMed] [Google Scholar]
- 41.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996;271:5961–4. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 42.Wei LH, Kuo ML, Chen CA, Chou CH, Lai KB, Lee CN, et al. Interleukin-6 promotes cervical tumor growth by VEGF-dependent angiogenesis via a STAT3 pathway. Oncogene. 2003;22:1517–27. doi: 10.1038/sj.onc.1206226. [DOI] [PubMed] [Google Scholar]
- 43.Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, et al. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 overexpression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–5. [PubMed] [Google Scholar]
- 44.Wang D, Stravopodis D, Teglund S, Kitazawa J, Ihle JN. Naturally occurring dominant negative variants of Stat5. Mol Cell Biol. 1996;16:6141–8. doi: 10.1128/mcb.16.11.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita H, Iwase H, Toyama T, Fujii Y. Naturally occurring dominant-negative Stat5 suppresses transcriptional activity of estrogen receptors and induces apoptosis in T47D breast cancer cells. Oncogene. 2003;22:1638–52. doi: 10.1038/sj.onc.1206277. [DOI] [PubMed] [Google Scholar]
- 46.Yamashita H, Nishio M, Fujii Y, Iwase H. Dominant-negative Stat5 inhibits growth and induces apoptosis in T47D-derived tumors in nude mice. Cancer Sci. 2004;95:662–5. doi: 10.1111/j.1349-7006.2004.tb03326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahonen TJ, Xie J, LeBaron MJ, Zhu J, Nurmi M, Alanen K, et al. Inhibition of transcription factor Stat5 induces cell death of human prostate cancer cells. J Biol Chem. 2003;278:27287–92. doi: 10.1074/jbc.M304307200. [DOI] [PubMed] [Google Scholar]
- 48.Leong PL, Xi S, Drenning SD, Dyer KF, Wentzel AL, Lerner EC, et al. Differential function of STAT5 isoforms in head and neck cancer growth control. Oncogene. 2002;21:2846–53. doi: 10.1038/sj.onc.1205385. [DOI] [PubMed] [Google Scholar]
- 49.Jing N, Li Y, Xiong W, Sha W, Jing L, Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603–9. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 50.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angewandte Chemie (International ed 2004; 43:668-98. [DOI] [PubMed] [Google Scholar]
- 51.Jing N, Li Y, Xu X, Sha W, Li P, Feng L, et al. Targeting Stat3 with G-quartet oligodeoxynucleotides in human cancer cells. DNA Cell Biol. 2003;22:685–96. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]
- 52.Weerasinghe P, Garcia GE, Zhu Q, Yuan P, Feng L, Mao L, et al. Inhibition of Stat3 activation and tumor growth suppression of non-small cell lung cancer by G-quartet oligonucleotides. Int J Oncol. 2007;31:129–36. [PubMed] [Google Scholar]
- 53.McMicken HW, Bates PJ, Chen Y. Antiproliferative activity of G-quartet-containing oligonucleotides generated by a novel single-stranded DNA expression system. Cancer Gene Ther. 2003;10:867–9. doi: 10.1038/sj.cgt.7700652. [DOI] [PubMed] [Google Scholar]
- 54.Crinelli R, Bianchi M, Gentilini L, Magnani M. Design and characterization of decoy oligonucleotides containing locked nucleic acids. Nucleic Acids Res. 2002;30:2435–43. doi: 10.1093/nar/30.11.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dzau VJ. Transcription factor decoy. Circ Res. 2002;90:1234–6. doi: 10.1161/01.RES.0000025209.24283.73. [DOI] [PubMed] [Google Scholar]
- 56.Mann MJ, Dzau VJ. Therapeutic applications of transcription factor decoy oligonucleotides. J Clin Invest. 2000;106:1071–5. doi: 10.1172/JCI11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fang Y, Sun H, Zhai J, Zhang Y, Yi S, Hao G, et al. Antitumor activity of NF-kB decoy oligodeoxynucleotides in a prostate cancer cell line. Asian Pac J Cancer Prev. 2011;12:2721–6. [PubMed] [Google Scholar]
- 58.Ahn JD, Kim CH, Magae J, Kim YH, Kim HJ, Park KK, et al. E2F decoy oligodeoxynucleotides effectively inhibit growth of human tumor cells. Biochem Biophys Res Commun. 2003;310:1048–53. doi: 10.1016/j.bbrc.2003.09.124. [DOI] [PubMed] [Google Scholar]
- 59.Cho YS, Kim MK, Cheadle C, Neary C, Park YG, Becker KG, et al. A genomic-scale view of the cAMP response element-enhancer decoy: a tumor target-based genetic tool. Proc Natl Acad Sci U S A. 2002;99:15626–31. doi: 10.1073/pnas.242617799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leong PL, Andrews GA, Johnson DE, Dyer KF, Xi S, Mai JC, et al. Targeted inhibition of Stat3 with a decoy oligonucleotide abrogates head and neck cancer cell growth. Proc Natl Acad Sci U S A. 2003;100:4138–43. doi: 10.1073/pnas.0534764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roy SCPaI. Nucleic acid based therapeutic molecules. CRIPS 2008; 9. [Google Scholar]
- 62.Morishita R, Higaki J, Tomita N, Ogihara T. Application of transcription factor “decoy” strategy as means of gene therapy and study of gene expression in cardiovascular disease. Circ Res. 1998;82:1023–8. doi: 10.1161/01.RES.82.10.1023. [DOI] [PubMed] [Google Scholar]
- 63.Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24:970–9. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X, Zhang J, Wang L, Wei H, Tian Z. Therapeutic effects of STAT3 decoy oligodeoxynucleotide on human lung cancer in xenograft mice. BMC Cancer. 2007;7:149. doi: 10.1186/1471-2407-7-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Z, Yao Z, Liu S, Tang H, Yan X. An oligonucleotide decoy for Stat3 activates the immune response of macrophages to breast cancer. Immunobiology. 2006;211:199–209. doi: 10.1016/j.imbio.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 66.Chan KS, Sano S, Kiguchi K, Anders J, Komazawa N, Takeda J, et al. Disruption of Stat3 reveals a critical role in both the initiation and the promotion stages of epithelial carcinogenesis. J Clin Invest. 2004;114:720–8. doi: 10.1172/JCI21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen J, Li R, Li G. Inhibitory effects of decoy-ODN targeting activated STAT3 on human glioma growth in vivo. In Vivo. 2009;23:237–43. [PubMed] [Google Scholar]
- 68.Souissi I, Najjar I, Ah-Koon L, Schischmanoff PO, Lesage D, Le Coquil S, et al. A STAT3-decoy oligonucleotide induces cell death in a human colorectal carcinoma cell line by blocking nuclear transfer of STAT3 and STAT3-bound NF-κB. BMC Cell Biol. 2011;12:14. doi: 10.1186/1471-2121-12-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang X, Liu P, Zhang B, Wang A, Yang M. Role of STAT3 decoy oligodeoxynucleotides on cell invasion and chemosensitivity in human epithelial ovarian cancer cells. Cancer Genet Cytogenet. 2010;197:46–53. doi: 10.1016/j.cancergencyto.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 70.Sen M, Tosca PJ, Zwayer C, Ryan MJ, Johnson JD, Knostman KA, et al. Lack of toxicity of a STAT3 decoy oligonucleotide. Cancer Chemother Pharmacol. 2009;63:983–95. doi: 10.1007/s00280-008-0823-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lui VW, Boehm AL, Koppikar P, Leeman RJ, Johnson D, Ogagan M, et al. Antiproliferative mechanisms of a transcription factor decoy targeting signal transducer and activator of transcription (STAT) 3: the role of STAT1. Mol Pharmacol. 2007;71:1435–43. doi: 10.1124/mol.106.032284. [DOI] [PubMed] [Google Scholar]
- 72.Crinelli R, Bianchi M, Gentilini L, Palma L, Sørensen MD, Bryld T, et al. Transcription factor decoy oligonucleotides modified with locked nucleic acids: an in vitro study to reconcile biostability with binding affinity. Nucleic Acids Res. 2004;32:1874–85. doi: 10.1093/nar/gkh503. [DOI] [PMC free article] [PubMed] [Google Scholar]