Abstract
Owing to their sessile nature, plants have evolved sophisticated genetic and epigenetic regulatory systems to respond quickly and reversibly to daily and seasonal temperature changes. However, our knowledge of how plants sense and respond to warming ambient temperatures is rather limited. Here we show that an increase in growth temperature from 22 °C to 30 °C effectively inhibited transgene-induced posttranscriptional gene silencing (PTGS) in Arabidopsis. Interestingly, warmth-induced PTGS release exhibited transgenerational epigenetic inheritance. We discovered that the warmth-induced PTGS release occurred during a critical step that leads to the formation of double-stranded RNA (dsRNA) for producing small interfering RNAs (siRNAs). Deep sequencing of small RNAs and RNA blot analysis indicated that the 22–30 °C increase resulted in a significant reduction in the abundance of many trans-acting siRNAs that require dsRNA for biogenesis. We discovered that the temperature increase reduced the protein abundance of SUPPRESSOR OF GENE SILENCING 3, as a consequence, attenuating the formation of stable dsRNAs required for siRNA biogenesis. Importantly, SUPPRESSOR OF GENE SILENCING 3 overexpression released the warmth-triggered inhibition of siRNA biogenesis and reduced the transgenerational epigenetic memory. Thus, our study reveals a previously undescribed association between warming temperatures, an epigenetic system, and siRNA biogenesis.
Global warming has greatly affected plant growth and development and poses a serious threat to the global food supply (1, 2), highlighted by a discovery estimating that global warming caused an ∼10% reduction in rice yield for every 1-°C temperature increase (3). Knowledge of plant responses to extreme heat and cold has been accumulating; however, our understanding of plant responses to moderate temperature increases is limited (4, 5), despite recent discoveries implicating a histone variant in warmth-regulated gene expression and a basic helix–loop–helix protein in the thermoactivation of flowering and warmth-triggered hypocotyl elongation (6–8).
Recent studies suggest that environmental stresses can override diverse epigenetic regulation. For instance, heat shock and UV-B stress could change the epigenetic control of a silent reporter gene (transcriptional gene silence; TGS), with alterations in histone occupancy and histone 3 acetylation but not in DNA methylation in Arabidopsis (9). Similarly, several repetitive elements subjected to epigenetic regulation by TGS were reactivated by prolonged heat stress, with loss of nucleosomes but only minor changes in histone modifications (10). Plants can also change physiological processes to cope with severe environmental stresses with epigenetic transgenerational effects in offspring. For example, pathogen infection could stimulate somatic recombination in Arabidopsis (11). Similarly, somatic homologous recombination of a transgenic reporter was increased in a population treated with UV-C or flagellin and in the subsequent, untreated generations (12). The heat- and UV-B–mediated release of TGS also displayed heritable but limited effects (9). Recently, an explosion of publications elaborated on the role of small interfering RNAs (siRNAs) in transgenerational inheritance in a non-Mendelian manner in different organisms including Caenorhabditis elegans (13) and Arabidopsis (14), suggesting a conserved mechanism in epigenetic memory in coping with severe stresses. However, it remains unclear whether plant responses to mild stresses, such as moderate increases in ambient temperature, also involve siRNA-mediated epigenetic systems in having long-lasting effects on subsequent generations.
A sense transgene can sometimes lead to its cosuppression with its corresponding endogenous gene, a phenomenon first observed in Petunia when overexpressing chalcone synthase gene (15, 16), and was later recognized as the cause of posttranscriptional gene silencing (PTGS) (17), which could also be induced by antisense sequences (A-PTGS), inverted repeats (IR-PTGS), and virus infection (VIGS) (18, 19). These PTGS pathways involve the formation of double-stranded RNAs (dsRNAs) via different mechanisms and subsequent cleavage by RNase III-like endonuclease [DICER or DICER-Like (DCL)] into siRNAs that target complementary mRNAs (19).
In this report, we study a warm temperature-mediated mechanism of PTGS release by which the transgene-induced cosuppression of the Arabidopsis brassinosteroid (BR) receptor BRASSINOSTEROID-INSENSITIVE 1 (BRI1) gene is inhibited by a mild increase in ambient temperature. Interestingly, such a warmth-induced PTGS inhibition can be observed in subsequent generations grown under normal conditions. We show that warmth-released RNA silence is a general PTGS mechanism that likely involves the SUPPRESSOR OF GENE SILENCING 3 (SGS3) protein that functions in dsRNA formation for siRNA biogenesis.
Results
Cosuppression of BRI1 by Overexpressing BRI1–XA21 Chimeric Receptors.
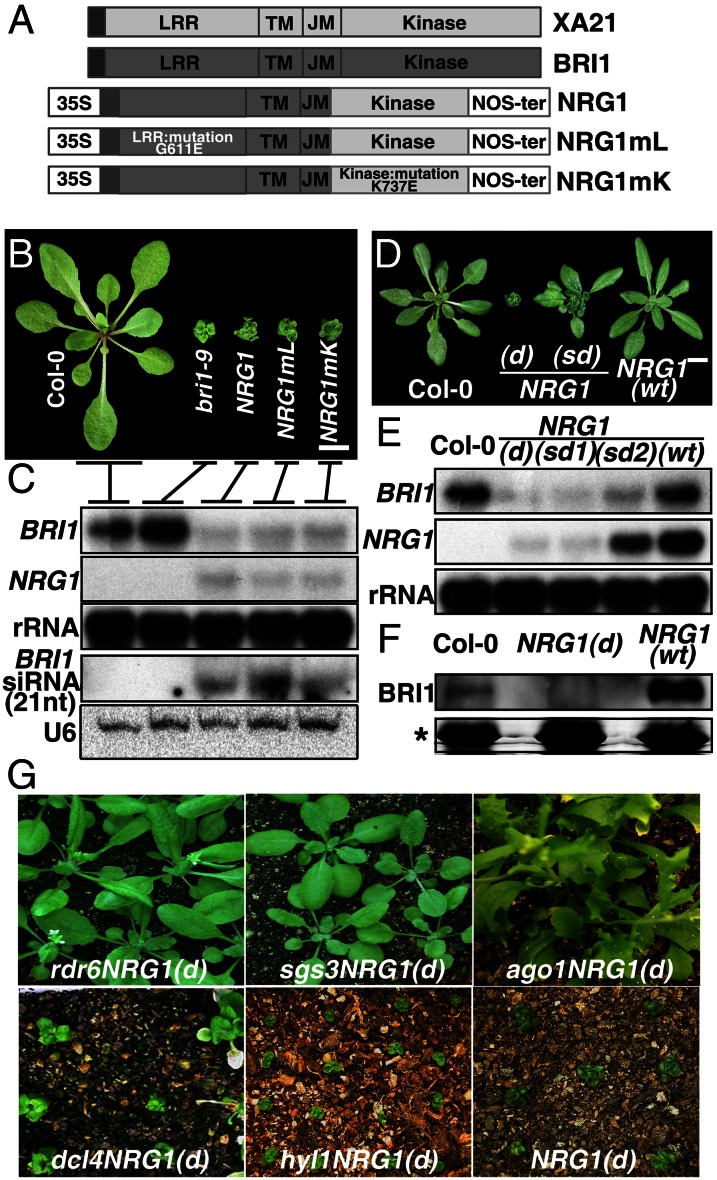
Previously, we demonstrated that a chimeric receptor-like kinase, novel resistance gene 1 (NRG1), consisting of the extracellular and transmembrane domains of the Arabidopsis BR receptor BRI1 and the kinase domain of the rice pattern-recognition receptor XA21 (XA as the rice resistance gene family to Xanthomonas oryzae pv. Oryzae), activated an XA21-specific plant-defense response in a BR-dependent manner in cell culture (20). To test whether this chimeric receptor also functions in planta, we generated transgenic Arabidopsis plants that expressed either the functional or the mutant NRG1 (Fig. 1A). Interestingly, ∼30% of the resulting transgenic lines were morphologically similar to the bri1-9 mutant (21). RNA blot analysis indicated that sense transgene-mediated PTGS (S-PTGS) caused this bri1-like phenotype (Fig. 1 B and C). The dwarfed NRG1 plants accumulated much less BRI1 and NRG1 transcripts and BRI1 protein compared with wild-type (WT) or WT-like NRG1 plants (Fig. 1 C–F). A BRI1 probe detected the presence of siRNAs (21-nt) in the dwarfed NRG1 plants but not in the bri1 mutant or WT (Fig. 1C). We chose a semidwarf T1 (T as transgenic plants) NRG1 line for further analysis, which produced dwarf (d) and semidwarf offspring in the T2 generation in a non-Mendelian manner (Fig. 1D), and discovered that the expression of NRG1 and BRI1 was cosilenced in the dwarf plants but varied among individual T2 semidwarfs (Fig. 1E). Further support for the S-PTGS of BRI1 was provided by our finding that loss-of-function mutations in SGS3, RNA-DEPENDENT RNA POLYMERASE 6 (RDR6), and ARGONAUTE1 (AGO1), all of which are known to be required for S-PTGS, suppressed the dwarf phenotype of the NRG1(d) line (Fig. 1G). Surprisingly, a loss-of-function mutation in DCL4, which cleaves dsRNAs into 21-nt siRNAs (22, 23), had little effect on the morphology of the NRG1(d) line (Fig. 1G). This result might be explained by the ability of other DCLs to cut DCL4 substrates in a dcl4 mutant (22, 24).
Fig. 1.
Dwarf phenotype of NRG1(d) is caused by PTGS of the endogenous BRI1 gene. (A) Schematic diagram of NRG1 and its mutant variants, NRG1mL and NRG1mK (20). LRR, leucine-rich repeat; TM, transmembrane domain; JM, juxtamembrane domain; NOS-ter, termination sequence of the nopaline synthase gene. (B) Transgenic plants grown at 22 °C expressing NRG1 or its mutant variants. (Scale bar, 1 cm.) (C) RNA blot analysis of BRI1 and NRG1 transcripts and BRI1-specific siRNAs. Reprobing the filters with a 32P-labeled 18S rRNA cDNA or 32P-labeled U6 was used to control equal loading. (D) Four-week-old Col (WT), strong (d), and semidwarfed (sd) segregates of a semidwarf NRG1 line and WT-like (wt) NRG1 seedlings. (Scale bar, 1 cm.) (E) RNA blot analysis of BRI1 and NRG1 transcripts in Col-0, three segregates of the semidwarf NRG1 line, and a WT-like NRG1 line (wt). The filter was rehybridized with an 18S rRNA probe to control equal loading. (F) Immunoblot analysis of BRI1 abundance in 4-wk-old plants. Coomassie blue staining of the protein gel (*) served as a loading control. (G) Four-week-old rdr6/NRG1(d), sgs3/NRG1(d), and hyponastic leaves (hyl1)/NRG1(d) plants, 6-wk-old ago1/NRG1(d) and dcl4/NRG1(d) homozygous filial generation 3 (F3) plants, and 4-wk-old control NRG1(d) plants grown in soil.
Warm Temperatures Inhibit PTGS as a General Epigenetic Mechanism.
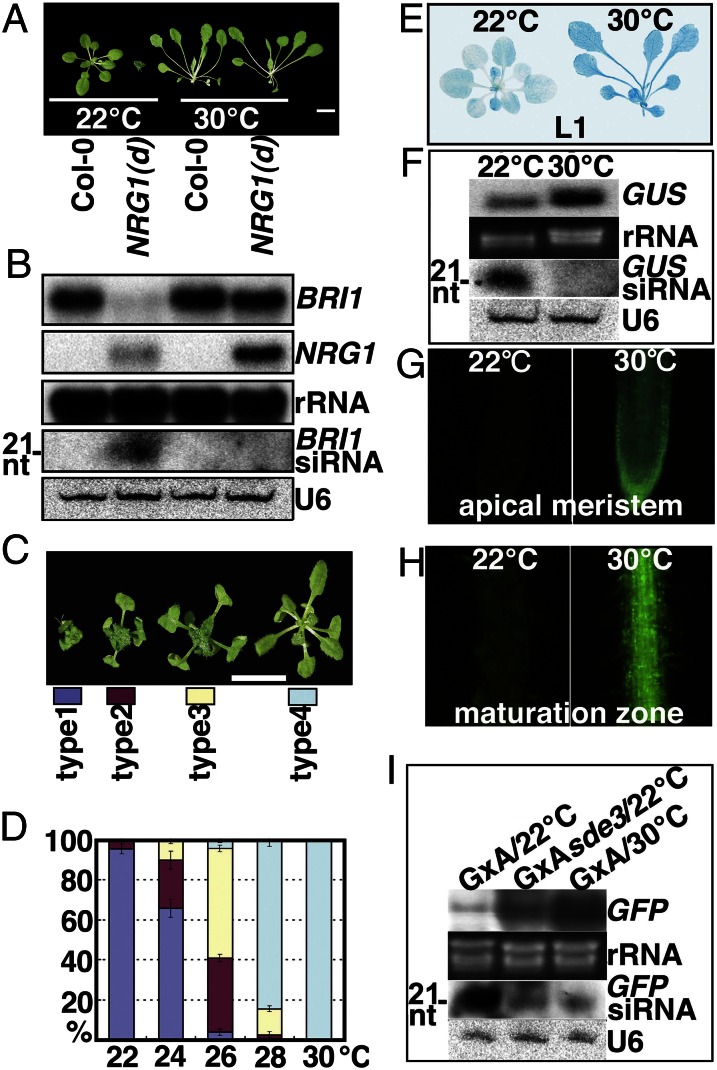
Several studies have shown that temperature changes could affect VIGS and A-PTGS (25, 26), both involving formation of dsRNAs to trigger siRNA-mediated RNA degradation. To investigate whether temperature changes could also affect S-PTGS, we examined the effect of temperature change on a stable NRG1(d) line that did not segregate out a single wild-type–looking plant when grown and propagated at 22 °C with a population size of >150 at each generation for >20 generations. NRG1(d) plants grown at 15 °C remained dwarfs (Fig. S1A), indicating that the low temperature, which effectively inhibited A-PTGS of a transgenic line (27) carrying an antisense construct of glutamyl-cysteine synthetase 1 (GSH1) (Fig. S1 B–E), had no detectable effect on the S-PTGS of BRI1. The obvious differential effects of the low temperature suggest a potential mechanistic difference between A-PTGS and S-PTGS. Surprisingly, when grown at 30 °C, a temperature known to alter plant morphology (5), the NRG1(d) plants were no longer dwarfs but became all WT-looking (Fig. 2A). This morphological change was not caused by activation of BR signaling, because growth at 30 °C was unable to rescue the dwarf phenotype of two known BR mutants, deetiolated2 (det2) and bri1-9 (Fig. S2A). RNA blot analysis revealed the accumulation of both BRI1 and NRG1 transcripts and the concomitant disappearance of BRI1-specific siRNAs at 30 °C (Fig. 2B), indicating that the morphological change was due to the 30 °C-induced S-PTGS release. Additional experiments revealed that this PTGS release was specific for the temperature increase because treatment with the stress hormone abscisic acid or drought/salt stress failed to suppress the dwarf phenotype of NRG1(d) (Fig. S2 B–D).
Fig. 2.
Release of S-PTGS by warming ambient growth temperatures. (A) Four-week-old Col-0 and NRG1(d) plants grown at 22 and 30 °C. (Scale bar, 1 cm.) (B) RNA blot analysis of BRI1 and NRG1 transcripts and BRI1-specific siRNAs. (C and D) Percentage of four different types of NRG1(d) plants at different growth temperatures. About 100 plants at each temperature were analyzed. (Scale bar, 1 cm.) The average (± SD) values from 3 repeats of the analysis are shown (D). (E) Histochemical staining of GUS activity in 4-wk-old L1 seedlings grown at 22 and 30 °C. (F) RNA blot analysis of GUS mRNA and GUS-specific siRNAs in 22/30 °C-grown 4-wk-old L1 plants. (G and H) Fluorescent microscopic examination of the GFP signal in the apical root meristem (G) and maturation zone (H) of 4-wk-old GxA seedlings grown at 22 and 30 °C. (I) RNA blot analysis of GFP mRNA and GFP-specific siRNAs. Rehybridization with a 32P-labeled 18S rRNA cDNA (B), a 32P-labeled U6 probe (B, F, and I), or ethidium bromide staining of the rRNAs (F and I) served as loading controls.
To determine the threshold temperature for releasing S-PTGS, we grew NRG1(d) plants at 22–30 °C. To aid our analysis, we phenotypically classified four types of NRG1 plants (Fig. 2C). As the temperature increased, the percentage of severely dwarfed NRG1(d) plants decreased from 95% at 22 °C to 65% at 24 °C, 4% at 26 °C, 1% at 28 °C, and 0% at 30 °C, but the percentage of WT-looking plants increased from 0% at 22–24 °C to 5% at 26 °C, 84% at 28 °C, 100% at 30 °C (Fig. 2D), and 100% in continuous generations (>5) at 30 °C. Thus, warmth-induced S-PTGS release is a dose-dependent stochastic process between 24 and 28 °C but becomes deterministic at 30 °C. Experiments examining the minimum duration of 30-°C growth required for S-PTGS release revealed stochastic inhibition between 7 and 11 d and deterministic suppression after 13 d (Fig. S2E).
To investigate whether 30 °C-induced S-PTGS inhibition is a general phenomenon, we analyzed two well-studied S-PTGS systems, low expresser 1 (L1) carrying a direct repeat of the 35S-GUS (β-glucuronidase) transgene (28) and GxA (GFP142 x Amp243 as the widely used transgenic lines for post-transcriptional gene silence) containing a 35S-GFP transgene and a 35S-potato virus X (PVX):GFP amplicon (29). L1 plants grown at 30 °C exhibited higher GUS activity and increased GUS expression than those grown at 22 °C (Fig. 2 E and F). GUS-specific siRNAs consistently accumulated in plants grown at 22 °C but became undetectable at 30 °C (Fig. 2F). Similarly, a strong GFP signal was easily observed in the roots of GxA plants grown at 30 °C but was weak at 22 °C (Fig. 2 G and H). As expected, GFP transcript accumulation also recovered as a result of PTGS release at 30 °C, whereas GFP-specific siRNAs were significantly reduced, similar to the effects observed in the presence of the silencing defective 3 (sde3) mutation (Fig. 2I), which inhibits S-PTGS (30). Taken together, these results demonstrate that warmth-induced S-PTGS release is a general epigenetic phenomenon, which was recently exploited to avoid S-PTGS when studying shoot meristem development (31).
Temperature-Induced PTGS Release Exhibits Transgenerational Inheritance.
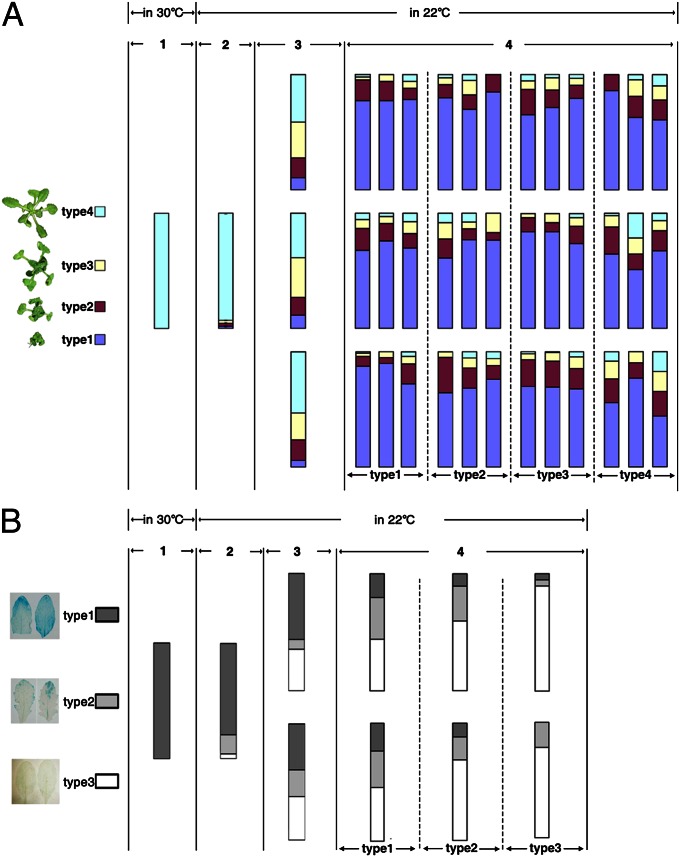
Surprisingly, the 30 °C-induced S-PTGS release exhibited a transgenerational inheritance pattern in subsequent generations grown at 22 °C. Approximately 93% of the offspring derived from 30 °C-grown NRG1(d) plants were morphologically WT-looking, even when grown at 22 °C (Fig. 3A). Importantly, 38–53% of third-generation offspring of WT-looking NRG1(d) plants retained WT-like morphology, which was still present in fourth-generation offspring, albeit at a much lower percentage (Fig. 3A). Intriguingly, a small number of WT-looking plants were segregated out in the fourth generation from severely dwarfed parents (type 1 in Fig. 3A), likely caused by epigenetic metastability. The 30 °C-induced transgenerational inheritance of the inhibitory effect on S-PTGS was also observed in the L1 line (Fig. 3B). These results indicated the successive transgenerational heritability of warmth-induced S-PTGS release. This discovery and stochastic occurrence of warmth-induced S-PTGS release suggested involvement of epigenetic changes. We therefore examined DNA methylation changes that are often associated with an epigenetic phenomenon (32). DNA blot analysis and bisulfite sequencing not only detected transgene DNA methylation in NRG1(d) and L1 lines but also revealed warmth-induced DNA methylation changes in the two transgenes (Fig. S3). However, there seems no clear and consistent correlation between detected methylation changes and warmth-induced S-PTGS release or its transgenerational inheritance.
Fig. 3.
Transgenerational inheritance of warmth-induced release of PTGS. (A) Each vertical bar represents one line with ∼100 individual plants, and each color bar indicates the percentage of plants within a given NRG1(d) population exhibiting the four color-coded morphological phenotypes shown (Left). Seeds from 22 °C-grown NRG1(d) dwarf plants were germinated and grown at 30 °C, producing all phenotypically WT-looking plants (first generation). Seeds from a randomly chosen WT-like NRG1 plant were germinated and grown at 22 °C to produce the second generation. The offspring of three individual second-generation WT-like NRG1 plants and three individual third-generation plants of each phenotypic group grown at 22 °C were independently analyzed. (B) Transgenerational inheritance of warmth-triggered PTGS in the L1 line. More than 100 offspring of two individual second-generation WT-like plants and two individual third-generation plants of each of the three phenotypic groups were analyzed.
Temperature-Induced PTGS Release Involves the dsRNA Formation Step.
To determine the likely PTGS step impeded by 30-°C growth, we analyzed the temperature effect on IR-PTGS. IR-PTGS and S-PTGS share many common components but differ in how dsRNAs are formed. S-PTGS requires RDR6, an RNA-dependent RNA polymerase, to convert aberrant single-stranded RNAs into dsRNAs (33, 34), whereas IR-PTGS generates dsRNAs through production of self-complementary transcripts (18). We generated several dwarf lines using an inverted-repeat BRI1 transgene (BRI1-RNAi) and found that the temperature increase did not suppress but instead enhanced the dwarf phenotype of the BRI1-RNAi lines (Fig. 4 A and B). Such a warmth-enhanced dwarfism is unlikely caused by increased gene silencing, because the 22–30 °C change had no significant effect on the BRI1 transcript level but caused similar morphological changes in the two known BR mutants det2 and bri1-9 (Fig. S2A). We thus concluded that the 30-°C growth failed to suppress IR-PTGS, which was further confirmed in the Arabidopsis At-PDSi transgenic line (35) carrying a chemically inducible IR-RNAi construct of the phytoene desaturase gene (PDS) (Fig. S4A). The differential effects of 30-°C growth on S-PTGS and IR-PTGS suggested that the warmer temperatures most likely inhibit a step leading to the formation of stable dsRNAs involving RDR6 and SGS3, a unique dsRNA-binding protein that recognizes 5′ overhangs (36).
Fig. 4.
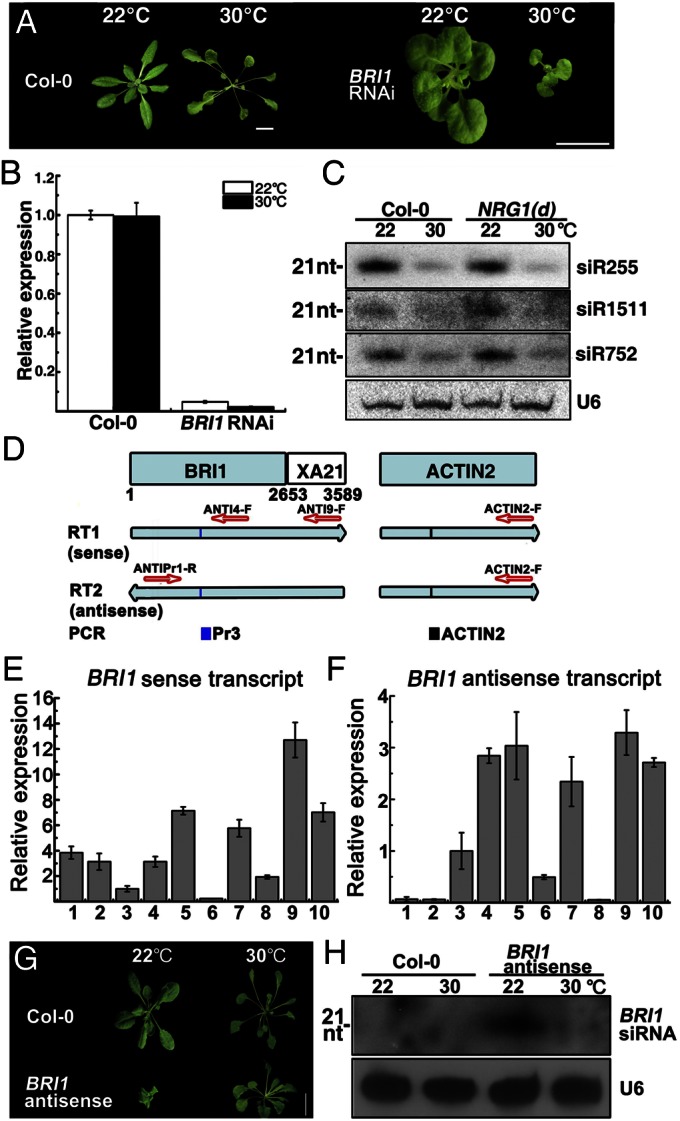
Growth at 30 °C likely inhibits dsRNA formation. (A) Four-week-old plants of WT (Left) and a BRI1-RNAi line (Right) grown at 22 and 30 °C. Two independent stable lines with similar phenotype were analyzed. (Scale bars, 1 cm.) (B) A qPCR analysis of relative BRI1 mRNA levels. (C) RNA blot analysis of three tasiRNAs in 22/30 °C-grown NRG1(d) and WT seedlings. (D) Schematic representation of the assay for sense and antisense transcripts of BRI1 (see Materials and Methods for details). (E and F) qPCR analysis of sense (E) and antisense (F) BRI1 transcripts in 22/30 °C-grown seedlings of each of the following lines: 1, 22 °C-grown Col-0; 2, 30 °C-grown Col-0; 3, 22 °C-grown NRG1(d); 4, 30 °C-grown NRG1(d); 5, 22 °C-grown second-generation type 4 plants; 6, 22 °C-grown third-generation type 1 plants; 7, 22 °C-grown third-generation type 4 plants; 8, 22 °C-grown NRG1(d)/rdr6; 9, 22 °C-grown NRG1(d)/sgs3; and 10, 22 °C-grown NRG1(d)/ago1. (G) Four-week-old WT and BRI1 antisense plants grown at 22 or 30 °C. (Scale bar, 1 cm.) Eight independent stable lines with similar phenotype were analyzed. (H) RNA blot analysis of BRI1-derived siRNAs in WT and BRI1 antisense plants grown at 22 or 30 °C. For RNA blot analysis, reprobing with a U6 probe (C and H) was used to serve as a loading control, whereas qPCR assays used the ACTIN2 signal for normalization. The average (±SD) values from three biological repeats are shown (B, E, and F).
Recent studies have revealed that plants contain at least three classes of endogenous siRNAs, including trans-acting siRNAs (tasiRNAs), natural antisense siRNAs (natsiRNAs), and chromatin-associated siRNAs (37, 38). Both tasiRNAs and natsiRNAs are derived from dsRNA triggers, which are formed via an RDR6/SGS3-mediated process. If RDR6/SGS3-mediated dsRNA formation is inhibited at 30 °C, a reduced accumulation of endogenous siRNAs would also be observed. Indeed, besides the dramatic reduction of NRG1/BRI1-derived siRNAs (Fig. S4B), deep RNA sequencing revealed that the abundance of many tasiRNAs derived from eight Arabidopsis Trans-acting siRNA (TAS) loci was significantly and consistently reduced in both WT and NRG1(d) plants by the 30-°C growth (Fig. S4C and Dataset S1). The 30 °C-induced reduction of TAS1-derived siR255 and siR752 plus the TAS2-derived siR1511 was confirmed by RNA blot analysis (Fig. 4C). Consistently, the transcript levels of TAS2 and several tasiRNA-target genes were increased at 30 °C in both the WT and NRG1(d) line, with the latter exhibiting greater inductions that could be caused by increased TAS2 transcription and/or tasiRNA production in 22 °C-grown plants (Fig. S4 D and E). Taken together, these data showed that the 22–30 °C temperature increase also inhibits an endogenous RDR6/SGS3-mediated siRNA pathway.
SGS3 Is Crucial for Temperature-Induced PTGS Inhibition.
To determine whether 30-°C growth directly inhibits the RDR6-dependent production of antisense transcripts, we analyzed accumulation of both sense and antisense transcripts of NRG1 and BRI1 in the NRG1(d) line and WT (Fig. 4D). As expected, sense transcripts were accumulated in 30 °C-grown NRG1(d) plants and their 22 °C-grown WT-looking offspring (Fig. 4E). Surprisingly, antisense transcript levels were also elevated in these plants (Fig. 4F). Accumulation of both sense and antisense transcripts was also detected in NRG1(d)/sgs3 and NRG1(d)/ago1 transgenic mutant lines but not in the NRG1(d)/rdr6 line, which accumulated only sense, but not antisense, NRG1 transcripts. These results not only indicated that the warm temperature did not inhibit the RDR6-dependent production of antisense transcripts but also suggested that SGS3 and/or AGO1 are the likely targets of the 22–30 °C temperature increase.
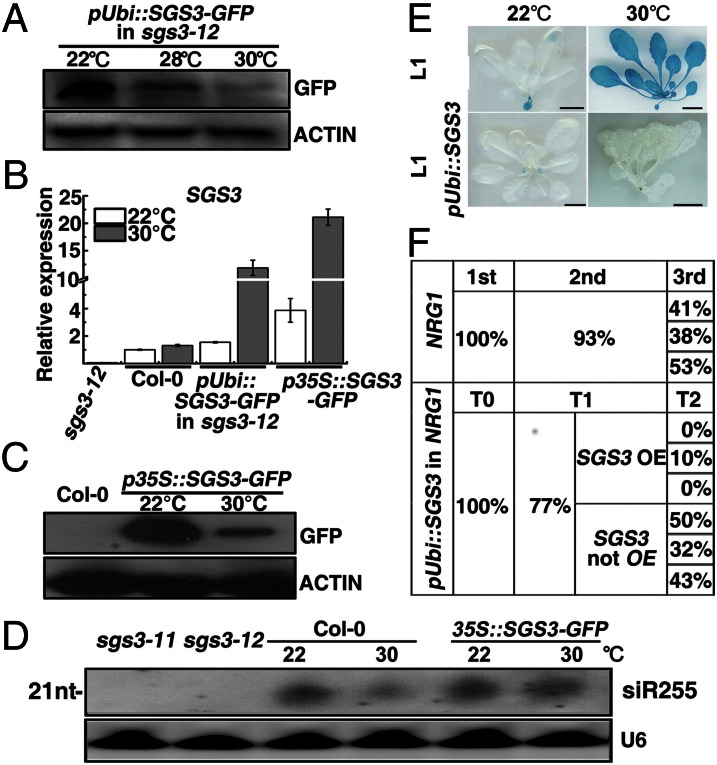
Consistent with our analysis of sense and antisense transcripts, immunoblot and enzyme assays showed that 30-°C growth had little effect on the protein abundance or enzyme activity of RDR6 (Fig. S5 A and C). A similar immunoblotting assay also failed to detect a warmth-induced change in AGO1 protein abundance (Fig. S5B). However, immunoblot analysis of a phenotypically rescued transgenic sgs3-12 mutant (Fig. S6A) carrying a pUbi::SGS3-GFP transgene driven by a maize ubiquitin promoter (pUbi) showed that the warmer temperatures (28 and 30 °C) significantly and reproducibly reduced the steady-state level of SGS3-GFP (Fig. 5A) despite an approximately sevenfold increase of the SGS3-GFP transcript (Fig. 5B), implying that the endogenous SGS3 protein could be reduced to a much lower level. Taken together, our results not only revealed that SGS3 is the most likely target of the 22–30 °C increase but also suggested that SGS3 likely acts at a step downstream of antisense transcript production. Consistent with our interpretation, the 30-°C growth also inhibited A-PTGS of BRI1 (Fig. 4 G and H and Fig. S6F) and GSH1 (Fig. S6 B–E), in which antisense transcripts are directly transcribed from the corresponding transgenes by RNA polymerase II.
Fig. 5.
SGS3 overexpression releases warmth-triggered inhibition of siRNA biogenesis and reduces transgenerational epigenetic memory. (A) Immunoblotting of SGS3-GFP in a pUbi::SGS3-GFP sgs3-12 transgenic line grown at different temperatures. The same amounts of total proteins extracted from 4-wk-old plants were separated by 10% SDS/PAGE and analyzed by immunoblotting with a commercial anti-GFP antibody, and the same blots were reprobed with an anti-ACTIN antibody to control equal loading. (B) qPCR analysis of SGS3 transcript levels that were normalized to the ACTIN2 signals. The average values (±SD) from three replicates of the qPCR experiments are shown. (C) Immunoblot analysis of SGS3-GFP abundance in the p35S::SGS3-GFP line. (D) RNA blot analysis of siR255 in 22/30 °C-grown plants of sgs3-11, sgs3-12, a p35S::SGS3-GFP transgenic line, and the WT. A U6 blot was used as a loading control. (E) Histochemical staining of the GUS activity of 4-wk-old 22/30 °C-grown seedlings. (Scale bars, 1 cm.) (F) Percentages of WT-looking plants in offspring of 22 °C-grown NRG1(d) and pUbi::SGS3 NRG1(d) lines. Approximately 100 independent T1 lines (equivalent of the second generation in Fig. 3) and 30 T2 plants (equivalent of the third generation in Fig. 3) of six independent pUbi::SGS3 NRG1(d) lines [three being SGS3 overexpressors (OE) and the other with low SGS3 transcript levels (not OE)] were analyzed.
To confirm that the reduced SGS3 abundance was responsible for the warmth inhibition of siRNA production, we further analyzed the effect of 30-°C growth on tasiRNA biogenesis with a stable p35S::SGS3-GFP line using siR255 as a convenient readout. Quantitative (q)PCR analysis showed that the SGS3-GFP transcript levels in this line were approximately twofold higher than that of the pUbi::SGS3-GFP sgs3-12 line at both 22 and 30 °C (Fig. 5B). Although 30-°C growth still reduced SGS3-GFP abundance (Fig. 5C), the remaining amount of SGS3-GFP in this line, presumably above the critical threshold required for siRNA biogenesis, was able to neutralize the inhibitory effect of the 30-°C growth on the siR255 level (Fig. 5D). Consistently, the transcript levels of the two tasiRNA-target genes At1g62950 and At4g29770 remained low at 30 °C in the p35S::SGS3-GFP line (Fig. S7A). Similarly, SGS3 overexpression driven by the pUbi promoter was able to maintain low GUS activity of 30 °C-grown L1 seedlings (Fig. 5E and Fig. S7B). These results suggested that a threshold level of SGS3 is required to maintain dsRNA formation and subsequent siRNA biogenesis. This interpretation is consistent not only with an earlier discovery that a small reduction of SGS3 activity could have a major impact on S-PTGS efficiency (39) but also with our observed stochasticity–determinism transition of the warmth-induced S-PTGS release at 30 °C. Interestingly, transformation of the pUbi::SGS3 transgene into 30 °C-grown NRG1(d) plants (first generation or T0) significantly decreased the percentage of WT-looking plants in the subsequent generation (second generation or T1) grown at 22 °C. Importantly, whereas the percentages of T2 (equivalent to third-generation) WT-looking plants of three independent SGS3-overexpressing NRG1(d) T1 lines were 0–10%, three other independent pUbi::SGS3 NRG1(d) T1 lines with low SGS3 transcript levels segregated out the same percentage of WT-looking offspring similar to second-generation NRG1(d) plants (Fig. 5F and Fig. S7C), suggesting that SGS3 overexpression could also decrease the transgenerational memory.
Discussion
Our current study has revealed an interesting association between warmer ambient temperatures, a plant epigenetic system, and siRNA biogenesis. We showed that a moderate temperature hike from 22 to 30 °C reduced the abundance of SGS3 protein and inhibited the dsRNA formation step in the A-PTGS, S-PTGS, and tasiRNA pathways. Our experiments also suggested that low temperature inhibits A-PTGS at an A-PTGS–specific step instead of a temperature-sensitive DCL complex as previously suggested based on differential effects of low temperatures on the biogenesis of siRNAs and microRNAs (miRNAs) (26). Given the cross-regulatory interactions among siRNAs, miRNAs, and mRNAs (40, 41), warmth-induced inhibition of SGS3/RDR6-mediated siRNA biogenesis might also contribute to other temperature-influenced processes, such as plant defense against pathogens (42) and sexual reproduction (43). Our discovery also suggests a potential problem for the agricultural application of RNAi technology at warming crop-growth temperatures, as these techniques often require the RDR6/SGS3-mediated production of transitive siRNAs to amplify the gene silencing response (37). However, it remains to be determined whether the reduced abundance of SGS3 was caused by decreased biosynthesis or increased degradation, and to investigate whether the inhibitory effect of the 30-°C growth on SGS3 protein abundance is controlled by the same epigenetic system responsible for the observed transgenerational memory. Further genetic, genomic, and biochemical studies of the NRG1(d) dwarf line could lead to identification of proteins or RNAs responsible for SGS3 regulation and/or transgenerational epigenetic memory.
Our discovery of a moderate temperature hike having a transgenerational inhibitory effect on S-PTGS/siRNA biogenesis provides another example of transgenerational epigenetic inheritance of environmentally induced traits, which has attracted substantial interest in recent years (32). Unlike the previously reported pathogen/UV light-triggered epigenetic memory, which persisted for several generations with slowly increasing strengths (12), warmth-induced epigenetic memory was maintained for at least three generations with rapidly declining strength. The meiotically heritable “memory of warmth” is also different from the well-studied “memory of cold” involved in vernalization, a plant flowering regulatory process whereby a prolonged exposure to cold results in the mitotically heritable but meiotically intransmissible epigenetic transcriptional silencing of a floral suppressor gene (44). It is quite possible that the transgenerational inheritance of the warmth-induced inhibition of SGS3/RDR6-mediated siRNA biogenesis could offer an adaptive advantage or genomic flexibility for better fitness in warmer environments. It is interesting to note that the memory of warmth could be recovered with low frequency from some severely dwarfed NRG1(d) plants that had apparently lost such a memory (Fig. 3A). This observation is consistent with the well-known metastable characteristics of an environmentally altered epigenome (32). It is quite possible that a few germ-line cells revert to the “warmth-triggered epigenetic state” responsible for S-PTGS release.
We observed that the 22–30 °C temperature increase changed the DNA methylation status of the transgenes; however, there seems to be no general correlation between the observed DNA methylation changes of the target genes and the temperature-triggered release of S-PTGS or its subsequent transgenerational inheritance in the NRG1(d) line and the L1 PTGS model line. However, it remains a formal possibility that some of the observed changes in DNA methylation at certain sites of a silenced target gene are responsible for the transgenerational inheritance of warmth-induced S-PTGS release. Further studies with Arabidopsis mutants defective in specific types of DNA methylation or DNA methylation inhibitors could investigate such a possibility.
Materials and Methods
Plant Materials and Growth.
Arabidopsis ecotype Col-0 was used to generate various transgenic plants. NRG1 transgenic plants were generated by transformation with p35S::NRG1 chimeric constructs (20; Fig. 1A), and stable PTGS lines were obtained. Plants were grown at different temperatures (see SI Materials and Methods for details).
Generation of Transgenic Plants.
Various transgenes were introduced into WT, L1, and sgs3-12 plants by Agrobacterium-mediated transformation (see detailed descriptions in SI Materials and Methods).
RNA Analysis.
Total RNAs were isolated for RNA blot and real-time PCR (qPCR) analysis (see SI Materials and Methods, Table S1 for details).
Deep Sequencing of Small RNAs.
Small RNAs were isolated from total RNAs, reverse-transcribed, and amplified for Solexa sequencing. The entire sequencing data can be obtained from the National Center for Biotechnology Information Gene Expression Omnibus database under accession nos. GSE25738 and GSM632205–GSM632209 (see SI Materials and Methods for details).
Protein Analysis.
Total proteins were extracted from 4-wk-old seedlings and analyzed by immunoblot according to a previously described procedure (45) using antibodies against BRI1 (45), GFP (Torrey Pines Biolabs), AGO1 (46), and anti-HA (CWBIO). After stripping, the same membranes were reblotted with an ACTIN antibody (Abmart) for loading controls.
RDR6 Enzyme Activity Assay.
RDR6 enzymatic activity was assayed as described (47) (see SI Materials and Methods for details).
Analysis of DNA Methylation.
DNA blotting with radiolabeled probes and bisulfite sequencing were used to examine the DNA methylation of transgenes in NRG1(d) and L1 lines (see SI Materials and Methods for details).
Supplementary Material
Acknowledgments
We thank Drs. D. Baulcombe, X. M. Chen, R. S. Poethig, N. H. Chua, David J. Oliver, and H. S. Guo for mutant and transgenic lines, and Drs. J. K. Zhu and Z. X. Xie for helpful discussions. This work was supported by grants from the National Research Program of China (2011CB100700), the National Natural Science Foundation of China (91117018 and 30730064), by the Chinese Academy of Sciences (to Z.-H.H.) and National Science Foundation (IOS1121496).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession nos. GSE25738 and GSM632205–GSM632209).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219655110/-/DCSupplemental.
References
- 1.Long SP, Ort DR. More than taking the heat: Crops and global change. Curr Opin Plant Biol. 2010;13(3):241–248. doi: 10.1016/j.pbi.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Lobell DB, Field CB. Global scale climate–crop yield relationships and the impacts of recent warming. Environ Res Lett. 2007;2(1):014002. [Google Scholar]
- 3.Peng S, et al. Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci USA. 2004;101(27):9971–9975. doi: 10.1073/pnas.0403720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blázquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nat Genet. 2003;33(2):168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- 5.Lin L, Zhong SH, Cui XF, Li J, He ZH. Characterization of temperature-sensitive mutants reveals a role for receptor-like kinase SCRAMBLED/STRUBBELIG in coordinating cell proliferation and differentiation during Arabidopsis leaf development. Plant J. 2012;72(5):707–720. doi: 10.1111/j.1365-313X.2012.05109.x. [DOI] [PubMed] [Google Scholar]
- 6.Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140(1):136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 7.Koini MA, et al. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol. 2009;19(5):408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 8.Kumar SV, et al. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484(7393):242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang-Mladek C, et al. Transgenerational inheritance and resetting of stress-induced loss of epigenetic gene silencing in Arabidopsis. Mol Plant. 2010;3(3):594–602. doi: 10.1093/mp/ssq014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pecinka A, et al. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22(9):3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lucht JM, et al. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet. 2002;30(3):311–314. doi: 10.1038/ng846. [DOI] [PubMed] [Google Scholar]
- 12.Molinier J, Ries G, Zipfel C, Hohn B. Transgeneration memory of stress in plants. Nature. 2006;442(7106):1046–1049. doi: 10.1038/nature05022. [DOI] [PubMed] [Google Scholar]
- 13.Rechavi O, Minevich G, Hobert O. Transgenerational inheritance of an acquired small RNA-based antiviral response in C. elegans. Cell. 2011;147(6):1248–1256. doi: 10.1016/j.cell.2011.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calarco JP, et al. Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell. 2012;151(1):194–205. doi: 10.1016/j.cell.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell. 1990;2(4):279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Krol AR, Mur LA, Beld M, Mol JN, Stuitje AR. Flavonoid genes in petunia: Addition of a limited number of gene copies may lead to a suppression of gene expression. Plant Cell. 1990;2(4):291–299. doi: 10.1105/tpc.2.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Depicker A, Montagu MV. Post-transcriptional gene silencing in plants. Curr Opin Cell Biol. 1997;9(3):373–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- 18.Waterhouse PM, Graham MW, Wang MB. Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc Natl Acad Sci USA. 1998;95(23):13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baulcombe D. RNA silencing in plants. Nature. 2004;431(7006):356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- 20.He ZH, et al. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288(5475):2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 21.Jin H, Yan Z, Nam KH, Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol Cell. 2007;26(6):821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102(36):12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37(12):1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 24.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15(16):1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 25.Chellappan P, Vanitharani R, Ogbe F, Fauquet CM. Effect of temperature on geminivirus-induced RNA silencing in plants. Plant Physiol. 2005;138(4):1828–1841. doi: 10.1104/pp.105.066563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szittya G, et al. Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J. 2003;22(3):633–640. doi: 10.1093/emboj/cdg74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiang C, Werner BL, Christensen EM, Oliver DJ. The biological functions of glutathione revisited in Arabidopsis transgenic plants with altered glutathione levels. Plant Physiol. 2001;126(2):564–574. doi: 10.1104/pp.126.2.564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elmayan T, et al. Arabidopsis mutants impaired in cosuppression. Plant Cell. 1998;10(10):1747–1758. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dalmay T, Hamilton A, Mueller E, Baulcombe DC. Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell. 2000;12(3):369–379. doi: 10.1105/tpc.12.3.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalmay T, Horsefield R, Braunstein TH, Baulcombe DC. SDE3 encodes an RNA helicase required for post-transcriptional gene silencing in Arabidopsis. EMBO J. 2001;20(8):2069–2078. doi: 10.1093/emboj/20.8.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gagne JM, Clark SE. The Arabidopsis stem cell factor POLTERGEIST is membrane localized and phospholipid stimulated. Plant Cell. 2010;22(3):729–743. doi: 10.1105/tpc.109.068734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauser MT, Aufsatz W, Jonak C, Luschnig C. Transgenerational epigenetic inheritance in plants. Biochim Biophys Acta. 2011;1809(8):459–468. doi: 10.1016/j.bbagrm.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000;101(5):543–553. doi: 10.1016/s0092-8674(00)80864-8. [DOI] [PubMed] [Google Scholar]
- 34.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101(5):533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 35.Guo HS, Fei JF, Xie Q, Chua NH. A chemical-regulated inducible RNAi system in plants. Plant J. 2003;34(3):383–392. doi: 10.1046/j.1365-313x.2003.01723.x. [DOI] [PubMed] [Google Scholar]
- 36.Fukunaga R, Doudna JA. dsRNA with 5′ overhangs contributes to endogenous and antiviral RNA silencing pathways in plants. EMBO J. 2009;28(5):545–555. doi: 10.1038/emboj.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brodersen P, Voinnet O. The diversity of RNA silencing pathways in plants. Trends Genet. 2006;22(5):268–280. doi: 10.1016/j.tig.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Ramachandran V, Chen XM. Small RNA metabolism in Arabidopsis. Trends Plant Sci. 2008;13(7):368–374. doi: 10.1016/j.tplants.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elmayan T, et al. A neomorphic sgs3 allele stabilizing miRNA cleavage products reveals that SGS3 acts as a homodimer. FEBS J. 2009;276(3):835–844. doi: 10.1111/j.1742-4658.2008.06828.x. [DOI] [PubMed] [Google Scholar]
- 40.Vaucheret H. Post-transcriptional small RNA pathways in plants: Mechanisms and regulations. Genes Dev. 2006;20(7):759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- 41.Hollick JB. Sensing the epigenome. Trends Plant Sci. 2008;13(7):398–404. doi: 10.1016/j.tplants.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Zhu Y, Qian W, Hua J. Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog. 2010;6(4):e1000844. doi: 10.1371/journal.ppat.1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fitter AH, Fitter RS. Rapid changes in flowering time in British plants. Science. 2002;296(5573):1689–1691. doi: 10.1126/science.1071617. [DOI] [PubMed] [Google Scholar]
- 44.Schmitz RJ, Amasino RM. Vernalization: A model for investigating epigenetics and eukaryotic gene regulation in plants. Biochim Biophys Acta. 2007;1769(5-6):269–275. doi: 10.1016/j.bbaexp.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 45.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410(6826):380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 46.Qi Y, Denli AM, Hannon GJ. Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell. 2005;19(3):421–428. doi: 10.1016/j.molcel.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 47.Curaba J, Chen XM. Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J Biol Chem. 2008;283(6):3059–3066. doi: 10.1074/jbc.M708983200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.