Abstract
Inactivation of the ubiquitin ligase E6 associated protein (E6AP) encoded by the UBE3A gene has been associated with development of the Angelman syndrome. Recently, it was reported that in mice, loss of E6AP expression results in increased levels of the synaptic protein Arc and a concomitant impaired synaptic function, providing an explanation for some phenotypic features of Angelman syndrome patients. Accordingly, E6AP has been shown to negatively regulate activity-regulated cytoskeleton-associated protein (Arc) and it has been suggested that E6AP targets Arc for ubiquitination and degradation. In our study, we provide evidence that Arc is not a direct substrate for E6AP and binds only weakly to E6AP, if at all. Furthermore, we show that down-regulation of E6AP expression stimulates estradiol-induced transcription of the Arc gene. Thus, we propose that Arc protein levels are controlled by E6AP at the transcriptional rather than at the posttranslational level.
Posttranslational modification of proteins by ubiquitin and ubiquitin-like (UBL) proteins plays a prominent role in the regulation of many eukaryotic processes (1, 2). In recent years, components of the respective conjugation systems have emerged as potential targets in the treatment of human diseases because their deregulation has been associated with the development of distinct disorders or because they control pathways that, for instance, are of fundamental importance for the proliferative potential of cancer cells (3, 4). An impressive example for the latter is represented by the E3 ubiquitin ligase E6 associated protein (E6AP), which is encoded by the UBE3A gene on chromosome 15q11-13 (5, 6) and has been linked to three distinct disorders. Firstly, E6AP was originally isolated as an interacting protein of the E6 protein of oncogenic human papillomaviruses (HPVs) (7, 8). In complex with E6, E6AP targets proteins for degradation [e.g., the tumor suppressor protein p53] that are normally not recognized by E6AP, thereby contributing to HPV-induced cervical carcinogenesis (9, 10). Secondly, loss of E6AP expression or function results in the development of Angelman syndrome (AS), a neurodevelopmental disorder (11–14). Finally, deregulation of E6AP expression has been associated with autism spectrum disorders (15, 16), and studies with transgenic mice suggest that amplification of the Ube3a gene results in increased E6AP levels that contribute to autistic phenotypes (17).
In general, E3 proteins mediate the specific recognition of substrates of the ubiquitin-conjugation system (1, 2, 18). Thus, identification of substrate proteins of E6AP should provide insights into cellular processes/pathways that are controlled by E6AP and whose deregulation contributes to the different pathologic conditions. Several potential substrates of E6AP have been reported, including human homolog of Rad23 A (HHR23A) and HHR23B, amplified in breast cancer 1 protein (AIB1), Promyelocytic leukemia protein (PML), α-Synuclein, and really interesting new gene 1b (Ring1b) (19–23). However, the pathophysiologic relevance of these interactions remains unclear. Ube3a knockout mice are valuable tools for uncovering the mechanism(s) underlying AS development, because in many aspects, the phenotype of such mice resembles that of AS patients (24, 25). By analyzing transgenic mice expressing HA-tagged ubiquitin crossed with wild-type mice or Ube3a knockout mice, Sacsin was recently identified as a protein whose ubiquitination status is altered in the absence of E6AP (26). Sacsin is a giant protein of 4,579 aa, and, thus, obtaining evidence that Sacsin is a direct substrate of E6AP is difficult, if not impossible, by available means. Like the HHR23 proteins, Sacsin contains an XPC domain (27). Because the activity-regulated cytoskeleton-associated protein (Arc), which mediates endocytosis of AMPA receptors at excitatory synapses, may also contain an XPC-like domain (26), it was hypothesized that XPC domains represent binding modules for E6AP. In support of this hypothesis, evidence was provided to suggest that Arc is a substrate of E6AP (26).
At the time Arc was reported to be a substrate of E6AP, we had evidence that the XPC domain is not required for HHR23A to bind E6AP. Hence, we revisited the interactions of E6AP with HHR23A, Sacsin, and Arc and found that (i) E6AP binds to the UBL domain of HHR23A and of Sacsin rather than to the respective XPC domain, (ii) E6AP does not or only poorly bind to Arc or its proposed XPC domain, and (iii) E6AP does not or only inefficiently target Arc for ubiquitination and degradation. Finally, we report that RNA interference-mediated knockdown of E6AP expression stimulates estradiol-induced transcription of the Arc gene, providing an alternative mechanism by which E6AP controls Arc protein levels.
Results
E6AP Binds to the UBL Domains of HHR23A and Sacsin.
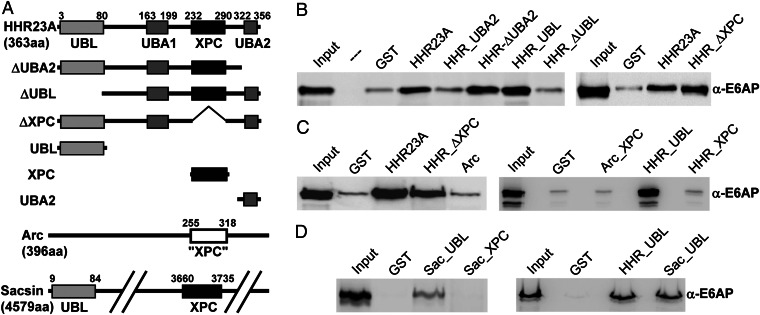
HHR23A belongs to the family of UBL and ubiquitin-associated (UBA) domain-containing proteins that act as bridging factors between ubiquitinated proteins and the 26S proteasome (because the UBL, UBA, and XPC domains of HHR23A and HHR23B are highly similar, with more than 75% identity, we limited our analysis to HHR23A) (28–30). To delineate the region(s) of HHR23A that serve(s) as binding site(s) for E6AP (19), we expressed HHR23A and several deletion mutants (Fig. 1A) as GST fusion proteins in Escherichia coli (Fig. S1) and tested their ability to bind to E6AP in coprecipitation experiments. This revealed that an HHR23A variant devoid of the UBL domain does not, or only poorly, bind E6AP, whereas the presence of the XPC domain is not required for E6AP binding (Fig. 1B). Furthermore, the UBL domain but not the XPC domain is sufficient to bind E6AP (Fig. 1C, Right).
Fig. 1.
E6AP binds to the UBL domain of HHR23A and Sacsin but not to XPC domains. (A) Schematic structure of the proteins used. (B–D) HHR23A and its various deletion mutants, Arc and its XPC domain (Arc_XPC), and the XPC domain and the UBL domain of Sacsin (SAC_XPC, SAC_UBL) were bacterially expressed as GST fusion proteins. Upon purification and adjustment of protein amounts (Fig. S1), the various fusion proteins were incubated with baculovirus-expressed E6AP. After 4 h at 4 °C, the amount of E6AP bound to the various proteins was determined by SDS/PAGE followed by Western blot analysis. Input corresponds to 20% of E6AP used for binding reactions.
Because the results obtained with HHR23A did not support the notion that XPC domains represent E6AP-binding modules (26), we extended the interaction studies to Arc and Sacsin. However, neither the proposed XPC domain of Arc (26) nor the XPC domain of Sacsin (27) bound to E6AP above background (Fig. 1 C and D). Importantly, an interaction between full-length Arc and E6AP was also not observed (Fig. 1C, Left). Because of its size (4,579 aa), in vitro binding experiments with full-length Sacsin could not be performed. However, in addition to the XPC domain, Sacsin contains an N-terminal UBL domain (UniProtKB entry Q9NZJ4; Fig. 1A). Indeed, similar to HHR23A, the UBL domain of Sacsin was able to bind E6AP (Fig. 1D).
Arc Is Not a Substrate for E6AP Within Cells.
The coprecipitation data indicate that binding of E6AP to HHR23A and Sacsin is mediated by their UBL domain rather than their XPC domain and that Arc does not bind or poorly binds to E6AP. The possibility that E6AP has a low affinity for Arc is supported by the observation that as reported (26), Arc can be ubiquitinated by E6AP in vitro (Fig. S2A). This finding is reminiscent of the interaction of E6AP with p53 in the absence of the E6 oncoprotein: E6AP does not detectably bind to p53 in coprecipitation assays and is not involved in p53 degradation in HPV-negative cells (31–34), and yet it is ubiquitinated by E6AP in vitro (35) (Fig. S2A). Furthermore, although we have no evidence that the E3 ligase Human homolog of Mdm2 (Hdm2) binds to Arc, Arc is ubiquitinated by Hdm2 in vitro (Fig. S2B). Taken together, these data indicate that results obtained in in vitro ubiquitination reactions have to be interpreted with caution and cannot be readily extrapolated to the situation in cells (for further discussion, see SI Results and Discussion).
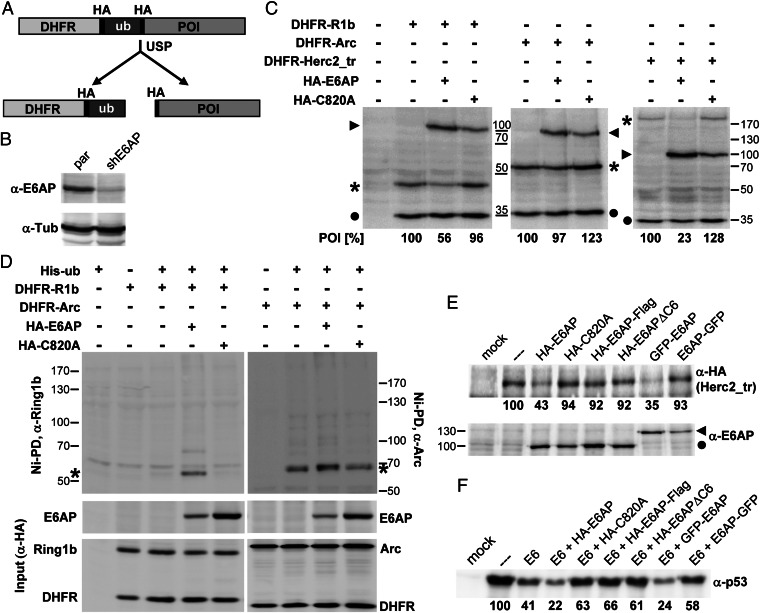
To study whether Arc can serve as substrate for E6AP in cells, we used the dehydrofolate reductase (DHFR)–ubiquitin fusion protein system (36–38). This system takes advantage of the fact that ubiquitin fusion proteins are cotranslationally cleaved by ubiquitin-specific proteases in a quantitative manner resulting in two separate proteins (Fig. 2A). In brief, the cDNA encoding the protein of interest (POI) is fused in frame to a cDNA encoding a DHFR–ubiquitin fusion protein, resulting in a construct expressing a single mRNA encoding a DHFR–ubiquitin–POI fusion protein. Because upon translation and concomitant cleavage, two separate proteins are generated from a common precursor (Fig. 2A), comparison of the relative levels of the POI and DHFR–ubiquitin (a mutant form of ubiquitin, in which Lys-48 is replaced by Arg, is used in this system to avoid degradation of DHFR–ubiquitin) provides a direct measure to determine the effect of an E3 ligase on the turnover rate and the ubiquitination status of a POI.
Fig. 2.
Arc does not represent a substrate for E6AP in cells. (A) Schematic of the DHFR–HA–ubiquitin fusion protein system. For details, see Results. (B) Extracts were prepared from HEK293T cells (par) and HEK293T cells, in which endogenous E6AP expression is stably down-regulated by RNA interference (shE6AP), and subjected to Western blot analysis with antibodies against E6AP and tubulin (Tub). (C) HEK293T-shE6AP cells were cotransfected with expression constructs for HA-tagged E6AP or the inactive mutant E6AP-C820A (HA-C820A), and DHFR–HA–ubiquitin fusion proteins of HA-tagged Ring1b-I53S (R1b), HA-tagged Arc, or HA-tagged HERC2_tr. Protein extracts were prepared 24 h after transfection. Levels of the various proteins were determined by Western blot analysis using an anti-HA antibody and quantified. The relative ratio of HA-tagged Ring1b-I53S, Arc, or HERC2_tr to DHFR–HA–ubiquitin is indicated, with the ratio of HA-tagged Ring1b-I53S, Arc, or HERC2_tr to DHFR–HA–ubiquitin in the absence of E6AP set to 100%. Running positions of DHFR–HA–ubiquitin are indicated by closed circles; those of HA-tagged Ring1b-I53S, Arc or HERC2_tr are indicated by asterisks, and those of E6AP and E6AP-C820A are indicated by arrowheads. Running positions of molecular mass markers (kDa) are marked. Ring1b-I53S was used as substrate, because it cannot ubiquitinate itself (23). Thus, its degradation by E6AP can be easily monitored. HERC2_tr is an N-terminal truncation mutant of HERC2 (amino acids 2958–4834), and, unlike full-length HERC2 (39), it is an efficient substrate for E6AP. (D) Transfections were performed as in C but in the presence of a construct encoding His-tagged ubiquitin (His-ub). Protein extracts were prepared and ubiquitinated proteins were isolated by Ni2+-affinity chromatography (Ni-PD). Levels of ubiquitinated Ring1b-I53S (R1b) and Arc were determined by Western blot analysis with antibodies against Ring1b or Arc. Running positions of molecular mass markers (kDa) are indicated. * indicates (presumably mono) ubiquitinated forms of Ring1b-I53S and Arc. Input corresponds to 15% of the extracts used for affinity purification. Running positions of E6AP, Ring1b-I53S, Arc, and DHFR–ubiquitin are indicated. (E) H1299 cells, in which endogenous E6AP expression is stably down-regulated (45), were cotransfected with constructs encoding HA-tagged HERC2_tr, various forms of E6AP as indicated, and β-galactosidase (to determine transfection efficiency). Twenty-four hours after transfection, extracts were prepared and levels of HERC2_tr (Upper) and of the various forms of E6AP (Lower) were determined by Western blot analysis and quantified; mock, mock transfected cells; HA-E6AP-Flag, HA-E6AP with a C-terminal Flag tag; HA-E6APΔC6, C-terminally truncated E6AP (deletion of the C-terminal 6 amino acids) with an N-terminal HA tag; GFP-E6AP, GFP fused to the N terminus of E6AP; E6AP-GFP, GFP fused to the C terminus of E6AP. Running positions of the HA-tagged forms and GFP fusions of E6AP are indicated by a closed circle or an arrowhead, respectively. Running positions of molecular mass markers (kDa) are indicated. (F) As in E, but parental H1299 cells were used for transfection and p53 was used as substrate for the E6AP-HPV E6 complex. Note that endogenous E6AP is sufficient for E6-mediated degradation of p53 (45). Thus, the finding that coexpression of inactive forms of E6AP (HA-E6APΔC6, HA-C820A) and of C-terminal fusions of E6AP (HA-E6AP-Flag, E6AP-GFP) interferes with E6-mediated degradation of p53 indicates that these proteins are not only inactive but, moreover, act as dominant-negative mutants. Experiments shown in C–F were repeated at least three times with similar results.
To determine the effect of E6AP on Arc stability, Arc was expressed as a DHFR–ubiquitin fusion protein in the absence or presence of E6AP or a catalytically inactive E6AP mutant (E6AP-C820A) in HEK293T cells in which endogenous E6AP expression is knocked down by RNA interference (Fig. 2B). As controls, Ring1b-I53S (an inactive form of the E3 ligase Ring1b), which is a substrate for E6AP (23), and an N-terminally truncated form of HECT domain and RCC1-like domain-containing protein 2 (HERC2_tr) (39), which serves as an artificial substrate of E6AP (for further details, see the legend to Fig. 2C), were used. This revealed that coexpression of E6AP had no significant effect on Arc levels (Fig. 2C). In contrast, levels of Ring1b-I53S and HERC2_tr were significantly reduced in the presence of E6AP but not in the presence of E6AP-C820A (Fig. 2C). Similarly, coexpression of E6AP had no significant effect on the levels of ubiquitinated Arc, whereas it facilitated (mainly) monoubiquitination of Ring1b-I53S (Fig. 2D). Thus, the results obtained indicate that within cells, Ring1b-I53S and HERC2_tr, but not Arc, are targeted by E6AP for ubiquitination and degradation.
Knockdown of E6AP Expression Stimulates Estradiol-Induced Transcription of the Arc Gene.
It was recently reported that transgenic mice carrying two additional Ube3a alleles show autism-like features (17). The additional Ube3a alleles were engineered such that they express E6AP with a C-terminal Flag tag. To provide evidence that “E6AP-Flag” still has E3 activity, it was shown that in the transgenic mice, Arc levels are significantly decreased in brain-derived lysates (17). However, it was previously shown that fusing a C-terminal extension (e.g., Flag tag) to E6AP or deleting the six C-terminal residues (E6APΔC6) result in ubiquitination-defective proteins (40). Indeed, like E6APΔC6 and the E6AP-C820A mutant, E6AP-Flag as well as E6AP-GFP (Flag tag or GFP fused to the C terminus of E6AP) did not target HERC2_tr for degradation within cells, whereas GFP-E6AP (GFP fused to the N terminus of E6AP) was active (Fig. 2E). Similarly, E6AP-Flag and E6AP-GFP were not able to facilitate HPV E6-mediated degradation of p53 but rather acted as dominant-negative mutants (Fig. 2F), supporting the notion that fusing a C-terminal extension to E6AP impairs its E3 activity. Thus, the autism-like features of Ube3a-Flag transgenic mice (17) do not appear to be caused by an increase in E3 activity but rather by inhibition of the E3 activity of endogenous E6AP and/or by an increase in E3-independent properties of E6AP.
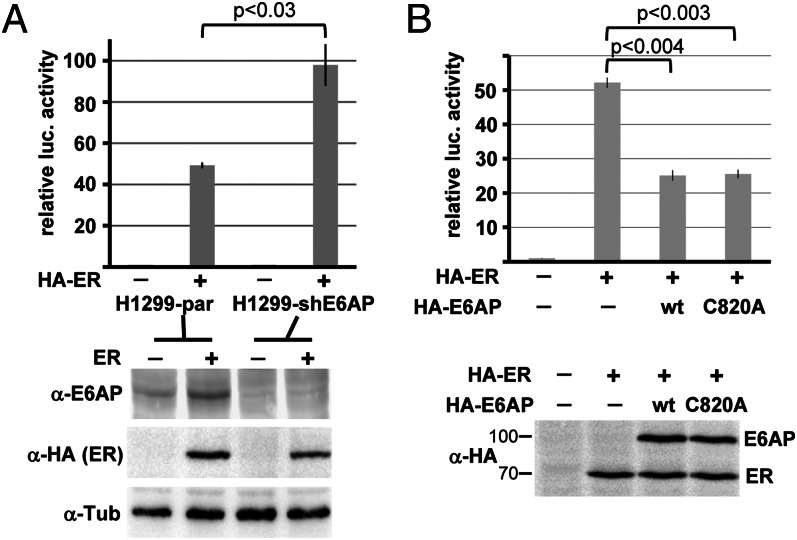
How can the data obtained so far be reconciled with the observations that mice overexpressing E6AP-Flag have reduced Arc levels (17) and that under certain conditions, Arc levels are increased in brain lysates derived from Ube3a-null mice (26)? E6AP was reported to affect nuclear hormone receptor-mediated transcription by E3-independent and E3-dependent mechanisms (41–43). Furthermore, expression of the human Arc gene is increased by estradiol signaling (44). We, therefore, hypothesized that loss of E6AP results in increased estrogen receptor (ER)-mediated transcription and thereby Arc expression. To address this possibility, we first studied ER-induced transcription of a respective reporter construct in H1299-shE6AP cells, in which E6AP expression is down-regulated by RNA interference (45) and in parental H1299 cells. Indeed, ER-induced expression of the reporter gene was approximately twofold higher in H1299-shE6AP cells than in H1299 cells (Fig. 3A). In contrast, ectopic expression of E6AP decreased ER-induced transcription by a factor of two to three, and this repressive effect was not dependent on E3 activity (Fig. 3B).
Fig. 3.
E6AP affects ER-mediated transcription. (A) H1299 cells (H1299-par) and H1299 cells, in which endogenous E6AP expression is stably down-regulated (H1299-shE6AP) (45), were transfected with an ER-responsive reporter construct encoding luciferase and a construct encoding β-galactosidase (to determine transfection efficiency) in the absence (−) or presence (+) of a construct encoding HA-tagged ERα (ER). Extracts were prepared 24 h after transfection, luciferase activity was determined as a measure of ER activity, and the relative values obtained were adjusted for transfection efficiency. Twenty percent of the extracts were subjected to Western blot analysis with antibodies against E6AP, HA tag (ER), and tubulin (Tub). (B) Transfection of parental H1299 cells was performed as in A but in the presence of HA-tagged E6AP (wt) or the inactive mutant E6AP-C820A (C820A). Analysis was performed as in A. Running positions of molecular mass markers (kDa) are indicated. Error bars represent the SD from at least three independent experiments. P values (independent two-sample t test) are depicted.
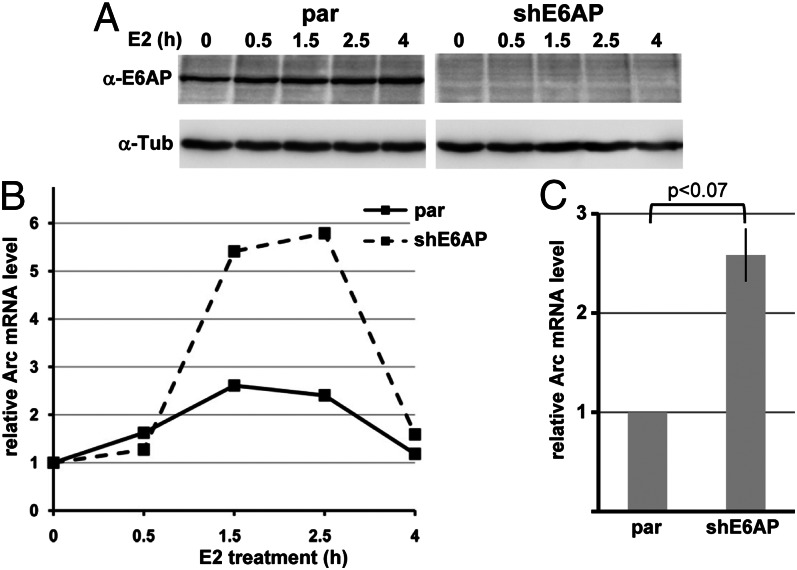
Having shown that loss of E6AP expression stimulates ER-induced transcription of a reporter gene, we then determined the effect of estradiol treatment on endogenous Arc mRNA levels in neuroblastoma-derived SH-SY5Y cells and in SH-SY5Y–shE6AP cells, in which E6AP expression is down-regulated by RNA interference (Fig. 4A). As reported (44), treatment of parental SH-SY5Y cells with estradiol resulted in a transient increase of Arc mRNA levels (Fig. 4B). In line with the results obtained with the ER-responsive reporter construct, this increase was 2–3 times higher in SH-SY5Y–shE6AP cells (Fig. 4 B and C). In conclusion, our data indicate that at least under certain conditions, E6AP constrains estradiol-induced transcription of the Arc gene.
Fig. 4.
Knockdown of E6AP expression stimulates estradiol-induced transcription of the Arc gene. SH-SY5Y cells (par) and SH-SY5Y cells, in which endogenous E6AP expression is stably down-regulated by RNA interference (shE6AP), were treated with 10 nM estradiol (E2), and cells were harvested at the times indicated. (A) Ten percent of the cells were used to determine levels of E6AP (α-E6AP) and, as loading control, tubulin (α-Tub). (B) The remaining cells were used to isolate total RNA followed by cDNA synthesis and determination of relative Arc mRNA levels by real-time PCR. Simultaneous determination of Actin mRNA levels served as reference. The ratio between Arc mRNA and Actin mRNA at time 0 was set to 1. Real-time PCR was performed in four independent experiments with similar results, but the time course of estradiol-induced transcription of the Arc gene varied slightly between individual experiments. The graph shows a representative example. (C) Summary of the data obtained in four independent experiments. The maximum level of Arc mRNA observed upon addition of estradiol to parental cells (e.g., mRNA level at 1.5 h in B) was set to “1” for each individual experiment and the relative increase of Arc mRNA levels observed in E6AP knockdown cells (shE6AP) was correlated accordingly. SD (± 0.27) is indicated. P value (independent two-sample t test) is depicted.
Discussion
All of the known genetic abnormalities associated with AS development result in loss of E6AP expression or expression of E6AP mutants with reduced E3 activity (11, 12, 46, 47), indicating that constitutive or transient increases in the level of substrate proteins of E6AP play a major role in AS development. Arc is a crucial player in synaptic plasticity, whose deregulation contributes to the manifestation of AS (24, 48, 49). Thus, the finding that under certain conditions, Arc levels are elevated in E6AP-null mice (26) likely explains some of the symptoms of AS patients. Although our data support the notion that loss of E6AP expression results in deregulation of Arc, we found that E6AP negatively affects Arc expression at the transcriptional rather than at the posttranslational level, as proposed previously (26).
In contrast to a previous report (26), we were not able to obtain evidence that E6AP binds to Arc above background (Fig. 1C). A possibility to explain these different results is that E6AP binds to Arc with rather low affinity and, thus, detection of this interaction depends on the conditions used. The finding that E6AP ubiquitinates Arc in vitro (Fig. S2) (26) may support this possibility. However, as indicated above (also see SI Results and Discussion), we propose that E6AP-mediated ubiquitination of Arc in vitro is a nonspecific process. In other words, in vitro ubiquitination studies are important to support the notion that a protein is a substrate for E6AP; however, additional criteria need to be met to conclude that the ubiquitination observed in vitro is physiologically relevant.
An important cornerstone in the line of arguments that a certain E3 ligase regulates the stability of a given protein is the verification that the E3 ligase affects the ubiquitination status and the turnover rate of the respective protein in cells in coexpression experiments. Similar to our data (Fig. 2D), it was reported that ubiquitination of Arc is observed in the absence of ectopically expressed E6AP (26). In contrast to our data, however, coexpression of E6AP and Arc resulted in somewhat increased levels of ubiquitinated Arc, whereas ubiquitinated Arc species were not detected in the presence of an inactive E6AP mutant. A potential drawback of cotransfection experiments is that transfection efficiencies can vary between individual transfections and, thus, transfection efficiencies need to be considered when interpreting results obtained in such experiments. To do so, the DHFR–ubiquitin fusion protein system (36–38, 45) is ideally suited, because the relative transfection efficiency can be readily determined by monitoring the level of DHFR–ubiquitin (Fig. 2A). Using this system, we showed that E6AP has neither a significant effect on the ubiquitination status nor on the level of ectopically expressed Arc (Fig. 2 C and D). The latter result is again in contrast to published data (26). However, it should be noted that in the respective study, data from proteasome inhibitor or half-life experiments to indicate that the decrease in Arc levels is indeed attributable to enhanced degradation rather than to effects on, for instance, the transcriptional or translational level were not provided (26). Such experiments are not required when using the DHFR–ubiquitin fusion protein system, because DHFR–ubiquitin and the POI are expressed as a single polyprotein from the same mRNA. Thus, determination of the relative ratio between the POI and DHFR–ubiquitin provides unequivocal evidence as to whether a protein is preferentially degraded or not.
In support of the notion that Arc is an E6AP substrate, it was reported that upon treatment of hippocampal cells with kainate, Arc protein levels but not Arc mRNA levels are increased in E6AP-null cells (26). However, it is known that in response to various stimuli, Arc transcription is induced but only transiently (e.g., refs. 44, 50, and 51). For example, similar to our results (Fig. 4), it was reported that treatment of SH-SY5Y cells with estradiol or carbachol results in a rapid but transient induction of Arc mRNA levels and that this transient increase in mRNA levels is mirrored in a transient increase in Arc protein levels but with different kinetics (44, 50). Because in the study with kainate (26), the analysis was performed at a single time point (and not in a time course experiment), we propose that the increase of Arc protein levels observed in E6AP-null cells was attributable to a transient increase in mRNA levels rather than a prolonged half-life of the Arc protein.
Besides challenging the previously proposed mechanism by which E6AP regulates Arc levels (26), our data suggest that the importance of the connection between E6AP and nuclear hormone receptors for development of AS (and, possibly, autism spectrum disorders; Results) has been underestimated so far. Estrogen was reported to affect Arc transcription (44), as well as various neuronal processes including synaptic plasticity (52–56). Furthermore, ∼5–10% of AS patients do not harbor a detectable defect in the UBE3A gene (12). Thus, although we do not yet know how E6AP interferes with estradiol-induced transcription of the Arc gene [E6AP can affect transcriptional processes by E3-dependent and E3-independent mechanisms (41–43)], it is tempting to speculate that some AS patients carry defects along the respective signaling pathway(s). Furthermore, analysis of transgenic mice, in which E6AP expression is ablated in mammary tissue, identified a few genes, the estradiol-induced expression of which is down-regulated in the absence of E6AP (57). Similarly, preliminary data (Fig. S3) suggest that in contrast to the Arc gene, down-regulation of E6AP expression in SH-SY5Y cells results in a decrease in estradiol-induced transcription of the cathepsin D gene, an established estradiol-responsive gene (58, 59). Taken together, these data support the notion that E6AP has both positive and negative effects on estradiol-mediated transcription. Thus, delineation of the mechanism(s) involved in this phenomenon and identification of the genes whose expression is altered in the absence of E6AP should contribute to the elucidation of the pathways involved in AS development.
Materials and Methods
For cell lines, plasmids, transfection procedures, antibodies, coprecipitation, and luciferase reporter assays, see SI Materials and Methods.
Ubiquitination and Degradation Assays.
For ubiquitination assays, one 6-cm plate of cells was transfected with expression constructs encoding E6AP or E6AP-C820A (2.5 µg), His-tagged ubiquitin (1.5 µg), and DHFR–HA–ubiquitin–HA-Ring1B–I53S or DHFR–HA–ubiquitin–HA–Arc (1 µg). Twenty-four hours after transfection, 30% of the cells were lysed under nondenaturing conditions (30) to determine levels of E6AP, E6AP-C820A, DHFR–HA–ubiquitin, HA-Ring1B-I53S, and HA-Arc. The remaining cells were lysed under denaturing conditions and ubiquitinated proteins were purified as described (30).
Degradation assays with DHFR–ubiquitin fusion proteins were performed similar to ubiquitination assays, except that 3.5 and 1 µg of the constructs encoding E6AP and E6AP-C820A, respectively, were used and the construct for His–ubiquitin was omitted. Quantification of the intensity of the signals was performed with the Aida 4.08 software package (Raytest). HPV E6-induced degradation of p53 was monitored as described (45).
Estradiol Treatment and Real-Time PCR.
Cells were starved overnight in phenol red-free medium containing 5% (vol/vol) charcoal-stripped FBS. Medium was exchanged and cells were treated with 10 nM 17β-estradiol (Sigma). Approximately 5× 106 cells were harvested at the times indicated (Fig. 4), and RNA was isolated by using TRIzol Reagent (Invitrogen) and a Genejet RNA purification column (Fermentas). Reverse transcription was performed using the SuperScript III Reverse Transcriptase Kit (Invitrogen). The resulting cDNA was purified using the GeneJet PCR purification kit (Fermentas). Quantitative PCR analysis was performed using the Maxima SYBR Green/ROX qPCR Master Mix (Fermentas) or FastStart Universal SYBR Green Master Mix (ROX) (Roche). mRNA levels were compared using the ΔΔ−Ct method (60, 61). Actin mRNA was chosen as reference target. Primers used were as follows: Arc forward, CGCGAGGTGTTCTAC; Arc reverse, AGCCAGTACTCCTCAG; Actin forward, GCTCCGGCATGTGCAA; and Actin reverse, TGGCACCACACCTTCTAC.
Supplementary Material
Acknowledgments
We thank Thomas Kapitza and Susanne Schmid for excellent technical assistance, Armin Günther (Trenzyme) for help with real-time PCR, and Thomas U. Mayer for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grants SCHE 346/6-1 and SFB 969 B2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302792110/-/DCSupplemental.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem. 2012;81:167–176. doi: 10.1146/annurev-biochem-051910-094049. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz AL, Ciechanover A. Targeting proteins for destruction by the ubiquitin system: Implications for human pathobiology. Annu Rev Pharmacol Toxicol. 2009;49:73–96. doi: 10.1146/annurev.pharmtox.051208.165340. [DOI] [PubMed] [Google Scholar]
- 4.Edelmann MJ, Nicholson B, Kessler BM. Pharmacological targets in the ubiquitin system offer new ways of treating cancer, neurodegenerative disorders and infectious diseases. Expert Rev Mol Med. 2011;13:e35. doi: 10.1017/S1462399411002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nicholls RD, Knepper JL. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu Rev Genomics Hum Genet. 2001;2:153–175. doi: 10.1146/annurev.genom.2.1.153. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto Y, Huibregtse JM, Howley PM. The human E6-AP gene (UBE3A) encodes three potential protein isoforms generated by differential splicing. Genomics. 1997;41(2):263–266. doi: 10.1006/geno.1997.4617. [DOI] [PubMed] [Google Scholar]
- 7.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10(13):4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13(2):775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaudenon S, Huibregtse JM. HPV E6, E6AP and cervical cancer. BMC Biochem. 2008;9(Suppl 1):S4. doi: 10.1186/1471-2091-9-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheffner M, Huibregtse JM, Vierstra RD, Howley PM. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75(3):495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 11.Clayton-Smith J, Laan L. Angelman syndrome: A review of the clinical and genetic aspects. J Med Genet. 2003;40(2):87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagli A, Buiting K, Williams CA. Molecular and clinical aspects of Angelman syndrome. Mol Syndromol. 2012;2(3-5):100–112. doi: 10.1159/000328837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15(1):70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 14.Matsuura T, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15(1):74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 15.Glessner JT, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogart A, Wu D, LaSalle JM, Schanen NC. The comorbidity of autism with the genomic disorders of chromosome 15q11.2-q13. Neurobiol Dis. 2010;38(2):181–191. doi: 10.1016/j.nbd.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith SE, et al. (2011) Increased gene dosage of Ube3a results in autism traits and decreased glutamate synaptic transmission in mice. Sci Transl Med 3(103):103ra197. [DOI] [PMC free article] [PubMed]
- 18.Metzger MB, Hristova VA, Weissman AM. HECT and RING finger families of E3 ubiquitin ligases at a glance. J Cell Sci. 2012;125(Pt 3):531–537. doi: 10.1242/jcs.091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Talis AL, Howley PM. Identification of HHR23A as a substrate for E6-associated protein-mediated ubiquitination. J Biol Chem. 1999;274(26):18785–18792. doi: 10.1074/jbc.274.26.18785. [DOI] [PubMed] [Google Scholar]
- 20.Mulherkar SA, Sharma J, Jana NR. The ubiquitin ligase E6-AP promotes degradation of alpha-synuclein. J Neurochem. 2009;110(6):1955–1964. doi: 10.1111/j.1471-4159.2009.06293.x. [DOI] [PubMed] [Google Scholar]
- 21.Louria-Hayon I, et al. E6AP promotes the degradation of the PML tumor suppressor. Cell Death Differ. 2009;16(8):1156–1166. doi: 10.1038/cdd.2009.31. [DOI] [PubMed] [Google Scholar]
- 22.Mani A, et al. E6AP mediates regulated proteasomal degradation of the nuclear receptor coactivator amplified in breast cancer 1 in immortalized cells. Cancer Res. 2006;66(17):8680–8686. doi: 10.1158/0008-5472.CAN-06-0557. [DOI] [PubMed] [Google Scholar]
- 23.Zaaroor-Regev D, et al. Regulation of the polycomb protein Ring1B by self-ubiquitination or by E6-AP may have implications to the pathogenesis of Angelman syndrome. Proc Natl Acad Sci USA. 2010;107(15):6788–6793. doi: 10.1073/pnas.1003108107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang YH, et al. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21(4):799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 25.Miura K, et al. Neurobehavioral and electroencephalographic abnormalities in Ube3a maternal-deficient mice. Neurobiol Dis. 2002;9(2):149–159. doi: 10.1006/nbdi.2001.0463. [DOI] [PubMed] [Google Scholar]
- 26.Greer PL, et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamionka M, Feigon J. Structure of the XPC binding domain of hHR23A reveals hydrophobic patches for protein interaction. Protein Sci. 2004;13(9):2370–2377. doi: 10.1110/ps.04824304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dantuma NP, Heinen C, Hoogstraten D. The ubiquitin receptor Rad23: At the crossroads of nucleotide excision repair and proteasomal degradation. DNA Repair (Amst) 2009;8(4):449–460. doi: 10.1016/j.dnarep.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol. 2009;10(10):659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glockzin S, Ogi FX, Hengstermann A, Scheffner M, Blattner C. Involvement of the DNA repair protein hHR23 in p53 degradation. Mol Cell Biol. 2003;23(24):8960–8969. doi: 10.1128/MCB.23.24.8960-8969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beer-Romero P, Glass S, Rolfe M. Antisense targeting of E6AP elevates p53 in HPV-infected cells but not in normal cells. Oncogene. 1997;14(5):595–602. doi: 10.1038/sj.onc.1200872. [DOI] [PubMed] [Google Scholar]
- 32.Hengstermann A, et al. Growth suppression induced by downregulation of E6-AP expression in human papillomavirus-positive cancer cell lines depends on p53. J Virol. 2005;79(14):9296–9300. doi: 10.1128/JVI.79.14.9296-9300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y, Cairns MJ, Marouga R, Sun LQ. E6AP gene suppression and characterization with in vitro selected hammerhead ribozymes. Cancer Gene Ther. 2003;10(9):707–716. doi: 10.1038/sj.cgt.7700623. [DOI] [PubMed] [Google Scholar]
- 34.Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273(11):6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 35.Hengstermann A, Whitaker NJ, Zimmer D, Zentgraf H, Scheffner M. Characterization of sequence elements involved in p53 stability regulation reveals cell type dependence for p53 degradation. Oncogene. 1998;17(22):2933–2941. doi: 10.1038/sj.onc.1202282. [DOI] [PubMed] [Google Scholar]
- 36.Bachmair A, Finley D, Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986;234(4773):179–186. doi: 10.1126/science.3018930. [DOI] [PubMed] [Google Scholar]
- 37.Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005;399:777–799. doi: 10.1016/S0076-6879(05)99051-4. [DOI] [PubMed] [Google Scholar]
- 38.Matentzoglu K, Scheffner M. Ubiquitin-fusion protein system: A powerful tool for ectopic protein expression in mammalian cells. Biotechniques. 2009;46(1):21–22, 24, 26 passim. doi: 10.2144/000113023. [DOI] [PubMed] [Google Scholar]
- 39. Kuhnle S, et al. (2011) Physical and functional interaction of the HECT ubiquitin-protein ligases E6AP and HERC2. J Biol Chem 286(22):19410–19416. [DOI] [PMC free article] [PubMed]
- 40.Salvat C, Wang G, Dastur A, Lyon N, Huibregtse JM. The -4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. J Biol Chem. 2004;279(18):18935–18943. doi: 10.1074/jbc.M312201200. [DOI] [PubMed] [Google Scholar]
- 41.Nawaz Z, et al. The Angelman syndrome-associated protein, E6-AP, is a coactivator for the nuclear hormone receptor superfamily. Mol Cell Biol. 1999;19(2):1182–1189. doi: 10.1128/mcb.19.2.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li L, Li Z, Howley PM, Sacks DB. E6AP and calmodulin reciprocally regulate estrogen receptor stability. J Biol Chem. 2006;281(4):1978–1985. doi: 10.1074/jbc.M508545200. [DOI] [PubMed] [Google Scholar]
- 43.Ramamoorthy S, Dhananjayan SC, Demayo FJ, Nawaz Z. Isoform-specific degradation of PR-B by E6-AP is critical for normal mammary gland development. Mol Endocrinol. 2010;24(11):2099–2113. doi: 10.1210/me.2010-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chamniansawat S, Chongthammakun S. Estrogen stimulates activity-regulated cytoskeleton associated protein (Arc) expression via the MAPK- and PI-3K-dependent pathways in SH-SY5Y cells. Neurosci Lett. 2009;452(2):130–135. doi: 10.1016/j.neulet.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 45.Kuballa P, Matentzoglu K, Scheffner M. The role of the ubiquitin ligase E6-AP in human papillomavirus E6-mediated degradation of PDZ domain-containing proteins. J Biol Chem. 2007;282(1):65–71. doi: 10.1074/jbc.M605117200. [DOI] [PubMed] [Google Scholar]
- 46.Fang P, et al. The spectrum of mutations in UBE3A causing Angelman syndrome. Hum Mol Genet. 1999;8(1):129–135. doi: 10.1093/hmg/8.1.129. [DOI] [PubMed] [Google Scholar]
- 47.Cooper EM, Hudson AW, Amos J, Wagstaff J, Howley PM. Biochemical analysis of Angelman syndrome-associated mutations in the E3 ubiquitin ligase E6-associated protein. J Biol Chem. 2004;279(39):41208–41217. doi: 10.1074/jbc.M401302200. [DOI] [PubMed] [Google Scholar]
- 48.Bramham CR, Worley PF, Moore MJ, Guzowski JF. The immediate early gene arc/arg3.1: Regulation, mechanisms, and function. J Neurosci. 2008;28(46):11760–11767. doi: 10.1523/JNEUROSCI.3864-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yashiro K, et al. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12(6):777–783. doi: 10.1038/nn.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soulé J, et al. Balancing Arc synthesis, mRNA decay, and proteasomal degradation: Maximal protein expression triggered by rapid eye movement sleep-like bursts of muscarinic cholinergic receptor stimulation. J Biol Chem. 2012;287(26):22354–22366. doi: 10.1074/jbc.M112.376491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teber I, Köhling R, Speckmann EJ, Barnekow A, Kremerskothen J. Muscarinic acetylcholine receptor stimulation induces expression of the activity-regulated cytoskeleton-associated gene (ARC) Brain Res Mol Brain Res. 2004;121(1-2):131–136. doi: 10.1016/j.molbrainres.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 52.Casadesus G. Special issue on estrogen actions in the brain. Biochim Biophys Acta. 2010;1800(10):1029. doi: 10.1016/j.bbagen.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 53.Córdoba Montoya DA, Carrer HF. Estrogen facilitates induction of long term potentiation in the hippocampus of awake rats. Brain Res. 1997;778(2):430–438. doi: 10.1016/s0006-8993(97)01206-7. [DOI] [PubMed] [Google Scholar]
- 54.Kim HJ, Casadesus G. Estrogen-mediated effects on cognition and synaptic plasticity: What do estrogen receptor knockout models tell us? Biochim Biophys Acta. 2010;1800(10):1090–1093. doi: 10.1016/j.bbagen.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rissman EF, Heck AL, Leonard JE, Shupnik MA, Gustafsson JA. Disruption of estrogen receptor beta gene impairs spatial learning in female mice. Proc Natl Acad Sci USA. 2002;99(6):3996–4001. doi: 10.1073/pnas.012032699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Srivastava DP, et al. Rapid estrogen signaling in the brain: Implications for the fine-tuning of neuronal circuitry. J Neurosci. 2011;31(45):16056–16063. doi: 10.1523/JNEUROSCI.4097-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Catoe HW, Nawaz Z. E6-AP facilitates efficient transcription at estrogen responsive promoters through recruitment of chromatin modifiers. Steroids. 2011;76(9):897–902. doi: 10.1016/j.steroids.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 58.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 59.Johnsen SA, et al. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 2009;69(1):128–136. doi: 10.1158/0008-5472.CAN-08-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270(1):41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 61.Schmittgen TD, et al. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: Comparison of endpoint and real-time methods. Anal Biochem. 2000;285(2):194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.