Abstract
Analysis of correlates of risk of infection in the RV144 HIV-1 vaccine efficacy trial demonstrated that plasma IgG against the HIV-1 envelope (Env) variable region 1 and 2 inversely correlated with risk, whereas HIV-1 Env-specific plasma IgA responses directly correlated with risk. In the secondary analysis, antibody-dependent cellular cytotoxicity (ADCC) was another inverse correlate of risk, but only in the presence of low plasma IgA Env-specific antibodies. Thus, we investigated the hypothesis that IgA could attenuate the protective effect of IgG responses through competition for the same Env binding sites. We report that Env-specific plasma IgA/IgG ratios are higher in infected than in uninfected vaccine recipients in RV144. Moreover, Env-specific IgA antibodies from RV144 vaccinees blocked the binding of ADCC-mediating mAb to HIV-1 Env glycoprotein 120 (gp120). An Env-specific monomeric IgA mAb isolated from an RV144 vaccinee also inhibited the ability of natural killer cells to kill HIV-1–infected CD4+ T cells coated with RV144-induced IgG antibodies. We show that monomeric Env-specific IgA, as part of postvaccination polyclonal antibody response, may modulate vaccine-induced immunity by diminishing ADCC effector function.
The phase III RV144 ALVAC/AIDSVAX B/E HIV-1 vaccine efficacy trial in Thailand demonstrated 31.2% estimated vaccine efficacy through 42 mo of follow-up (1). Analysis of correlates of risk of infection indicated that envelope (Env)-specific plasma antibody responses were associated with a lower infection risk in vaccinees (2). Though plasma Env variable region 1 and 2 (V1/V2) IgG correlated with decreased infection risk, high levels of anti–HIV-1 Env plasma IgA correlated with increased infection risk (2). Interaction analyses demonstrated that, in the presence of low IgA Env antibodies, antibody-dependent cellular cytotoxicity (ADCC) responses inversely correlated with risk of infection, whereas in the presence of high IgA Env plasma antibodies, there was no correlation with risk of infection (2). Because there was no overall enhancement of infection risk in the trial (1), we hypothesized that Env IgA might block potentially protective effector functions of Env IgG antibodies.
Antibody function depends, in part, on ability to bind to Fc receptors (FcR) on effector cells. The antibody isotype and subclass influences its affinity for different cellular FcRs (3, 4). IgG antibodies that mediate ADCC through natural killer (NK) cells bind to FcγRIIIa (CD16). In contrast, IgA antibodies do not bind to FcγRIIIa, but, rather, have high affinity for FcαRI (CD89) expressed by monocytes/macrophages and polymorphonuclear cells (PMN). This differential profile of FcR binding by IgG and IgA antibodies impacts the effector function capabilities of these antibody isotypes.
Here, we examined plasma IgA and monomeric IgA monoclonal antibodies from RV144 vaccine recipients to test the hypothesis that some fraction of the vaccine-elicited IgA response could block IgG-mediated ADCC function. We found that a fraction of the HIV-1–specific IgA response was directed against conformational epitopes in the first constant (C1) region of glycoprotein 120 (gp120), a specificity of RV144 vaccine trial antibodies previously shown to mediate ADCC via NK cells (5, 6). Two IgA monoclonal antibodies, CH38 IgA2 and CH29 IgA2, were targeted to gp120 ADCC epitopes expressed on the surface of HIV-1–infected CD4 T cells. These IgA antibodies did not mediate ADCC via NK effector cells themselves, but rather blocked ADCC effector function on infected CD4+ T-cell targets by HIV-1 Env IgG. Thus, the RV144 vaccine-elicited polyclonal antibody response included IgA antibodies with specificities that blocked IgG-mediated ADCC.
Results
Env IgA/IgG Ratios in an RV144 Case Control Study.
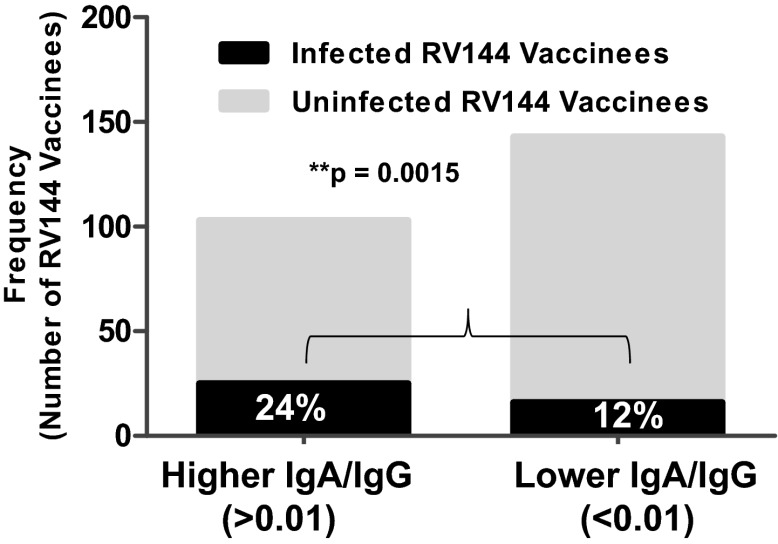
The RV144 vaccine regimen elicited polyclonal IgG and IgA responses to HIV-1 Env (2). To determine if some fraction of this polyclonal IgA response could block Env IgG-mediated ADCC activity, we first determined the ratio of Env-specific IgA to IgG in a follow-up analysis of the RV144 case control study (2). Notably, there was a significant enrichment of higher Env IgA/IgG ratios (Fig. 1; Fig. S1) among the infected vaccinees (cases) compared with uninfected vaccinees (controls). We have previously devised a magnitude and breadth score as a measurement of the quality of Env-specific antibody binding based on the magnitude of binding to a panel of HIV-1 envelope proteins representing clades A, AE, B, C, and G (2). For the combined magnitude and breadth score, there was an increased odds ratio (odds ratio is a descriptive statistic that herein indicates the strength of association between risk of HIV-1 infection and an antibody measurement) and lower P value for the IgA/IgG ratio antibody measurement compared with the IgA measurement alone (Table 1). Because there was no enhanced infection risk in RV144 vaccinees compared with placebos, this increased risk refers to decreased vaccine efficacy among those receiving the vaccine. Additionally, the IgA/IgG binding ratio response to the vaccine strain, AE.A244 gp120, directly correlated with infection risk. Not all Env IgA responses correlated with infection risk, as demonstrated by the Con6 Env gp120 IgA response (Table 1), which was similar to the HSV glycoprotein D (gD) IgA response that had no correlation with infection risk (neither decreased nor increased). Overall, by multiple measurements, Env IgA antibodies and the IgA/IgG ratio directly correlated with HIV-1 infection risk.
Fig. 1.
Enrichment of higher Env IgA/IgG ratio in infected RV144 vaccinees. The ratio of IgA/IgG HIV-1–specific Env binding was calculated for each sample and analyzed for differences between infected and uninfected vaccine recipients in the case control study. There was a significantly greater number of infected vaccinees with IgA/IgG ratio >1e-02 (A1 Congp140 Env) than uninfected vaccinees (weighted logistic regression using the method as reported in ref. 2; P = 0.0015).
Table 1.
IgA/IgG Env ratio significantly correlates with increased risk of infection (decreased vaccine efficacy)
| Envelope protein/peptide | Odds ratio |
P value |
||||
| IgA | IgG | IgA/IgG | IgA | IgG | IgA/IgG | |
| Vaccine strain clade AE.A244gp120* | 1.28 | 0.72 | 1.58 | ns | 0.03 | <0.01 |
| Env panel IgA primary score*,† | 1.39 | 0.78 | 1.59 | 0.05 | ns | <0.01 |
| CRF01 AE.C1 peptide | 1.69 | 0.85 | 1.79 | <0.001 | ns | <0.001 |
| Clade A.1ConEnv gp140 | 1.57 | 0.87 | 1.66 | <0.01 | ns | <0.01 |
| Con6 gp120 | 1.01 | 0.90 | 1.06 | ns | ns | ns |
| Non-HIV‡ HSV gD peptide | 1.01 | 1.02 | 1.01 | ns | ns | ns |
The odds ratio (univariate) for HIV-1 Env IgA and IgG binding to Env panel for the RV144 case control study are shown. As part of the RV144 correlates analyses, we previously reported the odds ratio and P values for the risk of infection for Env IgG and Env IgA measurements individually (2). For the two HIV-1 Env IgA responses with the strongest odds ratio for increased risk of infection (CRF01 AE.C1 peptide and A1.Con gp140), there was an increase in the odds ratio when the IgA/IgG ratio was measured (compared with Env IgA alone), although the P value was unchanged (P < 0.001; P = 0.0004 for both IgA alone and IgA/IgG ratio). Significant P values with increased odds ratio (increased risk of infection) are shown. ns, not significant.
Measurements that become significantly correlated with increased risk of infection when examined as a ratio of IgA to IgG Env binding.
Env IgA breadth score (univariate analysis) as reported (2). The multivariate analysis of this same breadth IgA score had an odds ratio of 1.54 (P = 0.03). Pink indicates significant correlation with increased risk of infection (decreased vaccine efficacy). Gray indicates significant correlation with decreased risk of infection.
HSV gD peptide that was present in the gp120 boost of vaccine.
Affinity of IgA and IgG Antibodies for Env Protein.
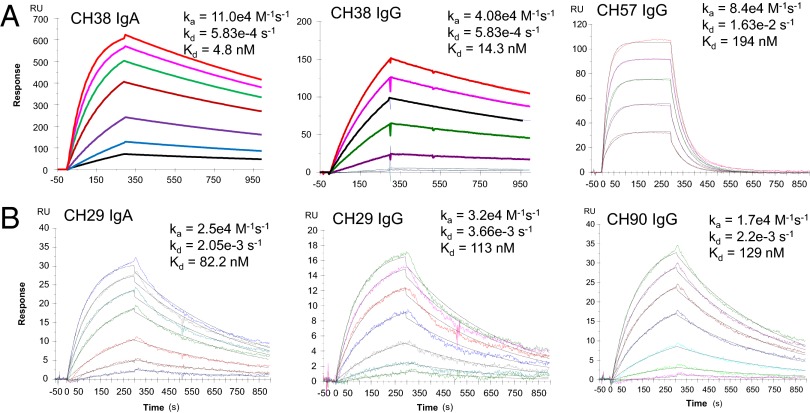
A dominant specificity of ADCC antibodies in RV144 was targeted to a conformational Env C1 region epitope as measured by blocking of ADCC with the fragment antigen-binding (Fab) of mAb A32 against this epitope (5, 6). Two of these ADCC-mediating mAbs, CH29 and CH38, were isolated from two ALVAC/AIDSVAX vaccine recipients and shown to mediate ADCC when expressed as IgG1 mAbs (5). The original natural isotypes in vivo of these two mAbs, however, were IgA1 and IgA2, respectively. Here, we expressed these antibody sequences as an IgA2 to test their functional properties. We first examined the binding Kd of these two IgA2 mAbs to the vaccine strain envelope AE.A244 gp120 and determined that CH38 IgA2 mAb bound 2.9-fold higher in both magnitude and affinity than CH38 IgG (4.8 nM compared with 14.3 nM; Fig. 2A). However, CH38 IgA2 bound to the vaccine strain Env with ∼40-fold greater affinity (4.8 nM compared with 194 nM) than the ADCC-mediating CH57 IgG mAb, which also targets the C1 conformational region and was isolated from the same vaccinee (5). In contrast, CH29 IgA2 and CH29 IgG both bound to HIV MN gp120 with similar affinities (but did not bind to AE.A244 gp120; Fig. 2B). CH90, a C1 conformational IgG mAb generated from the same vaccinee as CH29 (5), also had similar affinity to MNgp120 as CH29 IgA2 (Fig. 2B). These data demonstrated both the superior binding of CH38 IgA2 vs. IgG to AE.A244 gp120 Env protein and the heterogeneous nature of the RV144 vaccine-elicited IgA responses.
Fig. 2.
Different binding affinity of RV144 monoclonal antibodies of IgA and IgG origin. HIV-Env gp120 mAbs isolated from RV144 vaccinees were measured by SPR for binding to the vaccine strain envelope: AE.A244Δ11 gp120 (CH38, CH57) or MN gp120 (CH29, CH90). (A) The binding affinity to the vaccine strain AE.A244Δ11 gp120 is shown for CH38 IgA2, IgG, and CH57 IgG mAbs. CH38 and CH57 mAbs were generated from the same RV144 vaccinee. Data are representative of three measurements except for CH57 (Kd from two measurements were 163 and 194 nM). (B) The binding affinity to the vaccine strain MN gp120 is shown for CH29 IgA2, CH29 IgG, and CH90 IgG mAbs. CH29 and CH90 mAbs were generated from the same vaccinee. Data are each representative of three measurements, and mean Kd values for CH29 IgA2, CH29 IgG, and CH90 IgG were 82.1, 126, and 128 nM, respectively.
Cross-Blocking of C1 Region-Specific IgA and IgG mAbs.
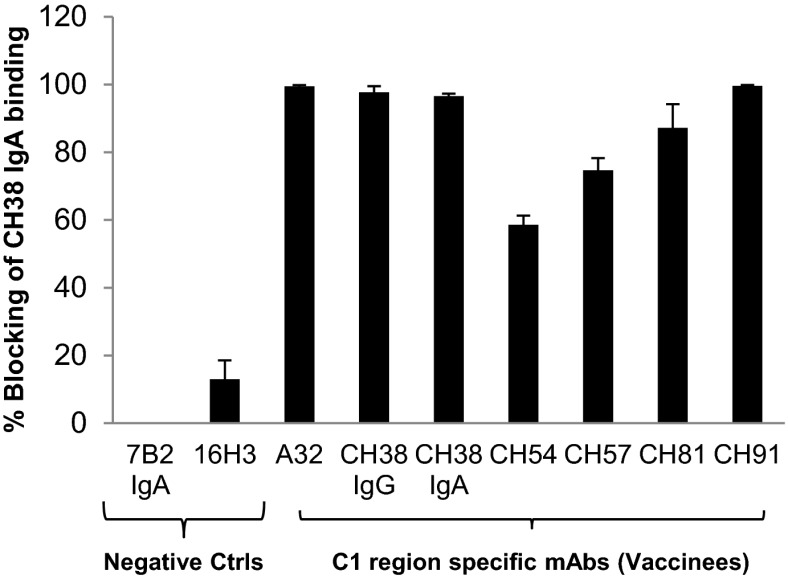
We examined the ability of C1 conformational IgG mAbs from four different RV144 vaccinees to compete with CH38 IgA2 mAb Env binding (Fig. 3). CH38 IgA2 binding to A244 gp120 Env was blocked by C1 region-specific mAbs (CH54, CH57, CH81, and CH91 mAbs), indicating the overlapping specificities of C1 region conformational IgG and IgA antibodies elicited by RV144. Because the plasma concentration of IgG is considerably higher than plasma IgA, we tested whether the differences in affinity between CH38 IgA2 mAb and CH57 IgG mAb to the same epitope could result in a lower molar concentration of the IgA blocking higher molar concentrations of IgG (Fig. S2). Increasing concentrations of CH38 IgA2 (0–100 µg/mL) were used to block binding of a fixed concentration of a C1 conformational region mAb generated from RV144: CH57 IgG (100 µg/mL) to AE.A244 gp120 Env. At a molar ratio of IgA: IgG = 0.2, CH38 IgA2 blocked 50% of CH57 IgG binding. These data indicate that at ratios of IgA to IgG that are comparable to the IgA/IgG ratio in plasma, the affinity differences in the antibody isotypes for binding to the same or overlapping epitopes can enable blocking of IgG Env binding by IgA.
Fig. 3.
C1 region gp120-specific IgG mAbs block binding of CH38 IgA2 mAbs. HIV-1 Env (AE.A244Δ11 gp120) was incubated with the IgG mAbs representing C1 conformation mAbs from four different RV144 vaccinees. These antigen–antibody complexes were then flowed over the monoclonal antibody coupled to the sensor surface: CH38 mAb IgA2. CH38 IgA2 mAb was blocked for binding to gp120 Env by IgG antibodies with specificities for the C1 region of gp120. The percent blocking of mAb binding was calculated relative to the mAb binding in the presence of a control gp41 antibody (7B2). Means and SDs of percentage blocking are from four measurements.
C1 Conformational Region-Specific Plasma IgA.
We purified IgA from IgG-depleted plasma from RV144 vaccinees to quantify conformational C1 region-specific IgA antibodies. We chose vaccinees from the RV144 pilot study who had plasma Env antibody binding breadth and detectable ADCC activity. Ten of the 16 purified IgAs from vaccinee plasma were capable of inhibiting the C1-specific conformational IgG mAb A32 mAb binding to the AE.A244 gp120 (Fig. 4; Table S1). An average of 7 µg/mL CH38 IgA2 equivalent concentrations of purified IgA from plasma was present per mg/mL of IgA from these 10 RV144 vaccinees (Table S1; range 1.1–18.2 µg/mL CH38 equivalent per milligram per milliliter IgA).
Fig. 4.
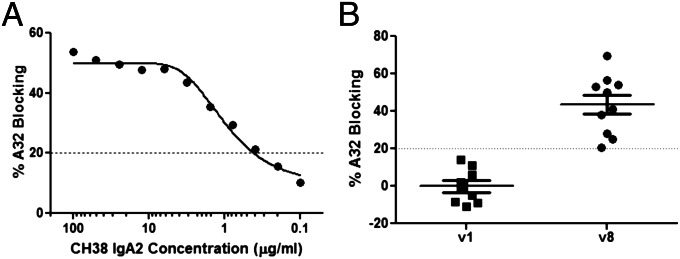
RV144-purified IgA inhibition of ADCC mediating A32 mAb binding to HIV-1 Env. Plasma IgA was purified by peptide M columns after IgG depletion. Purity of IgA was confirmed by HIV-1 binding antibody multiplex assays. Known concentration of purified IgA were tested for blocking biotinylated A32 IgG binding to HIV-1 Env. CH38 IgA2 was used as the positive control (A). Percentage blocking of IgA preparations (concentration range 0.5–1.3 mg/mL) at 1:2 dilution are shown (B).
Blocking of Plasma Anti-C1 mAb-Mediated ADCC by CH38 IgA2 mAb.
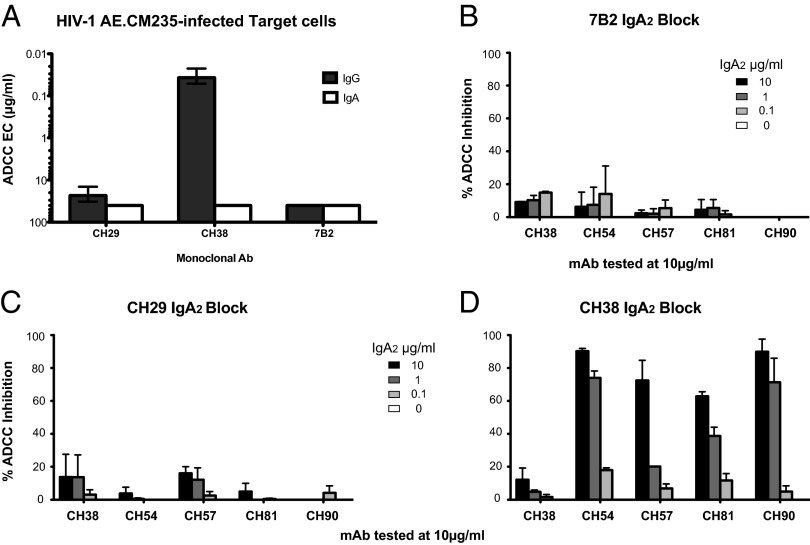
CH38 and CH29 mAbs expressed as IgG1 mediated ADCC against CM235-infected cells in the presence of NK effector cells at an endpoint concentration of 0.04 and 21.3 μg/mL, respectively (5) (Fig. 5A). These data are in agreement with the affinity data showing that CH38 mAb better recognize Env antigens expressed on the surface of HIV-1–infected cells than CH29 mAb. ADCC mediated by both mAbs expressed as IgG1s was blocked by the A32 Fab, indicating that the epitope recognized by the two mAbs was overlapping with that of the A32 C1 conformational epitope (5). The IgA2 versions of CH38 and CH29 were used to evaluate their ability to block ADCC mediated by other RV144 vaccinee IgG1 mAbs. Thus, we incubated HIV CM235-infected CD4+ target cells with CH38 or CH29 IgA2 mAbs and evaluated their ability to inhibit ADCC activity mediated by the RV144 mAbs CH54, CH57, CH81, and CH90. As a negative control, we tested 7B2 IgA mAb (with specificity to a gp41 epitope that is not present in the RV144 vaccine) and, as expected, this mAb did not significantly inhibit ADCC of any of the gp120 mAbs (Fig. 5B). Similar results were observed for CH29 IgA2 (Fig. 5C). In contrast, CH38 IgA2 mAb significantly inhibited ADCC mediated by RV144 CH54, CH57, CH81, and CH90 mAbs in a dose-dependent manner (Fig. 5D; 65–90% blocking). These data indicated that some but not all vaccine-induced IgA Env antibodies can effectively block IgG antibody-mediated ADCC effector function.
Fig. 5.
CH38 IgA2 does not mediate ADCC, but can inhibit IgG-mediated ADCC. (A) ADCC endpoint concentration (EC in micrograms per milliliter) of the IgG1 (black bars) and IgA2 (white bars) version of the CH29, CH38, and 7B2 mAb. The 7B2 mAb was used as control. HIV-1 AE.CM235-infected CEM.NKRCCR5 cells were used as target cells. (B–D) Blocking of ADCC by 7B2 IgA, CH29 IgA2, and CH38 IgA2. IgA mAbs were tested at four different concentrations to inhibit the ADCC mediated by 10 µg/mL of the IgG-mAbs CH38, CH54, CH57, CH81, and CH90. Each bar represents one of the four used IgA mAb concentrations. HIV-1 AE.CM235-infected target cells were used in the GranToxiLux assay. The 7B2 IgA2 was used as negative control. Each mAb combination was tested in duplicate, and each bar represents the average results from duplicate experiments ± SEM. The results are reported as a percentage of inhibition of the percentage of the granzyme activity observed in absence of preincubation with the CH38 IgA2 and CH29-IgA2 mAbs.
Blocking of Purified IgG-Mediated ADCC from RV144 Vaccinee Plasma by CH38 IgA2 and CH29 IgA2.
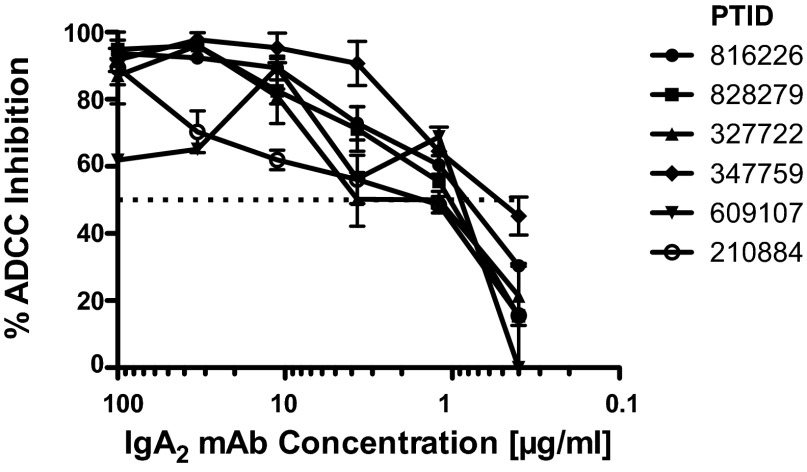
To determine whether the CH38 IgA2 and CH29 IgA2 mAbs could block ADCC mediated by plasma IgG, we purified IgG from the plasma of vaccine recipients with known ADCC activity. HIV-1 Env gp120-coated target cells were preincubated with serial concentrations of IgA mAbs, and then purified IgG from vaccinees was added at concentrations where peak ADCC was previously measured. The combination of CH38 IgA2 and CH29 IgA2 was capable of inhibiting 50% of the ADCC responses at ∼1 μg/mL (range 0.9–1.2 μg/mL; Fig. 6). These data demonstrate that purified IgA2 mAbs derived from RV144 vaccinee B cells can directly block binding of C1-specific conformational IgG antibodies that mediate ADCC.
Fig. 6.
CH38 plus CH29 IgA2 mAbs can inhibit ADCC mediated by purified plasma IgG from RV144 vaccinees. Purified IgG preparations were purified from the RV144 vaccine recipients (each line is one vaccinee). Each preparation was tested at the initial dilution of 1:250 at which the maximum ADCC activity was observed. The combination of CH29 and CH38 IgA2 mAbs was added starting at 100 μg/mL with a threefold serial dilution. The dotted line represents the 50% ADCC inhibition.
Discussion
We have shown that HIV-1 Env IgA antibodies elicited by the RV144 vaccine can interfere with binding and functional activity of vaccine-induced IgG responses. First, we demonstrate that the HIV-1 Env-specific IgA/IgG ratio directly correlated with infection risk, suggesting that for some HIV-1 Env specificities, the balance of Env IgA and Env IgG might influence vaccine efficacy. Second, we demonstrated that IgA mAbs isolated from RV144 vaccinees can both inhibit Env binding and block ADCC function of vaccine-induced IgGs that target the same epitope as the vaccine-induced IgAs. Thus, here we have identified a potential mechanism by which vaccine IgA might attenuate protective vaccine efficacy. The findings provide a rationale to test this concept in nonhuman primate passive protection studies with RV144 mAbs, and to include the measurement of Env IgA/IgG ratios, including C1 conformational-specific IgA/IgG ratios, in the evaluation of future HIV-1 vaccine studies.
The correlation of the IgA/IgG ratio with infection risk could be due to HIV-1–specific plasma IgA interfering with potentially protective IgG effector functions. Interference of IgG function by IgA antibodies has been reported for host immunity to bacteria (7, 8), for the regulation of autoantibodies (9), and for ADCC activity of EBV-infected target cells in the setting of nasopharyngeal cancer (10). However, this mechanism of action has not been previously reported for vaccine-elicited IgA antibodies. Here, we report, that a portion of the Env-specific IgA response in RV144 was capable of inhibiting ADCC activity of vaccine-elicited IgG.
In this study, two mAbs, CH38 and CH57, were generated from the same vaccine recipient. CH38 IgA2 mAb had significantly higher affinity for the vaccine strain Env protein than CH57 IgG1 mAb, and both bound to the C1 region of gp120. CH38 IgA2 also had higher affinity for HIV-1 Env than the corresponding CH38 IgG. Our finding that an IgA could have higher affinity for its epitope than an IgG of the same epitope specificity is consistent with previous reports. It was recently reported that 2F5 IgA (IgA2) mAb had higher affinity to its epitope than 2F5 IgG (11). Prior work by Pritsch et al. (12) also reported that an IgA isotype had ∼10-fold higher affinity than the IgG mAb (anti-tubulin mAb from serum of a lymphoma patient). These data indicate that antibody isotype can influence affinity and suggest that there may be differences both in the secondary structure of the antibody constant region CH1 domain and, potentially, interactions with VH (13) as well as in the length of the hinge region (14) that may influence antibody binding properties.
In contrast to IgG, human IgA can exist in multiple forms as monomeric, dimeric, and secretory IgA. Moreover, there are two different IgA subclasses (IgA1 and IgA2) that can be influenced by the nature of the antigen stimulation. IgA1 is predominant in the serum, whereas IgA2 is higher in mucosal secretions. Recombinant mAbs are generally made as an IgA2 due to the longer hinge region of IgA1 that makes IgA1 more susceptible to bacterial proteinases (reviewed in ref. 15). CH29 was originally an IgA1, and thus further studies are merited to determine whether HIV-1 vaccines can elicit differential IgA subclasses with particular specificities and function.
Plasma IgA is predominantly monomeric, whereas mucosal IgA is either predominantly dimeric or polymeric with secretory component (16). Effector functions of IgA are different from IgG due to the differences in antibody binding to FcRα vs. FcRγ receptors (3), and the lack of binding of IgA to C1q complement. Dimeric and monomeric IgA can mediate ADCC or phagocytosis by monocytes and PMNs, but in contrast to IgG cannot mediate ADCC via NK cells. Thus, other effector functions of vaccine-elicited IgA with antiviral activity (either through engaging different effector cells or through presence at mucosal sites) are plausible. Mucosal samples were not collected as part of the RV144 efficacy trial, so mucosal IgA levels and functions could not be evaluated as correlates of infection risk. Notably, we have found that plasma antibody responses do not predict mucosal responses, indicating a dichotomy between systemic and mucosal antibody responses (17, 18). In the VAX004 efficacy trial, Env-specific plasma IgA was detected (∼60% response rate), but there were no cervicovaginal or gingival IgA elicited (19). It is important to emphasize that our current study only addresses the ability of plasma (monomeric) IgA to block IgG NK-mediated effector function, and does not address the issue of vaccine-induced mucosal IgA or IgG (nor does it address effector functions mediated by cells other than NK cells). Follow-up studies using the RV144 ALVAC prime/AIDSVAX B/E boost vaccine regimen are in progress (RV305, RV306) to search for the presence of specific mucosal antibody responses with the RV144 vaccine regimen. Our data indicate that Env-specific monomeric IgA may block vaccine-induced IgG ADCC activity. Thus, it will be important to determine IgA/IgG ratios of antibodies induced to potentially protective epitopes in new vaccine trials. Understanding how plasma IgA antibodies are induced and how they may be modulated by vaccine immunogens or adjuvants is a new and important area of vaccine development research.
Materials and Methods
Plasma and Cellular Samples from Vaccine Recipients.
All trial participants gave written informed consent as described for both studies (1, 20). Samples were collected and tested according to protocols approved by institutional review boards at each site involved in these studies. Plasma samples were obtained from volunteers enrolled in the phase I/II clinical trial (20) and in the community-based, randomized, multicenter, double-blind, placebo-controlled phase III efficacy trial (1); both trials tested the prime–boost combination of vaccines containing ALVAC-HIV (vCP1521) (Sanofi Pasteur) and AIDSVAX B/E (Global Solutions for Infectious Diseases, South San Francisco, CA). Peripheral blood mononuclear cells (PBMCs) from two vaccine recipients enrolled in the phase II (T141449) and phase III (347759) trials whose plasma showed ADCC activity were used for isolation of memory B cells and mAbs. Both subjects had negative serology for HIV-1 infection at the time of collection.
Statistical Analyses.
In a follow-up analysis of the IgA responses in the RV144 case-control study, we measured the HIV-1–specific IgA/IgG ratio to determine if this calculation improved the prediction for risk of infection. To combine the ratio of IgA/IgG binding to a panel of HIV-1 Env proteins representing clades A, AE, B, C, and G into a single measure, we considered two different weighting schemes. Because the focus here is more on IgA binding, we chose the first set of weights based on the magnitude of IgA binding to the panel, and it was designed to down-weight antigens that are overrepresented and up-weight antigens that are underrepresented to achieve uniform representation of the epitope diversity. Details for weight construction algorithm can be found in ref. 2. We also considered a simple equally weighted combination of ratios for the purpose of sensitivity analysis. To create a binary variable for a discrete representation of IgA/IgG ratio, we chose to dichotomize at 0.01 given the preprocessing of the IgA and IgG readouts based on positive response calls (2). The statistical analyses of correlates of infection risk were performed as described in ref. 2, which were based on multiple logistic regression models with inverse probability weighting.
Isolation of ADCC-Mediating Monoclonal Antibodies.
Monoclonal antibodies were isolated either from memory B cells that bound to HIV-1 group M consensus gp140 (ConS gp140) Env sorted by flow cytometry (21) or from IgG+ memory B cells cultured at near clonal dilution for 14 d (22) followed by sequential screenings of culture supernatants for HIV-1 gp120 Env binding and ADCC activity. Vaccinee T141449 was tested using antigen-specific memory B-cell sorting as previously described (21) with the following modifications. Group M consensus gp140 (ConS gp140) Env labeled with Pacific Blue and Alexa Fluor 647 (Invitrogen) was used for sorting. Memory B cells were gated as Aqua Vital Dye−, CD3−, CD14−, CD16−, CD235a−, CD19+, and surface IgD−; memory B cells stained with gp140 (ConS gp140) in both colors were sorted as single cells as described (22). A total of 54,621 memory B cells from vaccinee T141449 were screened using this method. IgG+ memory B cells from vaccine 347759 were screened using two methods as previously described (5).
Production of Recombinant Anti–HIV-1 IgA Antibodies.
The variable regions of Ig heavy- and light-chain (VH and VL) genes of CH29 and CH38 Ab were isolated from clonal culture of memory B cells (5) by RT-PCR (23). For production of full-length recombinant CH29 and CH38 antibodies, the respective VH and VL genes were cloned into a pcDNA 3.1 expression vector expressing either human IgA2 constant region, generated by de novo synthesis based on the sequence from GenBank (accession no. BC073765) (24) or from the lambda light chain constant region gene (25). Recombinant CH29 and CH38 IgA2 antibodies were produced in 293F cells by cotransfection with CH29 and CH38 IgA2 heavy-chain and light-chain gene plasmids and purified by affinity chromatography using peptide M (InvivoGen). Recombinant 7B2 IgA was similarly produced and used as a negative control.
IgA Purification.
IgA purification from plasma was performed using peptide M/agarose resin (InvivoGen) following the manufacturer’s instructions with minor modifications. Briefly, purification columns were prepared by loading 400-µL peptide M agarose into 2-mL spin columns (Pierce/Thermo Scientific) and washing with PBS. Plasma samples were diluted 1:2 with PBS and loaded to prepared columns. After incubating at room temperature for 45 min, the flow-through fractions were collected and loaded onto fresh purification columns for an additional overnight incubation at 4 °C. Both purification columns were then washed with PBS, and IgAs were eluted off the columns using IgG elution buffer (Pierce/Thermo Scientific) and immediately neutralized with 1 M Tris (pH 8.8). The eluted fractions were subsequently buffer-exchanged against PBS using Amicon Ultra-15 filters with a 50-K cutoff (Millipore/Fisher Scientific), and concentrated. The IgA preps were then removed of contaminating IgG by incubating in Protein G Spin Columns (GE Healthcare). All IgA preps were tested in IgG binding antibody multiplex assays (26) to confirm the absence of IgG contamination before any subsequent assays/applications.
ELISA Antibody Blocking Assays.
The 384-well ELISA plates (Costar no. 3700) were coated with 30 ng per well A244Δ11 gp120 overnight at 4 °C and blocked with assay diluent [PBS containing 4% (wt/vol) whey protein/15% normal goat serum/0.5% Tween 20/0.05% sodium azide] for 1 h at room temp. All assay steps were conducted in assay diluent (except substrate step) and incubated for 1 h at room temperature followed by washing with PBS/0.1% Tween 20. Purified IgA from RV144 vaccinees were tested for blocking of biotinylated target mAb added at the EC50 (determined by a direct binding of biotinylated-mAb to JRFL). Biotin-mAb binding was detected with streptavidin alkaline phosphatase at 1:1,000 (Promega V5591) followed by substrate [carbonate bicarbonate buffer (CBC) buffer + 2 mM MgCl2 + 1 mg/mL 4-nitrophenyl phosphate di(2-amino-2-ethyl-1,3-propanediol) salt]. Plates were read with a plate reader at 405 nm at 45 min. Duplicate wells were background subtracted and averaged. Percent blocking was calculated as follows: 100 − (purified IgA triplicate mean/no inhibition control mean) × 100.
Surface Plasmon Resonance Kinetics and Kd Measurements.
Env gp120 binding Kd and rate constant for IgG mAbs were calculated on BIAcore 3000 instruments using an anti-human Ig Fc capture assay as described previously (27–29). Anti–respiratory syncytial virus mAb Synagis (Palivizumab) was captured on the same sensor chip as a control surface. Nonspecific binding of Env gp120 to the control surface and/or blank buffer flow was subtracted for each mAb–gp120 binding interactions. IgA antibodies were directly coupled via amine coupling chemistry to the sensor surfaces, and Env gp120 was flowed and data collected as above. All curve-fitting analyses were performed using global fit of multiple titrations to the 1:1 Langmuir model. Mean and SD of rate constants and Kd were calculated from at least three measurements on individual sensor surfaces with equivalent amounts of captured antibody. All data analysis was performed using BIAevaluation 4.1 analysis software (GE Healthcare).
Surface Plasmon Resonance Antibody Blocking Assay.
Surface plasmon resonance (SPR) antibody blocking was measured on BIAcore 3000 instruments by immobilizing the test mAb (IgG or IgA) on a CM5 sensor chip to ∼5,000–6,000 resonance units using standard amine coupling chemistry. ADCC-mediating blocking antibodies were preincubated with Env gp120 in solution at a molar ratio of Env to mAb at 1:3. The 1:3 molar ratio used gave binding to saturation level, which was predetermined by testing binding of the Env–mAb mixture to a blocking antibody immobilized on the chip. Following each binding cycle, surfaces were regenerated with a short injection (10–15 s) of either glycine⋅HCl (pH 2.0) or 100 mM phosphoric acid. Blocking percentages were calculated from the ratio of binding response as follows: [percent blocking = 1 – (response with gp120 + blocking mAb/response with gp120 + control mAb Synagis) × 100].
Competitive Blocking of Env gp120 Binding of IgG by IgA.
The RV144 PBMC-derived CH54 mAb was incubated at a fixed saturating concentration (100 μg/mL) with 20 μg of Env gp120 (A244 D11 gp120), and varying concentrations of CH38 IgA2 monomers (0–100 μg/mL) were added to the Env–IgG mixture, which was then incubated at either 4 °C overnight or at room temperature for ∼4 h. The percent blocking of IgG binding was calculated by measuring the concentration of unbound IgG in the antibody–antigen mixture with IgA, and following its capture and calibration on an anti-Fc IgG immobilized surface by SPR measurements. The molar ratio of IgA:IgG blocking was finally calculated from the above measurements.
Virus, Infectious Molecular Clones for ADCC GranToxiLux Assay.
The HIV-1 reporter virus used was replication-competent infectious molecular clones (IMC) designed to encode the CM235 (subtype A/E) Env genes in cis within an isogenic backbone that also expresses the Renilla luciferase reporter gene and preserves all viral ORFs (30). The Env-IMC-LucR viruses were subtype A/E NL-LucR.T2A-AE.CM235-ecto (IMCCM235; GenBank accession no. AF2699954; plasmid provided by J.H.K.). Reporter virus stocks were generated by transfection of 293T cells with proviral IMC plasmid DNA and titrated on TZM-bl cells for quality control.
ADCC GranToxiLux Assay.
ADCC activity was detected according to our previously described ADCC-GranToxiLux (GTL) procedure (31). CM235-infected CEM.NKRCCR5 (32) were used as target cells. Purified NK effector cells were obtained from PBMCs collected from an HIV-1 seronegative donor with the F/F phenotype of Fcγ-receptor 3A and used at the effector to target (E:T) ratio of 10:1. The mAb A32 (James Robinson, Tulane University, New Orleans), Palivizumab Synagis (MedImmune, LLC; negative control), and vaccine-induced mAbs were tested as 6 fourfold serial dilutions with a starting concentration of 40 µg/mL (range 40–0.039 µg/mL). The results are expressed as the endpoint concentration (EC) in μg/mL as previously described (5).
Inhibition of ADCC Activity by the IgA mAbs.
To demonstrate the blocking activity of the CH38 IgA2 and CH29 IgA2 mAbs on the IgG mAbs, we incubated (15 min) the IgA mAbs, as previously described (defined as primary mAb, at concentrations of 10, 1, 0.1, and 0 μg/mL with the target cells). We subsequently added IgG mAbs at 10 μg/mL and incubated with CM235-infected CEM.NKR.CCR5 target cells as described above. CH38 IgA2 and CH29 IgA2 were used together at 100, 33.3, 11.1, 3.7, 1.2, and 0.4 μg/mL to block ADCC mediated by the purified plasma IgG. Purified IgG was prepared as previously described (2). Each IgG preparation was tested against the A244Δ11 gp120-coated CEM.NKRCCR5 target cells at three concentrations representing the range of peak activity for each sample. The IgA version of the 7B2 mAb was used as negative control. The results are reported as percentage of ADCC inhibition, calculated as percentage of ADCC activity reduction based on the ADCC mediated by the IgG mAb or IgG preparation in absence of the IgA2 mAbs.
Supplementary Material
Acknowledgments
We are indebted to the volunteers and clinical staff who participated in the RV144 vaccine trial. We thank Drs. Merlin Robb, Robert O’Connell, Kelly A. Soderberg, and Charla Andrews for clinical trial and/or project management, Dr. Nicole L. Yates and R. Glenn Overman for technical expertise, and Dr. Sarzotti-Kelsoe for quality assurance oversight. This work was supported by the Center for HIV/AIDS Vaccine Immunology (CHAVI)–Immunogen Discovery Grant U01 AI067854; National Institutes of Health, National Institute of Allergy and Infectious Diseases Division of Acquired Immunodeficiency Syndrome (NIH/NIAID/DAIDS); NIH NIAID Grant AI07392; Bill and Melinda Gates Foundation Grants 1033098 and 1040758; Collaboration for AIDS Vaccine Discovery–Vaccine Immune Monitoring Consortium (CAVD-VIMC) Grant 3830913, CAVD-VIMC Grant CTVIMC OPP1032325, HIV-1 Vaccine Trials Network 5U01 AI46725-05; and the Duke University Center for AIDS Research Grant P30 AI64518. Funding was also provided by Interagency Agreement Y1-AI-2642-12 between the US Army Medical Research and Material Command and the NIAID through cooperative agreement W81XWH-07-2-0067 between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the US Department of Defense.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301456110/-/DCSupplemental.
References
- 1.Rerks-Ngarm S, et al. MOPH-TAVEG Investigators Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361(23):2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 2.Haynes BF, et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med. 2012;366(14):1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monteiro RC, Van De Winkel JG. IgA Fc receptors. Annu Rev Immunol. 2003;21:177–204. doi: 10.1146/annurev.immunol.21.120601.141011. [DOI] [PubMed] [Google Scholar]
- 4.Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275–290. doi: 10.1146/annurev.immunol.19.1.275. [DOI] [PubMed] [Google Scholar]
- 5.Bonsignori M, et al. Antibody-dependent cellular cytotoxicity-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J Virol. 2012;86(21):11521–11532. doi: 10.1128/JVI.01023-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferrari G, et al. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J Virol. 2011;85(14):7029–7036. doi: 10.1128/JVI.00171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilton JM. Suppression by IgA of IgG-mediated phagocytosis by human polymorphonuclear leucocytes. Clin Exp Immunol. 1978;34(3):423–428. [PMC free article] [PubMed] [Google Scholar]
- 8.Griffiss JM, Goroff DK. IgA blocks IgM and IgG-initiated immune lysis by separate molecular mechanisms. J Immunol. 1983;130(6):2882–2885. [PubMed] [Google Scholar]
- 9.Quan CP, Watanabe S, Forestier F, Bouvet JP. Human amniotic IgA inhibits natural IgG autoantibodies of maternal or unrelated origin. Eur J Immunol. 1998;28(12):4001–4009. doi: 10.1002/(SICI)1521-4141(199812)28:12<4001::AID-IMMU4001>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Mathew GD, Qualtiere LF, Neel HB, III, Pearson GR. IgA antibody, antibody-dependent cellular cytotoxicity and prognosis in patients with nasopharyngeal carcinoma. Int J Cancer. 1981;27(2):175–180. doi: 10.1002/ijc.2910270208. [DOI] [PubMed] [Google Scholar]
- 11.Tudor D, et al. Isotype modulates epitope specificity, affinity, and antiviral activities of anti-HIV-1 human broadly neutralizing 2F5 antibody. Proc Natl Acad Sci USA. 2012;109(31):12680–12685. doi: 10.1073/pnas.1200024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pritsch O, et al. Can isotype switch modulate antigen-binding affinity and influence clonal selection? Eur J Immunol. 2000;30(12):3387–3395. doi: 10.1002/1521-4141(2000012)30:12<3387::AID-IMMU3387>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 13.Janda A, Casadevall A. Circular dichroism reveals evidence of coupling between immunoglobulin constant and variable region secondary structure. Mol Immunol. 2010;47(7-8):1421–1425. doi: 10.1016/j.molimm.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furtado PB, et al. Solution structure determination of monomeric human IgA2 by X-ray and neutron scattering, analytical ultracentrifugation and constrained modelling: A comparison with monomeric human IgA1. J Mol Biol. 2004;338(5):921–941. doi: 10.1016/j.jmb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Kerr MA. The structure and function of human IgA. Biochem J. 1990;271(2):285–296. doi: 10.1042/bj2710285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woof JM, Mestecky J. Mucosal immunoglobulins. Immunol Rev. 2005;206:64–82. doi: 10.1111/j.0105-2896.2005.00290.x. [DOI] [PubMed] [Google Scholar]
- 17.Fouda GG, et al. Center for HIV/AIDS Vaccine Immunology HIV-specific functional antibody responses in breast milk mirror those in plasma and are primarily mediated by IgG antibodies. J Virol. 2011;85(18):9555–9567. doi: 10.1128/JVI.05174-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yates NL, et al. Mucosal Immunol. 2013. HIV-1 gp41 envelope IgA is frequently elicited after transmission but has an initial short half-life. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider JA, et al. Mucosal HIV-binding antibody and neutralizing activity in high-risk HIV-uninfected female participants in a trial of HIV-vaccine efficacy. J Infect Dis. 2007;196(11):1637–1644. doi: 10.1086/522232. [DOI] [PubMed] [Google Scholar]
- 20.Nitayaphan S, et al. Thai AIDS Vaccine Evaluation Group Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J Infect Dis. 2004;190(4):702–706. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 21.Gray ES, et al. Isolation of a monoclonal antibody that targets the alpha-2 helix of gp120 and represents the initial autologous neutralizing-antibody response in an HIV-1 subtype C-infected individual. J Virol. 2011;85(15):7719–7729. doi: 10.1128/JVI.00563-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonsignori M, et al. Analysis of a clonal lineage of HIV-1 envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J Virol. 2011;85(19):9998–10009. doi: 10.1128/JVI.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao HX, et al. Initial antibodies binding to HIV-1 gp41 in acutely infected subjects are polyreactive and highly mutated. J Exp Med. 2011;208(11):2237–2249. doi: 10.1084/jem.20110363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strausberg RL, et al. Mammalian Gene Collection Program Team Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99(26):16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao HX, et al. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J Virol Methods. 2009;158(1-2):171–179. doi: 10.1016/j.jviromet.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tomaras GD, et al. Initial B-cell responses to transmitted human immunodeficiency virus type 1: Virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J Virol. 2008;82(24):12449–12463. doi: 10.1128/JVI.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alam SM, et al. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J Immunol. 2007;178(7):4424–4435. doi: 10.4049/jimmunol.178.7.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam SM, et al. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: Antibody binding kinetics, induction, and potential for regulation in acute infection. J Virol. 2008;82(1):115–125. doi: 10.1128/JVI.00927-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alam SM, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci USA. 2009;106(48):20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edmonds TG, et al. Replication competent molecular clones of HIV-1 expressing Renilla luciferase facilitate the analysis of antibody inhibition in PBMC. Virology. 2010;408(1):1–13. doi: 10.1016/j.virol.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pollara J, et al. High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A. 2011;79(8):603–612. doi: 10.1002/cyto.a.21084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J Virol. 1999;73(11):8966–8974. doi: 10.1128/jvi.73.11.8966-8974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.