Abstract
The mitochondrial inner membrane organizing system (MINOS) is a conserved large hetero-oligomeric protein complex in the mitochondrial inner membrane, crucial for the maintenance of cristae morphology. MINOS has been suggested to represent the core of an extended protein network that controls mitochondrial function and structure, and has been linked to several human diseases. The spatial arrangement of MINOS within mitochondria is ill-defined, however. Using super-resolution stimulated emission depletion (STED) microscopy and immunogold electron microscopy, we determined the distribution of three known human MINOS subunits (mitofilin, MINOS1, and CHCHD3) in mammalian cells. Super-resolution microscopy revealed that all three subunits form similar clusters within mitochondria, and that MINOS is more abundant in mitochondria around the nucleus than in peripheral mitochondria. At the submitochondrial level, mitofilin, a core MINOS subunit, is preferentially localized at cristae junctions. In primary human fibroblasts, mitofilin labeling uncovered a regularly spaced pattern of clusters arranged in parallel to the cell growth surfaces. We suggest that this array of MINOS complexes might explain the observed phenomenon of largely horizontally arranged cristae junctions that connect the inner boundary membrane to lamellar cristae. The super-resolution images demonstrate an unexpectedly high level of regularity in the nanoscale distribution of the MINOS complex in human mitochondria, supporting an integrating role of MINOS in the structural organization of the organelle.
Keywords: MitOS, MICOS, membrane architecture, nanoscopy
Mitochondria are highly complex and dynamic organelles. Their shapes range from small oval fragments to interconnected networks of tubules that are constantly moving, fusing, and dividing (1). The innermost mitochondrial aqueous compartment, the matrix, is bounded by the highly convoluted inner membrane, which in turn is surrounded by the outer membrane. The inner membrane projects cristae into the matrix. Depending on the cell type and physiological conditions, the cristae can adopt a wide variety of shapes, ranging from simple tubular to lamellar (2, 3). The cristae membranes are connected by cristae junctions to the part of the inner membrane that parallels the outer membrane, the inner boundary membrane. Cristae junctions are relatively uniform tubular structures, typically 20–50 nm in diameter, that are important for maintaining the inner mitochondrial architecture (4). The inner boundary and the cristae membranes have distinct but overlapping protein compositions, which may be adapted to the cellular conditions (5–9).
Although the overall architecture of mitochondria has been described in detail, relatively little is known about the molecular components that determine the mitochondrial structure (10). Recently, several independent studies led to the identification of a large protein complex that is required for the establishment of inner membrane architecture (for review, see 11–13). This complex has been termed mitochondrial inner membrane organizing system (MINOS) (14, 15) or, alternatively, mitochondrial organizing structure (MitOS) (16) or mitochondrial contact site (MICOS) (17).
In the budding yeast Saccharomyces cerevisiae, MINOS is composed of at least six subunits: Fcj1, Mio10/Mos1/Mcs10, Aim13/Mcs19, Aim5/Mcs12, Aim37/Mcs27, and Mio27/Mos2/Mcs29. All of these subunits of MINOS are either integral or peripheral proteins of the inner membrane and expose their bulk domains to the intermembrane space. MINOS has been conserved in evolution; in humans, mitofilin/MINOS2 (Fcj1 in yeast), MINOS1 (Mio10 in yeast) and CHCHD3/MINOS3 (Aim13 in yeast) have been shown to be subunits of a large (>1 MDa) complex (15).
MINOS is crucial for the maintenance of the typical inner membrane morphology, and mutations affecting this protein complex induce a loss of cristae junctions, as well as detachment of cristae membranes from the inner boundary membrane. MINOS is involved in a multitude of physical and genetic interactions (11, 12). It has been shown to interact directly with several proteins of the outer membrane, including the translocase of the outer membrane (TOM) complex, the sorting and assembly machinery (SAM) complex (TOB), porin (VDAC), and the yeast fusion component Ugo1 (14-22). Moreover, MINOS is involved in the inheritance of mitochondrial DNA (23) and has been shown to transiently interact with the intermembrane space import receptor Mia40 (14). The genes of the MINOS complex show strong genetic interactions with ER-Mitochondria Encounter Structures (ERMES) and phospholipid biosynthesis genes (16). Given the multitude of processes in which MINOS is involved, it is perhaps not surprising that alterations of MINOS subunits are associated with a number of human diseases, including neurologic disorders and cardiomyopathy (12).
Despite its remarkable size (>1 MDa) (15), very little is known about the submitochondrial distribution of MINOS in human cells. In budding yeast, immunogold electron microscopy revealed that the MINOS subunits are enriched at cristae junctions (17, 24). Light microscopy using GFP-tagged MINOS seemed to suggest that MINOS acts as a skeletal structure, circumventing the mitochondrial inner membrane (16).
A major challenge in obtaining a comprehensive view on the localization of MINOS in human cells is that mitochondria have a diameter close to the resolution limit of light microscopy, largely precluding the use of conventional diffraction-limited microscopy to study submitochondrial protein distributions. Furthermore, cultured human cells are large and have a polarity toward the growth surface, which may affect the distribution of MINOS. In the present study, we investigated the distribution of MINOS in primary adult human fibroblasts using stimulated emission depletion (STED) super-resolution light microscopy, quantitative immunoelectron microscopy, and electron tomography. We found that MINOS is preferentially localized at cristae junctions, and that MINOS clusters frequently exhibit an ordered inner mitochondrial distribution that depends on the position of the mitochondrion within the cell and on the orientation of the growth surface.
Results
STED Super-resolution Microscopy Reveals Mitofilin Localized in Individual Clusters.
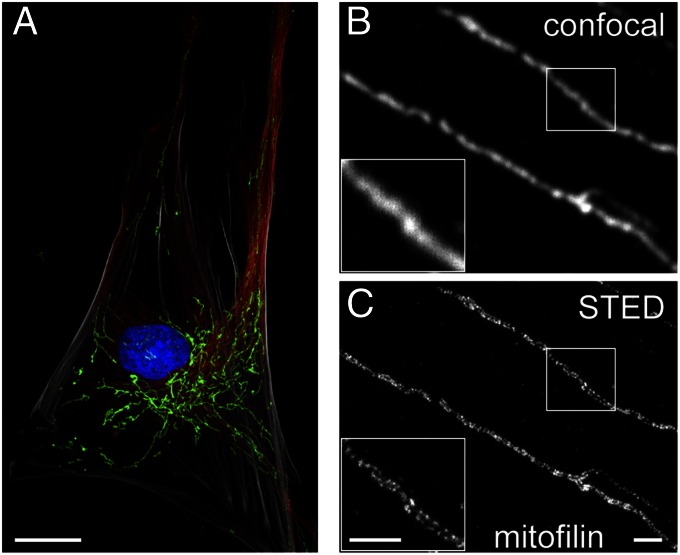
To visualize the distribution of mitofilin in primary adult human skin fibroblasts, we decorated chemically fixed cells with a polyclonal antiserum generated against full-length human mitofilin. This antiserum highlighted the entire mitochondrial network, demonstrating that in these primary fibroblasts, the mitochondria are primarily elongated tubular structures distributed throughout the entire cell (Fig. 1A). As generally observed in fibroblast cell lines (25), the mitochondrial network is denser in the perinuclear region than in the cell periphery. Diffraction-limited confocal microscopy suggested that mitofilin is largely homogenously distributed throughout the mitochondrial tubules (Fig. 1B). But mitochondria are small organelles, with diameters close to the resolution attainable with conventional optical microscopy (∼200 nm in the focal plane). To obtain insight into the nanoscale distribution of mitofilin in mitochondria of intact cells, we used STED super-resolution microscopy, which provides an optical resolution well below the diffraction limit (26). In STED images, mitofilin clearly was not homogenously distributed along the mitochondria, but rather was concentrated into individual clusters (Fig. 1C). Owing to the size of the antibodies used, any structure was presumably enlarged by ∼35 nm (27, 28), which is similar to the resolution of the STED microscope that we used (40–50 nm in the focal plane) (29). The antibody-decorated imaged clusters were substantially larger (∼85 nm); thus, we conclude that each cluster consists of multiple mitofilin proteins.
Fig. 1.
Mitofilin is localized in individual clusters in the mitochondria of primary adult human fibroblasts. (A) Cell overview. The mitochondrial network of a primary fibroblast is labeled with an antiserum against mitofilin (green). The microtubule cytoskeleton (red), the nucleus (blue) and the actin cytoskeleton (gray) are labeled as well. (B and C) STED super-resolution microscopy (C) reveals that mitofilin is localized in discrete clusters, which are blurred and not resolvable using diffraction-limited confocal microscopy (B) of the same region. (Insets) Magnification of the boxed areas. (Scale bars: 20 μm in A; 1 μm in B and C.)
Submitochondrial Distribution of Mitofilin Clusters Is Highly Ordered.
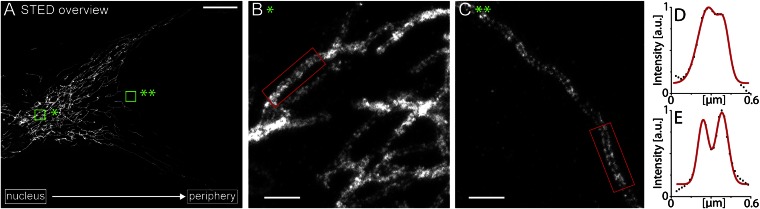
The mitofilin clusters were more numerous in the mitochondria close to the nucleus than in the mitochondria in the periphery (Fig. 2A–C and Fig. S1). In the perinuclear mitochondria, the clusters were often so densely packed that they were no longer unequivocally resolvable, whereas in the peripheral mitochondria, rather well-ordered arrangements with similar spacing between individual mitofilin clusters were frequently seen (Fig. 2 B and C).
Fig. 2.
Mitofilin cluster distribution is denser in the perinuclear mitochondria. (A) Overview image of a fibroblast labeled with an antiserum against mitofilin and imaged by STED microscopy. (B and C) Magnification of the boxed areas in A. (D and E) Averaged intensity profiles across the indicated mitochondrial tubule sections within the respective boxed areas in B and C. (Scale bars: 20 μm in A; 1 μm in B and C.)
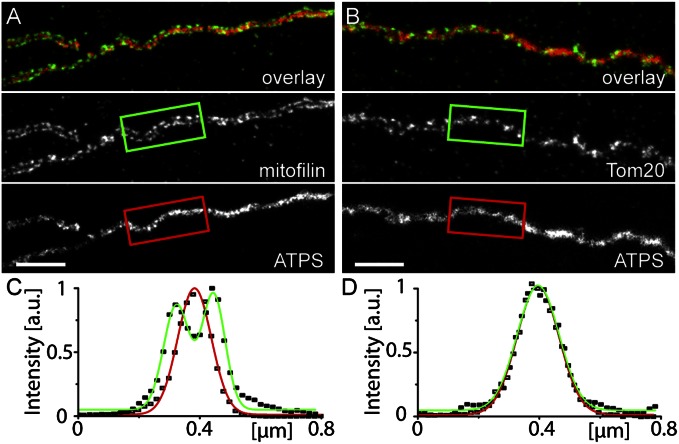
Intriguingly, the STED images revealed that in the peripheral mitochondria, the mitofilin clusters were often arranged at the sides of the mitochondrial tubules (Fig. 2 and Fig. S2). We dubbed this intriguing arrangement of the MINOS clusters a “discontinuous rail-like distribution”. This distribution was unexpected for two reasons. First, if the integral inner membrane protein mitofilin were evenly distributed over the highly convoluted inner membrane, then the proteins would be expected to be imaged also in the interior of the organelle. To experimentally test the assumption that mitofilin is more abundant at the rim (within or close to the inner boundary membrane) of the mitochondria, we also labeled the cells with antibodies against the beta subunit of the mitochondrial F1F0 ATPase (ATPS), which is enriched in the cristae membrane (7, 8). Dual-color STED imaging showed ATPS localized in the interior of the mitochondria, with mitofilin at the sides (Fig. 3 A and C). It can be concluded that the mitofilin complexes are preferentially localized at the rim of the mitochondria and are less abundant in the cristae membranes. Second, an enrichment of mitofilin at the rim of the mitochondria is necessary, but not sufficient, to explain the discontinuous rail-like distribution of the mitofilin clusters at the sides of the mitochondria, considering that the STED microscope used in these experiments had a high resolution in the optical plane but a diffraction-limited confocal resolution along the optical axis. The diameter of a mitochondrial tubule is small enough to fit into the rather long z-axis of the STED focus. Thus, the fluorescence signals from the upper and lower membranes are summed in the final image. As a result, clusters built by an outer membrane protein (e.g., Tom20), which are evenly distributed in the outer membrane are seen in the STED images not enriched at the mitochondrial rim (Fig. 3 B and D). Therefore, the observed discontinuous rail-like distribution of the mitofilin clusters is likely explained by a highly polarized (i.e., horizontal) distribution of rather constantly spaced MINOS clusters within or close to the inner boundary membrane.
Fig. 3.
Mitofilin clusters are localized at the horizontal sides of mitochondria. (A and B) Two-color STED microscopy of fibroblast mitochondria labeled with antisera against mitofilin and the beta-subunit of the mitochondrial F1Fo-ATPase (ATPS) (A) or with antisera against the outer membrane protein Tom20 and ATPS (B). In the overlay images, ATPS is shown in red, and mitofilin and Tom20 are in green. (C and D) Averaged intensity profiles across the indicated mitochondrial tubule sections within the boxed areas in A and B. The averaged ATPS intensity profile is shown in red, and the profiles of mitofilin and Tom20 are in green. (Scale bars: 1 µm.)
We also note that the regular arrangements were not always discernible in the peripheral mitochondria, possibly related to rotation of the mitochondrial tubule along its respective length axis. Likewise, the regularity was often not discernible at mitochondrial tips, possibly indicating a disturbance of regularity by fission and fusion events. The regularity was less obvious in the perinuclear mitochondria, possibly owing to the more abundant mitofilin clusters in these mitochondria, but also related to the fact that around the nucleus, numerous tubules were not strictly coaligned with the glass surface. Both factors may conceal an existing ordered arrangement of the clusters.
We propose that, at least in the peripheral mitochondria, the rather constantly spaced mitofilin clusters are enriched at or close to the inner boundary membrane, and the mitofilin clusters are frequently distributed parallel to the growth surface (the glass coverslip), resulting in the peculiar highly ordered discontinuous rail-like arrangement of the clusters revealed by STED microscopy.
Mitofilin, MINOS1, and CHCHD3 Show Similar Punctuate Localizations.
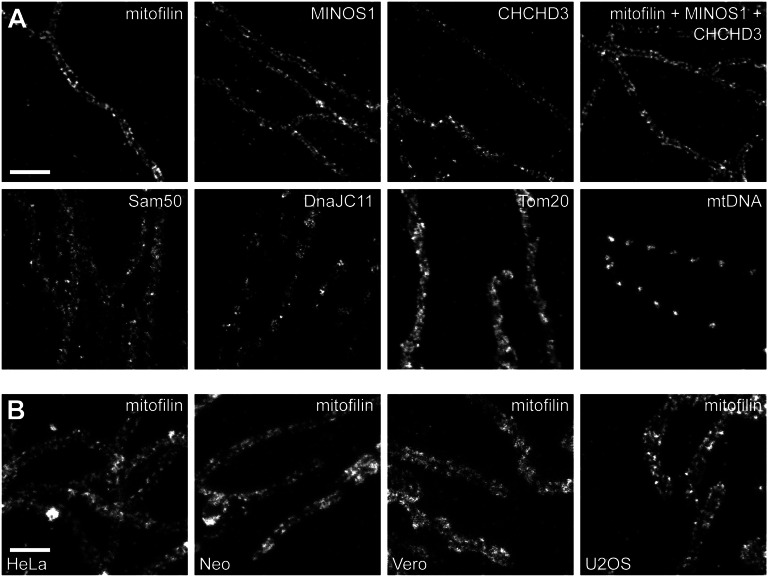
Among the six components of the budding yeast MINOS complex, three human homologs have been identified: mitofilin, MINOS1, and CHCHD3 (15), although the human complex may comprise further components, including CHCHD6/CHCM1 (18, 30). Mitofilin (Fcj1 in yeast) and MINOS1 (Mio10 in yeast) are considered to form the core MINOS complex to which the other proteins are attached (14, 15). To determine whether MINOS1 and CHCHD3 have the same distribution as mitofilin within mitochondria, we immunolabeled primary adult human fibroblast cells with polyclonal antisera against MINOS1 and CHCHD3 and imaged them with STED microscopy (Fig. 4A). We did not analyze the distribution of CHCHD6 because we lacked an appropriate antibody. Intriguingly, MINOS1 and CHCHD3 showed similar distribution patterns as mitofilin; this is, peripheral mitochondria frequently exhibited a regular pattern of clusters localized at the rim of the organelle. Contrary to some proposals (16), the high-resolution STED images provided no indication that any of these three proteins alone formed filamentous structures in human mitochondria. Dual-color STED microscopy revealed a high level of colocalization of mitofilin with CHCHD3 or MINOS1 (Fig. S3).
Fig. 4.
STED microscopy of MINOS. (A) Distribution of MINOS components in mitochondria of primary adult human fibroblasts. The cells were labeled with antibodies against the indicated proteins. (B) Localization of mitofilin in different mammalian cell lines [HeLa cells, neonatal human fibroblasts (Neo), Vero cells, and U2OS cells]. In each case, representative STED images of mitochondria located in the cell periphery are shown. (Scale bars: 1 µm.)
To test whether mitofilin, MINOS1, and CHCHD3 together form a larger, possibly filamentous hetero-oligomeric structure, we immunolabeled primary fibroblasts by blending the antisera against mitofilin, MINOS1, and CHCHD3 to label these three proteins simultaneously (Fig. 4A). Similar to our findings when using the antibodies separately, the peripheral mitochondria exhibited ordered arrays of immunolabeled clusters, but no indications of a larger superstructure formed by these proteins, as, for example, an arch, ring, or helical structure circling the matrix. The clusters labeled with antiserum against mitofilin were of a similar size (∼85 nm) as the clusters labeled with the three different antibodies together. We conclude that the core MINOS complex generally does not form a spatially extended superstructure in human fibroblasts.
The MINOS complex reportedly interacts with several other protein machineries. Human mitofilin interacts with Sam50, an essential component of the assembly machinery of the outer membrane (SAM complex) (15, 18–20, 31). Budding yeast Fcj1 (mitofilin) has been shown to interact with the TOM complex (14, 21, 31). In addition, the chaperone DnaJC11 interacts with mitofilin (18) and has been proposed to be part of the human MINOS complex (15).
To address the question of whether these MINOS-interacting components have a similar mitochondrial distribution as the core MINOS complex proteins, we immunolabeled human primary fibroblast cells with antisera against Sam50, DnaJC11, or Tom20, a peripheral receptor of the TOM complex (Fig. 4A). In accordance with previous reports (29), Tom20 was distributed in dense clusters all over the mitochondrial surface. The Sam50 clusters appeared to be less abundant than Tom20 clusters but were also evenly distributed over the mitochondria, whereas the distribution of DnaJC11 was rather patchy along the mitochondrial tubules. Notably, we did not find an ordered arrangement of these proteins at the rim of the mitochondria as was seen for mitofilin, MINOS1, and CHCHD3. In comparison, cells labeled with an antiserum against DNA showed the typical spaced distribution of the nucleoids along the length of the mitochondrial tubules (32).
We next addressed the question of whether the observed regular distribution of the core MINOS complex is specific to the primary skin fibroblast cells used or can be generally observed in cultured cells. To answer this question, we labeled HeLa cells (an immortal cell line derived from cervical cancer cells), neonatal human fibroblasts (Neo; derived from foreskin cells), Vero cells (a kidney epithelial cell line extracted from an African green monkey), and U2OS cells (a human osteosarcoma cell line) with antisera against mitofilin (Fig. 4B). We also observed a consistent discontinuous rail-like distribution of the mitofilin clusters in the peripheral mitochondria of HeLa, Neo, and Vero cells, whereas we found this peculiar distribution in only a subset of mitochondria in U2OS cells. Thus, such rail-like distributions of MINOS appear to be a frequent phenomenon in cultured adherent cells, although its distinctness may vary.
We conclude that the core MINOS subunits (mitofilin and MINOS1) and CHCHD3 form clusters that exhibit a regular discontinuous rail-like distribution in the mammalian cell lines tested. The mitochondrial distributions of Sam50, DnaJC11, and Tom20 as revealed by STED super-resolution microscopy are in disagreement with the assumption of a complete colocalization of any of these proteins with the core MINOS complex. We propose that only a subset of the Sam50, DnaJC11, and Tom20 pools are engaged in a physical interaction with the core MINOS complex at any one time.
Mitofilin Is Enriched at Cristae Junctions.
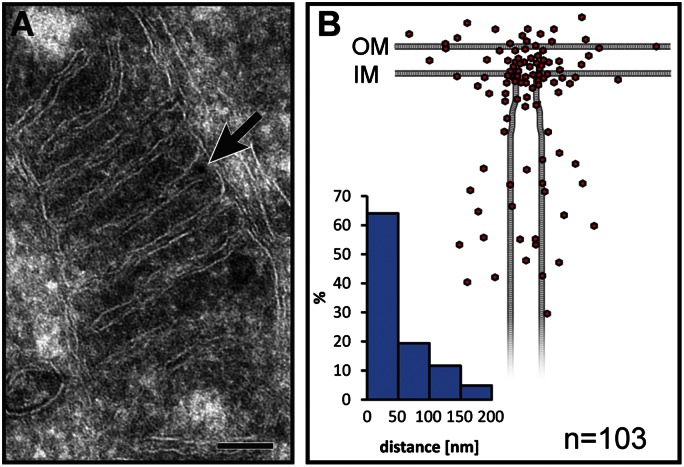
To gain further insight into the peculiar distribution of the core MINOS complex, we performed immunogold labeling of chemically fixed cryosectioned HeLa cells (Fig. 5). The sections were decorated with a primary antibody against mitofilin, followed by a secondary gold conjugate. To this end, the amount of antibodies used was kept at a very low level, to minimize unspecific labeling. Typically, a mitochondrion was decorated with one or two gold particles (Fig. 5A). For quantitative analysis, we determined the localization of each mitochondrial gold particle (n = 103) with respect to the inner boundary membrane and the closest cristae membrane and plotted its respective localization in a model (Fig. 5B). In the human fibroblast cells analyzed, the cristae membranes were generally connected to the inner boundary membrane by two or more cristae junctions; cristae tips were seen only rarely, and thus were omitted from the model. We found that the majority of all mitochondrial gold particles were enriched in the vicinity of cristae junctions to a similar extent as reported previously for its homolog Fcj1 and other components of the MINOS complex in yeast (17, 24). We conclude that in human cells, the core MINOS complex is localized predominantly at cristae junctions.
Fig. 5.
Submitochondrial localization of mitofilin using quantitative immunoelectron microscopy. (A) Immunogold labeling of mitofilin in HeLa cells. A representative mitochondrion is shown. The arrow points to the position of a gold particle. (B) Localizations of the gold particles (red) as determined by immunogold labeling of mitofilin plotted on a scheme representing a part of a mitochondrion. The histogram shows the fraction of gold particles within the indicated distance to the crista junction. The histogram and the graphical representation are based on the same measured gold particle localizations. OM, outer membrane; IM, inner membrane. (Scale bar: 100 nm.)
Cristae Junctions Are Frequently Aligned in Parallel to the Growth Substrate Surface.
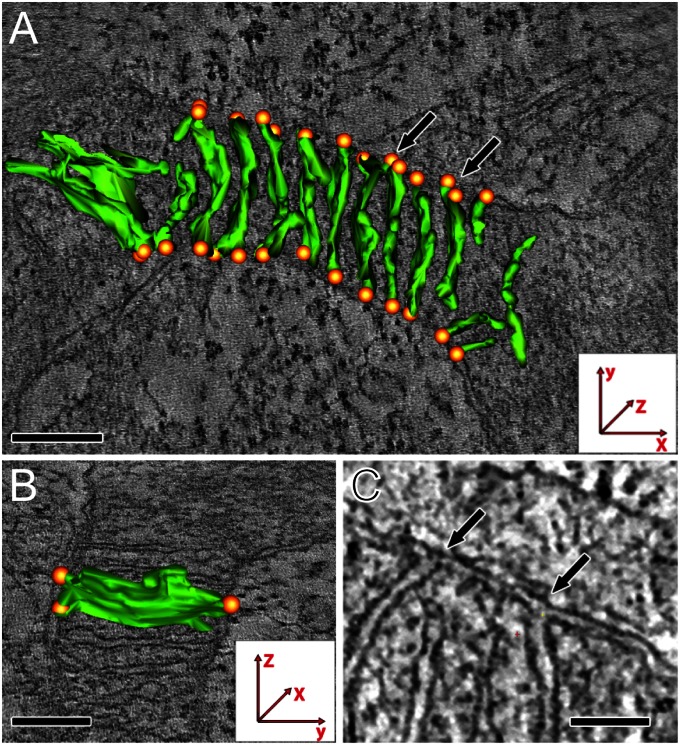
We next performed electron tomography to determine the arrangement of the cristae junctions in the mitochondria of the cultured primary human fibroblasts (Fig. 6). The cells were chemically fixed and embedded in epoxy resin; then 250-nm-thick sections were cut in parallel to the growth surface, because almost all mitochondria were coaligned to the surface in these cells. Dual-axis tomograms revealed that in most mitochondria the cristae were lamellar with a regular spacing. Most cristae featured two or more cristae junctions and were devoid of tips. Intriguingly, most of the cristae junctions were arranged in parallel to the growth surface. To verify this finding and to ensure that it was not related to the lower axial resolution, which potentially might conceal cristae junctions at the bottom and the top of the tomograms, we cut the fixed cells perpendicular to the growth surface. We observed a preferential arrangement of the cristae junctions in the horizontal plane in these perpendicular sections as well.
Fig. 6.
Cristae junction orientation and cristae morphology analyzed by dual-axis electron tomography. Primary adult human fibroblasts were grown on Aclar discs. (A and B) Single slices of the tomogram (grayscale) overlaid with reconstructions of segmented cristae membranes (green). Cristae junctions are denoted by orange spheres. Shown are a top view of a mitochondrion (A) (growth surface in plane with the tomogram) and a side view of the same mitochondrion (B). Note the lamellar cristae and the preferential orientation of the cristae junctions at the sides of the organelle, so that they are in parallel with the growth surface. (C) Magnification of a single slice of the tomogram (top view). The arrows point to the same cristae junctions as marked by the arrows in A. (Scale bar: 100 nm in A and C; 50 nm in B).
Taken together, our findings show that human mitofilin, a core component of MINOS, is enriched at cristae junctions. Electron tomography demonstrated that mitochondria of human fibroblasts often exhibit lamellar cristae that are attached to the inner boundary membrane by cristae junctions, which generally run in parallel to the growth surface, explaining the discontinuous rail-like distribution of the mitofilin clusters at the sides of the mitochondria. We conclude that MINOS is localized predominantly at cristae junctions, which in peripheral mitochondria of cultured mammalian cells are frequently evenly spaced and arranged in parallel to the growth surface.
Discussion
Using STED super-resolution microscopy, we have demonstrated that three components of the human MINOS complex (mitofilin, MINOS1, and CHCHD3) form clusters that in peripheral mitochondria of several mammalian adherent cell lines exhibit a highly organized periodic spatial distribution, which appears in superresolved immunofluorescence images as a discontinuous rail-like arrangement. Interestingly, we generally observed individual MINOS clusters, but no extended superstructures, such as arches or helices, around the mitochondrial tubules. Conventional wide-field imaging of yeast cells expressing functional GFP-fusion proteins of MINOS subunits, including Fcj1-GFP and Aim13-GFP, the yeast homologs of mitofilin and CHCHD3, suggested that these proteins form filamentous structures around the inner membrane in yeast. This observation, together with biochemical and genetic data, led to the proposal that MINOS acts as a mitochondrial skeletal structure in S. cerevisiae (16). It will be an important future challenge to clarify whether the yeast and the human MINOS complexes execute different functions, which may well be reflected by different submitochondrial distributions. To this end, it will be necessary to analyze the localization of the yeast MINOS with super-resolution microscopy. Likewise, it will be necessary to identify additional components of the human MINOS complex. It is possible that some other, as-yet unidentified components of the human MINOS complex may bridge the punctuate core components of the human MINOS clusters to an extended structure.
A striking finding revealed by STED microscopy of the human MINOS is the unexpected regular distribution of the MINOS clusters. In human fibroblast cells, the regular arrangement of the MINOS clusters was most often seen in the peripheral mitochondria outstretched in parallel to the growth surface. Interestingly, a recent study using electron tomography to analyze mitochondria in the calyx of Held adjacent to active zones demonstrated a polarized cristae structure and a greater density of cristae junctions in that part of the mitochondrion facing the presynaptic membrane (33). A high level of regularity in cristae structure, cristae junction and MINOS distribution might specifically occur in cells with a pronounced polarity. Cristae junctions are hypothesized to regulate the composition of the cristae membrane and to influence the ATP-synthesizing capability of mitochondria (4). Thus, the positioning of cristae junctions might be a pathway for fine-tuning mitochondrial function.
How the cell arranges the cristae junctions predominantly in parallel to the growth surface in the cultured human fibroblasts remains unknown. Presumably, additional cellular components are involved in conveying the positional information from the plasma membrane to the mitochondria. In mammalian cells, the mitochondrial tubules are often coaligned to the microtubule cytoskeleton and the endoplasmic reticulum (ER) (34–37). The large multisubunit MINOS has been suggested to act as a scaffold and interacting platform that not only is crucial for inner membrane architecture, but also interacts with protein complexes of the outer membrane. Stable isotope labeling with amino acids in cell culture (SILAC) immunoprecipitation experiments with antibodies against MINOS1 identified enrichment of three proteins (TUBG1, TUBGCP2, and FAM82B) associated with the microtubule cytoskeleton (15), possibly suggesting an indirect interaction of MINOS with microtubules. In budding yeast, the ER–mitochondria encounter structure (ERMES) tethers the mitochondria to the ER (38, 39). MINOS and ERMES are linked through strong genetic interactions (16), and MINOS even has been proposed to form a central part of an ER–mitochondria organizing network that controls mitochondrial membrane architecture and biogenesis (11). Although direct experimental evidence is lacking, this suggests the exciting possibility that positional cellular information influencing the inner mitochondrial architecture is mediated through MINOS, possibly via the ER or by cytoskeletal elements, thus adding another potential function to this multifaceted complex.
Methods and Materials
Cell Culture.
The following mammalian cell lines were used in this study: primary adult human fibroblasts (32), primary neonatal human fibroblasts (Invitrogen), immortal HeLa cells, immortal human osteosarcoma (U2OS) cells (European Collection of Cell Cultures) and immortal kidney epithelial cells (Vero) from the African green monkey Chlorocebus sp. The cells were cultivated in DMEM with Glutamax and 4.5% (wt/vol) glucose (Invitrogen), supplemented with 50 u/mL penicillin, 50 µg/mL streptomycin, 1 mM Na-pyruvate, and 10% (vol/vol) FCS (Invitrogen) at 37 °C and 7% (vol/vol) CO2.
Sample Preparation for Fluorescence Microscopy.
For immunolabeling, the samples were chemically fixed in 4% (wt/vol) prewarmed formaldehyde and prepared as described previously (40), using antibodies against mitofilin (Abcam), MINOS1/C1ORF151 (Abcam), CHCHD3 (Atlas Antibodies), Sam50 (Atlas Antibodies), DnaJC11 (Abnova), dsDNA (Progen), ATP synthase β-subunit (Abcam), and Tom20 (Santa Cruz Biotechnology). The primary antibodies were detected with secondary antibodies (sheep anti-mouse and goat anti-rabbit; Jackson Immuno Research) custom-labeled with ATTO590 (AttoTec) or KK114 (29). The samples were mounted in Mowiol mounting medium containing 0.1% 1,4-Diazabicyclo[2.2.2]octan (DABCO). The fixation, labeling, and embedding protocols were carefully optimized and controlled.
Fluorescence Microscopy.
Conventional diffraction-limited microscopy was performed with a Leica DM6000 epifluorescence microscope or a Leica TCS SP5 confocal microscope. Single-color STED and corresponding confocal microscopy were conducted using a custom-built STED microscope (29). Two-color STED images were recorded with a custom-built STED microscope, which combines two pairs of excitation and STED laser beams, all derived from a single supercontinuum laser source, as described previously (41). Using these microscopes, a resolution of ∼250 nm in the confocal images and 40–50 nm in the STED images was achieved. Imaging was performed essentially as described previously (29, 40). Except for contrast stretching, smoothing, and interpolation, no further image processing was applied.
Electron Microscopy.
For electron tomography, HeLa cells were grown on Aclar discs (Plano) to a confluency of ∼90% and fixed in prewarmed growth medium with 0.1% glutaraldehyde and 4% formaldehyde for 15 min. After additional fixation in 2.5% (wt/vol) glutaraldehyde in 0.1 M caccodylic buffer (pH 7.4) for 12 h at 4 °C, the cells were stained for 3 h in 1% (wt/vol) osmium tetroxide and for 30 min in uranyl acetate, then embedded in Agar 100 epoxid resin. Thin sections were counterstained with 1% uranyl acetate and lead citrate. Tilt series from 250-nm-thick sections of Agar 100-embedded cells were recorded with a Philips CM120 transmission electron microscope at 20,000× magnification with a TVIPS 2k × 2k slow-scan CCD camera. Orthogonal series were obtained from −64.5° to 64.5° in 3° Saxton intervals. The series were calculated using the IMOD software package (http://bio3d.colorado.edu/). Volume segmentation was performed with semiautomatic tracing of cristae boundaries using Imod software.
Immunoelectron Microscopy.
Cells were scraped off and harvested at a confluency of ∼90%. Ultrathin cryosections of the cells were prepared as described previously (6). The ultrathin sections (80 nm) were incubated with a polyclonal antiserum against mitofilin for 20 min, followed by incubation with 10 nm protein A gold (a gift from G. Posthuma, Utrecht University, Utrecht, The Netherlands) for 20 min. The cryosections were thoroughly washed, contrasted with uranyl acetate/methyl cellulose for 10 min on ice, embedded in the same solution, and finally examined with a Philips CM120 transmission electron microscope.
Supplementary Material
Acknowledgments
We thank Lars Kastrup for help with the two-color STED microscope, Jaydev Jethwa and Jonathan Melin for helpful comments on the manuscript, and Stefan W. Hell for discussions and advice. This work was supported by the Deutsche Forschungsgemeinschaft and by the Cluster of Excellence and Deutsche Forschungsgemeinschaft Research Center Nanoscale Microscopy and Molecular Physiology of the Brain (S.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301820110/-/DCSupplemental.
References
- 1.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 2.Frey TG, Mannella CA. The internal structure of mitochondria. Trends Biochem Sci. 2000;25(7):319–324. doi: 10.1016/s0968-0004(00)01609-1. [DOI] [PubMed] [Google Scholar]
- 3.Perkins G, et al. Electron tomography of neuronal mitochondria: Three-dimensional structure and organization of cristae and membrane contacts. J Struct Biol. 1997;119(3):260–272. doi: 10.1006/jsbi.1997.3885. [DOI] [PubMed] [Google Scholar]
- 4.Mannella CA. Structure and dynamics of the mitochondrial inner membrane cristae. Biochim Biophys Acta. 2006;1763(5-6):542–548. doi: 10.1016/j.bbamcr.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Wurm CA, Jakobs S. Differential protein distributions define two sub-compartments of the mitochondrial inner membrane in yeast. FEBS Lett. 2006;580(24):5628–5634. doi: 10.1016/j.febslet.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 6.Suppanz IE, Wurm CA, Wenzel D, Jakobs S. The m-AAA protease processes cytochrome c peroxidase preferentially at the inner boundary membrane of mitochondria. Mol Biol Cell. 2009;20(2):572–580. doi: 10.1091/mbc.E07-11-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel F, Bornhövd C, Neupert W, Reichert AS. Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol. 2006;175(2):237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilkerson RW, Selker JML, Capaldi RA. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003;546(2-3):355–358. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- 9.Stoldt S, et al. The inner-mitochondrial distribution of Oxa1 depends on the growth conditions and on the availability of substrates. Mol Biol Cell. 2012;23(12):2292–2301. doi: 10.1091/mbc.E11-06-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Neupert W. SnapShot: Mitochondrial architecture. Cell. 2012;149(3):722–722e1. doi: 10.1016/j.cell.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 11.van der Laan M, Bohnert M, Wiedemann N, Pfanner N. Role of MINOS in mitochondrial membrane architecture and biogenesis. Trends Cell Biol. 2012;22(4):185–192. doi: 10.1016/j.tcb.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 12.Zerbes RM, et al. Mitofilin complexes: Conserved organizers of mitochondrial membrane architecture. Biol Chem. 2012;393(11):1247–1261. doi: 10.1515/hsz-2012-0239. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann JM. MINOS is plus: A Mitofilin complex for mitochondrial membrane contacts. Dev Cell. 2011;21(4):599–600. doi: 10.1016/j.devcel.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 14.von der Malsburg K, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21(4):694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- 15.Alkhaja AK, et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23(2):247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoppins S, et al. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195(2):323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harner M, et al. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30(21):4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie J, Marusich MF, Souda P, Whitelegge J, Capaldi RA. The mitochondrial inner membrane protein mitofilin exists as a complex with SAM50, metaxins 1 and 2, coiled-coil-helix coiled-coil-helix domain-containing protein 3 and 6 and DnaJC11. FEBS Lett. 2007;581(18):3545–3549. doi: 10.1016/j.febslet.2007.06.052. [DOI] [PubMed] [Google Scholar]
- 19.Darshi M, et al. ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem. 2011;286(4):2918–2932. doi: 10.1074/jbc.M110.171975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott C, et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol Cell Biol. 2012;32(6):1173–1188. doi: 10.1128/MCB.06388-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zerbes RM, et al. Role of MINOS in mitochondrial membrane architecture: Cristae morphology and outer membrane interactions differentially depend on mitofilin domains. J Mol Biol. 2012;422(2):183–191. doi: 10.1016/j.jmb.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Körner C, et al. The C-terminal domain of Fcj1 is required for formation of crista junctions and interacts with the TOB/SAM complex in mitochondria. Mol Biol Cell. 2012;23(11):2143–2155. doi: 10.1091/mbc.E11-10-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hess DC, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5(3):e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rabl R, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185(6):1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakobs S, Stoldt S, Neumann D. Light microscopic analysis of mitochondrial heterogeneity in cell populations and within single cells. Adv Biochem Eng Biotechnol. 2011;124:1–19. doi: 10.1007/10_2010_81. [DOI] [PubMed] [Google Scholar]
- 26.Hell SW. Far-field optical nanoscopy. Science. 2007;316(5828):1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 27.Weber K, Rathke PC, Osborn M. Cytoplasmic microtubular images in glutaraldehyde-fixed tissue culture cells by electron microscopy and by immunofluorescence microscopy. Proc Natl Acad Sci USA. 1978;75(4):1820–1824. doi: 10.1073/pnas.75.4.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dyba M, Jakobs S, Hell SW. Immunofluorescence stimulated emission depletion microscopy. Nat Biotechnol. 2003;21(11):1303–1304. doi: 10.1038/nbt897. [DOI] [PubMed] [Google Scholar]
- 29.Wurm CA, et al. Nanoscale distribution of mitochondrial import receptor Tom20 is adjusted to cellular conditions and exhibits an inner-cellular gradient. Proc Natl Acad Sci USA. 2011;108(33):13546–13551. doi: 10.1073/pnas.1107553108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An J, et al. CHCM1/CHCHD6, novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J Biol Chem. 2012;287(10):7411–7426. doi: 10.1074/jbc.M111.277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bohnert M, et al. Role of mitochondrial inner membrane organizing system in protein biogenesis of the mitochondrial outer membrane. Mol Biol Cell. 2012;23(20):3948–3956. doi: 10.1091/mbc.E12-04-0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kukat C, et al. Super-resolution microscopy reveals that mammalian mitochondrial nucleoids have a uniform size and frequently contain a single copy of mtDNA. Proc Natl Acad Sci USA. 2011;108(33):13534–13539. doi: 10.1073/pnas.1109263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perkins GA, et al. The micro-architecture of mitochondria at active zones: Electron tomography reveals novel anchoring scaffolds and cristae structured for high-rate metabolism. J Neurosci. 2010;30(3):1015–1026. doi: 10.1523/JNEUROSCI.1517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxton WM, Hollenbeck PJ. The axonal transport of mitochondria. J Cell Sci. 2012;125(Pt 9):2095–2104. doi: 10.1242/jcs.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frederick RL, Shaw JM. Moving mitochondria: Establishing distribution of an essential organelle. Traffic. 2007;8(12):1668–1675. doi: 10.1111/j.1600-0854.2007.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Brito OM, Scorrano L. Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature. 2008;456(7222):605–610. doi: 10.1038/nature07534. [DOI] [PubMed] [Google Scholar]
- 37.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat Rev Mol Cell Biol. 2012;13(10):607–625. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc Natl Acad Sci USA. 2011;108(34):14151–14156. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stroud DA, et al. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011;413(4):743–750. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 40.Wurm CA, Neumann D, Schmidt R, Egner A, Jakobs S. Sample preparation for STED microscopy. Methods Mol Biol. 2010;591:185–199. doi: 10.1007/978-1-60761-404-3_11. [DOI] [PubMed] [Google Scholar]
- 41.Neumann D, Bückers J, Kastrup L, Hell SW, Jakobs S. Two-color STED microscopy reveals different degrees of colocalization between hexokinase-I and the three human VDAC isoforms. PMC Biophys. 2010;3(1):4. doi: 10.1186/1757-5036-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.