Abstract
Nucleotide signaling molecules are important messengers in key pathways that allow cellular responses to changing environments. Canonical secondary signaling molecules act through specific receptor proteins by direct binding to alter their activity. Cyclic diadenosine monophosphate (c-di-AMP) is an essential signaling molecule in bacteria that has only recently been discovered. Here we report on the identification of four Staphylococcus aureus c-di-AMP receptor proteins that are also widely distributed among other bacteria. Using an affinity pull-down assay we identified the potassium transporter-gating component KtrA as a c-di-AMP receptor protein, and it was further shown that this protein, together with c-di-AMP, enables S. aureus to grow in low potassium conditions. We defined the c-di-AMP binding activity within KtrA to the RCK_C (regulator of conductance of K+) domain. This domain is also found in a second S. aureus protein, a predicted cation/proton antiporter, CpaA, which as we show here also directly binds c-di-AMP. Because RCK_C domains are found in proteinaceous channels, transporters, and antiporters from all kingdoms of life, these findings have broad implications for the regulation of different pathways through nucleotide-dependent signaling. Using a genome-wide nucleotide protein interaction screen we further identified the histidine kinase protein KdpD that in many bacteria is also involved in the regulation of potassium transport and a PII-like signal transduction protein, which we renamed PstA, as c-di-AMP binding proteins. With the identification of these widely distributed c-di-AMP receptor proteins we link the c-di-AMP signaling network to a central metabolic process in bacteria.
Nucleotide signaling molecules control fundamental processes in all forms of life. There is now a large body of evidence linking nucleotides such as cAMP, cGMP, and guanosine tetra- (ppGpp) and pentaphosphate (pppGpp) to the control of fundamental metabolic pathways and stress response processes in eukaryotic and prokaryotic cells (1–3). Cyclic dinucleotides in particular have recently gained increased attention with the identification of additional nucleotides such as cyclic diadenosine monophosphate (c-di-AMP) and the hybrid c-AMP-GMP molecule in bacterial cells (4–6), as well as the discovery that cyclic dinucleotides are also produced by eukaryotic cells (7–9). The dinucleotide cyclic diguanosine monophosphate (c-di-GMP) and the molecular mechanisms by which it controls cellular pathways has been well characterized, and it is now recognized as a central regulator in bacterial cells that controls the switch from free-living planktonic to a sessile biofilm-associated lifestyles. In pathogenic organisms this is often linked to colonization of the human host and virulence (10). On the other hand, the function and the pathways controlled by the signaling nucleotide c-di-AMP are less clear, largely owing to a gap in our knowledge of specific receptor proteins.
Many Gram-positive bacteria, including the important human pathogens Staphylococcus aureus (11), Streptococcus pyogenes (12), Listeria monocytogenes (5), and Mycobacterium tuberculosis (13), produce c-di-AMP, and it is likely that c-di-AMP is also synthesized by several Gram-negative bacteria and a subset of archaea (14). c-di-AMP is synthesized by DisA_N domain-containing diadenylate cyclases DacA, DisA, and YojJ and degraded by the phosphodiesterase enzyme GdpP (4, 5, 11, 15–18). A variety of different phenotypes have been linked to altered c-di-AMP levels; an increase in c-di-AMP levels correlates with increased acid resistance (16, 19) and altered antibiotic resistance, including an increase in methicillin resistance in S. aureus (11, 18, 20). Most notable, however, are the findings that L. monocytogenes (5) and Bacillus subtilis (18) cannot grow in the absence of c-di-AMP, showing that in contrast to other signaling nucleotides, c-di-AMP controls essential cellular pathways. The molecular basis for this is currently not known, although it is assumed that, similar to other signaling molecules, c-di-AMP interacts with a specific set of target proteins and upon binding alters their activity or function. Currently only one bacterial c-di-AMP receptor protein, the transcription factor DarR, has been identified in Mycobacterium smegmatis (21). However, the absence of close DarR homologs in many organisms that likely produce c-di-AMP implies that additional c-di-AMP target proteins must exist.
In this study we have identified the potassium transporter-gating component KtrA as c-di-AMP target protein by using an affinity pull-down assay. KtrA is a member of the widely distributed RCK (regulator of conductance of K+) protein family, known to be involved in the gating of ion channels. Here we show that KtrA is required for the growth of S. aureus under potassium limiting conditions. Through subsequent binding studies we show that c-di-AMP specifically interacts with the C-terminal RCK_C domain of KtrA. A second S. aureus RCK_C domain-containing protein CpaA, a predicted cation/proton antiporter, was subsequently identified bioinformatically and its interaction with c-di-AMP confirmed experimentally. Last, using a genome-wide interaction screen, we identified the PII-like signal transduction protein PstA and the histidine kinase KdpD as additional c-di-AMP binding proteins. With the identification of these four widely distributed c-di-AMP binding proteins we provide a link between c-di-AMP and a fundamental cellular process in bacteria, namely ion transport.
Results
Identification of the c-di-AMP Target Protein KtrA.
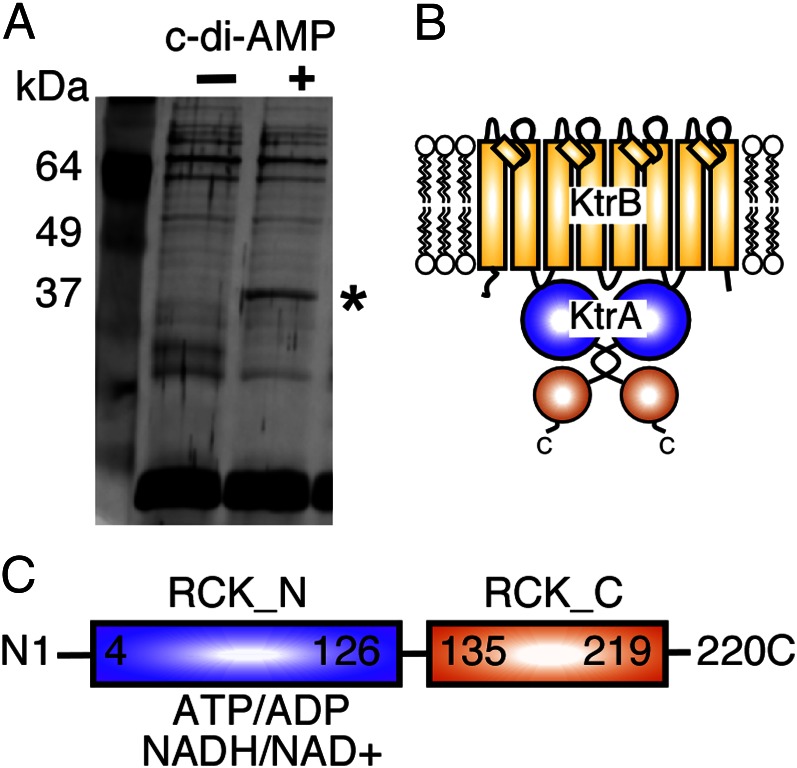
To identify c-di-AMP receptor proteins, we performed an affinity pull-down assay using c-di-AMP–coupled magnetic beads and protein extracts derived from the S. aureus strain LAC*. This strain is an erythromycin-sensitive derivative of the clinically relevant community-acquired methicillin-resistant USA300 strain LAC. One protein band was enriched in samples obtained from c-di-AMP–coupled beads (Fig. 1A) and identified by mass spectrometry as S. aureus protein SAUSA300_0988. This protein has high similarity to the B. subtilis proteins KtrABS (51% identity) and KtrCBS (63% identity) that, together with their respective membrane components KtrB and KtrD, form potassium transporters (Fig. 1B) (22). SAUSA300_0988 is the only KtrA/C-type protein in S. aureus and was renamed KtrASA. The cytoplasmic components of Ktr systems are part of the RCK protein family and play an important role in transporter gating (23, 24). KtrASA is a typical RCK protein, with an RCK_N domain and an RCK_C domain (Fig. 1C). On the basis of a structural model, it is likely that KtrASA assumes a similar two-lobed fold as the RCK domain in the potassium channel protein MthK of Methanobacterium thermoautotrophicus (Fig. S1) (25, 26). Interestingly, a nucleotide-binding site for ATP and other nucleotides has been identified previously in the RCK_N domain of the B. subtilis protein KtrA (23) and according to a structural model the RCK_N domain of the S. aureus protein is likely to assume the same fold with the conserved GxGxxG motif forming part of a nucleotide-binding site and with aspartic acid residues D32 and D52 acting as crucial nucleotide-binding residues (Fig. S1) (23).
Fig. 1.
Identification of S. aureus KtrASA as a potential c-di-AMP binding protein. (A) Silver-stained polyacrylamide gel of cytoplasmic S. aureus proteins retained on c-di-AMP-coupled (+) or uncoupled (-) beads. The protein band enriched in the c-di-AMP lane (asterisk) was identified by mass spectrometry as S. aureus protein SAUSA300_0988 (KtrASA). (B) Illustration of Ktr-type potassium transport systems, which are composed of a KtrB-type membrane component and a cytoplasmic KtrA-type gating component. (C) Schematic representation of the KtrASA domain structure with the RCK_N domain (amino acids 4–126) indicated in blue and RCK_C domain (amino acids 135–219) shown in orange. The RCK_N domain of the B. subtilis KtrA homolog is known to bind to nucleotides including ATP, ADP, NAD+, and NADH.
c-di-AMP Binds to the RCK_C Domain of KtrA.
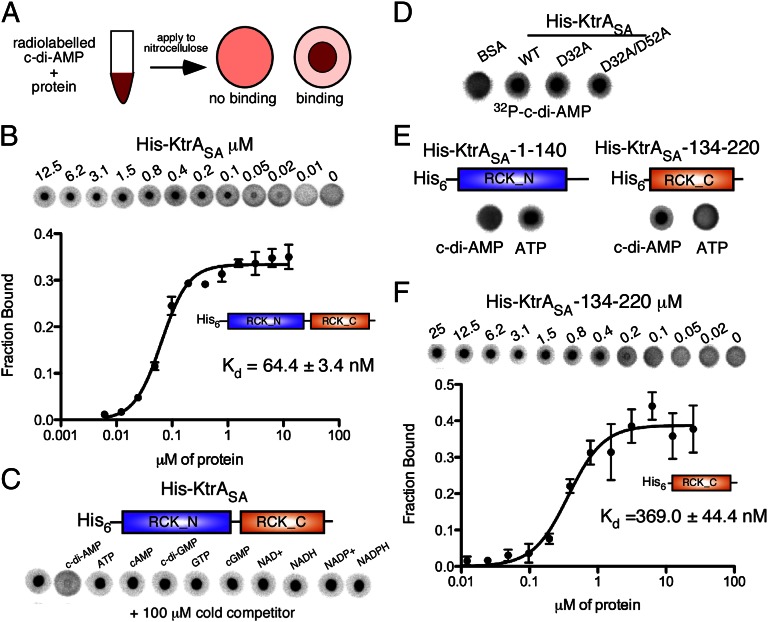
To confirm the interaction between KtrASA and c-di-AMP and to define more precisely the interaction domain, we adapted the differential radial capillary action of ligand assay (DRaCALA), which was previously used to study c-di-GMP-protein interactions (27). This assay is based on the principle that free nucleotides migrate outward when spotted on nitrocellulose membranes, whereas bound ligand is sequestered to the protein and immobilized in a tight spot on the membrane (Fig. 2A). The distribution of free and bound ligand can be readily visualized and quantified using radiolabeled nucleotides. To determine whether c-di-AMP–protein interactions could be measured with this assay, we produced 32P-labeled c-di-AMP (Fig. S2) and tested its interaction with purified S. aureus GdpP and B. subtilis DisA proteins, c-di-AMP degrading and synthesizing enzymes, respectively. c-di-AMP–specific binding to these control proteins was observed (Fig. S3), thus validating DRaCALA as a method to study c-di-AMP-protein interactions. Using this method, we next investigated the interaction between c-di-AMP and purified His-KtrASA protein and determined an interaction with a Kd of 64.4 ± 3.4 nM (Fig. 2B). Only an excess of unlabeled c-di-AMP, but not the other nucleotides tested, including ATP, competed for binding with labeled c-di-AMP (Fig. 2C). This also indicated that c-di-AMP does not bind to the previously described nucleotide-binding site in RCK_N. Furthermore, c-di-AMP bound to the KtrASA-D32A/D52A variant with alanine substitutions of the two key nucleotide-binding residues within RCK_N (Fig. 2D). To determine more specifically which portion of KtrASA interacts with c-di-AMP, the RCK_N and RCK_C domains were produced and purified separately. Although the RCK_N domain interacted, as expected, with ATP, it did not bind c-di-AMP (Fig. 2E). In contrast, the RCK_C domain bound c-di-AMP with a Kd of 369.0 ± 44.4 nM (Fig. 2 E and F), thus showing that the RCK_C domain is the receptor domain of c-di-AMP. To further validate the DRaCALA binding results, an interaction between c-di-AMP and KtrA or the RCK_C domain in the nM range was further confirmed by equilibrium dialysis (Fig. S4). Of note, a specific interaction between c-di-AMP and the RCK_C domain of KtrA was also obtained when DRaCALAs were performed using Escherichia coli extracts prepared from strains producing different KtrASA variants in place of purified proteins (Fig. S5). Furthermore, E. coli extracts containing the full-length B. subtilis KtrA protein, but not an N-terminal fragment lacking the RCK_C domain, interacted with c-di-AMP (Fig. S5). Taken together, these results show that KtrA is a bona fide bacterial c-di-AMP receptor protein and support a model whereby the two domains in Gram-positive KtrA-type proteins bind different nucleotides: ATP, ADP, NAD+, or NADH with the RCK_N and c-di-AMP within the RCK_C domain.
Fig. 2.
Characterization of the c-di-AMP/KtrASA interaction by DRaCALA. (A) Schematic representation of the DRaCALA to study c-di-AMP protein interactions. (B) Binding curve and Kd determination for c-di-AMP and purified His-KtrASA. Kd values were determined from the curve as previously described (27). (C) DRaCALAs with purified His-KtrASA protein and 32P-labeled c-di-AMP and an excess of cold competitor nucleotide as indicated above each spot. (D) DRaCALAs with purified His-KtrASA, His-KtrASA-D32A, or His-KtrASA-D32A/D52A and 32P-labeled c-di-AMP. (E) DRaCALAs with purified His-KtrASA-1-140 (RCK_N) or His-KtrASA-134-220 (RCK_C) and 32P-labeled c-di-AMP or 32P-labeled ATP as indicated below the spots. (F) Binding curves and Kd determination for c-di-AMP and purified His-KtrASA-134-220 protein containing only the RCK_C domain. The data were plotted, and the best-fit line was determined by nonlinear regression incorporating the hill equation using GraphPad Prism software.
KtrA Is Important for the Growth of S. aureus in Low Potassium.
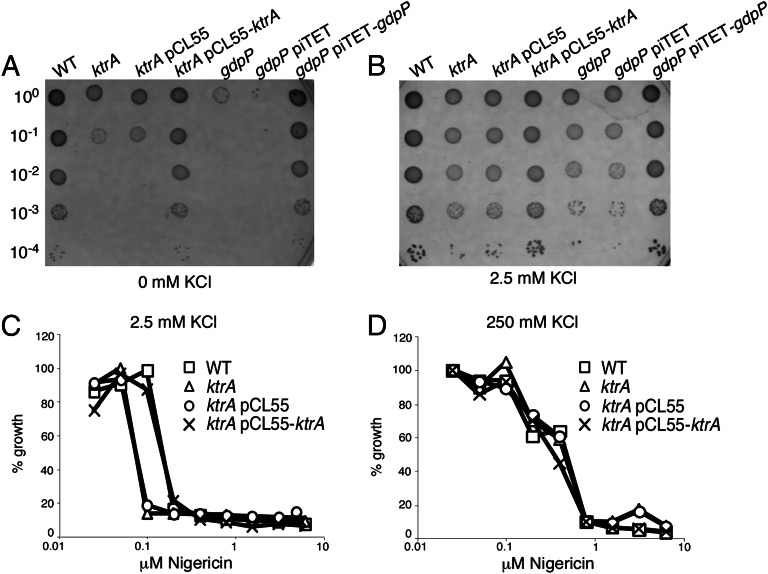
To investigate the involvement of KtrASA and c-di-AMP in the growth of S. aureus in low potassium conditions, the growth of ktrA and gdpP mutant strains was compared with that of the wild-type LAC* strain. The gdpP mutant strain has 15-fold higher levels of intracellular c-di-AMP (11), and therefore KtrA should be in the nucleotide-bound state under these conditions. Because potassium uptake is especially important during osmotic stress, the different S. aureus strains were grown on chemically defined medium (CDM) plates containing 0.75 M NaCl. Under these stress condition, a two to three log growth defect was observed for both the ktrA and gdpP mutant strains, which could be complemented either by the addition of potassium or by the introduction of a functional copy of ktrA or gdpP, respectively (Fig. 3 A and B). The ktrA mutant was also hyper-susceptible to the potassium ionophore nigericin, which causes an exchange of intracellular K+ for extracellular H+ (Fig. 3C). The hypersensitivity to nigericin could again be rescued by the addition of 250 mM potassium or by genetic complementation (Fig. 3 C and D). Similarly a ktrA mutant strain in the methicillin sensitive S. aureus strain background Newman was also more sensitive to nigericin and did not grow as well as the wild-type strain under the osmotic stress conditions unless potassium was added (Fig. S6). These results suggest a function for KtrASA in potassium uptake in S. aureus strains and that c-di-AMP binding to KtrASA might inactivate channel activity, because the gdpP mutant strain, which has greatly increased levels of c-di-AMP, displays a phenotype similar to the ktrA mutant.
Fig. 3.
Effect of potassium on growth of wild-type, ktrA, and gdpP S. aureus strains. (A and B) The indicated S. aureus strains were grown overnight in CDM containing 2.5 mM KCl. Next day serial dilutions of washed cells were spotted onto CDM agar plates containing 0.75 M NaCl and containing either 0 mM or 2.5 mM potassium. (C and D) Nigericin sensitivity curves of wild-type, ktrA mutant, and complemented S. aureus strains. The different strains were grown in 96-well plates in CDM medium supplemented with 2.5 mM or 250 mM potassium and nigericin at the indicated concentration. OD600 readings were determined after 24 h growth and plotted as % growth compared with the growth in the absence of nigericin. Experiments were repeated a minimum of five times. When grown in 2.5 mM KCl the ktrA mutant consistently showed a twofold reduced MIC for each experiment. The MIC for all of the strains varied between experiments from 0.1 to 0.8 µM for the wild-type and complemented strain and 0.05–0.4 µM for the mutant strains.
c-di-AMP Interacts with CpaA, a Second S. aureus RCK_C Domain-Containing Protein.
The identification of the RCK_C domain as a c-di-AMP interacting domain allows the bioinformatic prediction of other receptor proteins based on the presence of an RCK_C domain. In this manner we discovered the protein SAUSA300_0911 in S. aureus strain LAC*, which we rename CpaA. This protein is a predicted cation/proton antiporter that is composed of an N-terminal transmembrane region followed by an RCK domain (Fig. 4A). An interaction between its RCK_C domain and c-di-AMP was tested by performing DRaCALAs with E. coli extracts prepared from strains either containing the empty vector as a control, or expressing the complete RCK or the RCK_C domain of CpaA. 32P-labeled c-di-AMP interacted specifically with both the RCK and the RCK_C domain (Fig. 4B), thus showing that CpaA is a second c-di-AMP target protein.
Fig. 4.
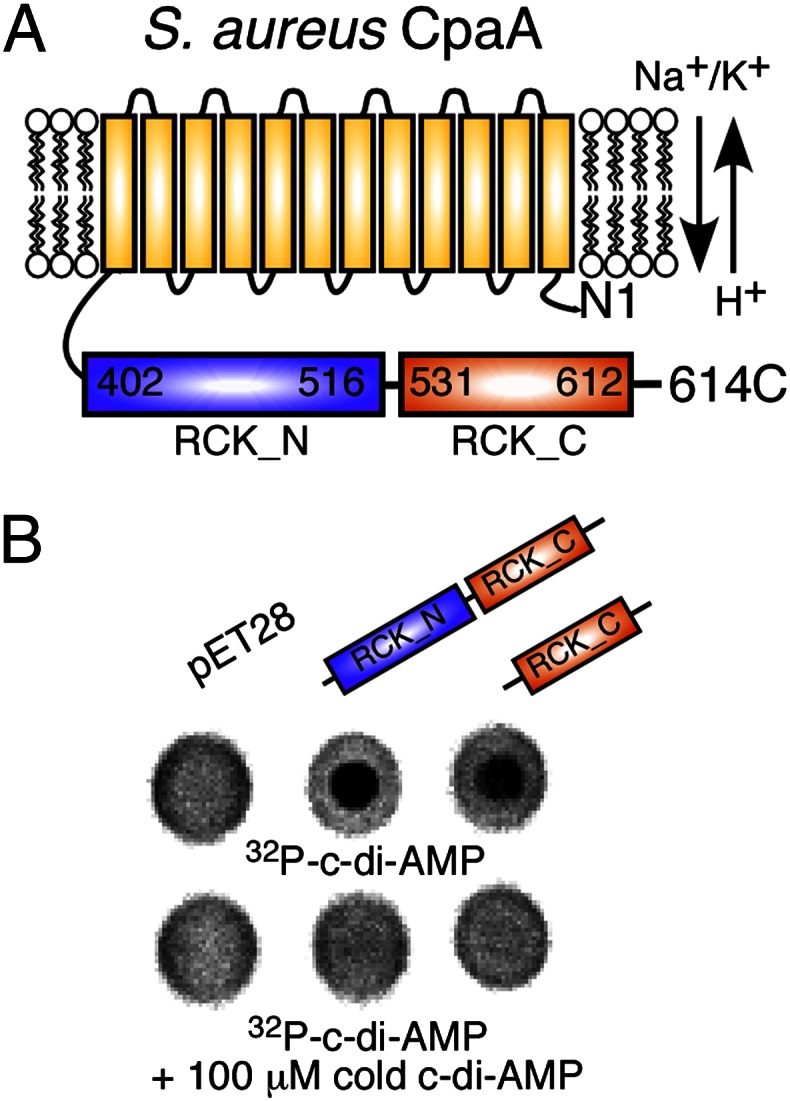
Identification of CpaA as an additional c-di-AMP target protein. (A) Schematic representation of the predicted K+ or Na+ antiporter CpaA (SAUSA300_0911), containing an N-terminal transmembrane (yellow) and cytoplasmically located RCK_N (blue) and RCK_C (orange) domains. (B) DRaCALAs with 32P-labeled c-di-AMP and E. coli extracts prepared from the vector control strain (pET28b) or strains overproducing His-CpaASA-402-614 (RCK_N and RCK_C) or His-CpaASA-513-614 (RCK_C). Cold c-di-AMP was added as a competitor where indicated.
Identification of PstA and KdpD as Specific di-AMP Binding Proteins Using a Genome-Wide ORF (ORFeome) DRaCALA Screen.
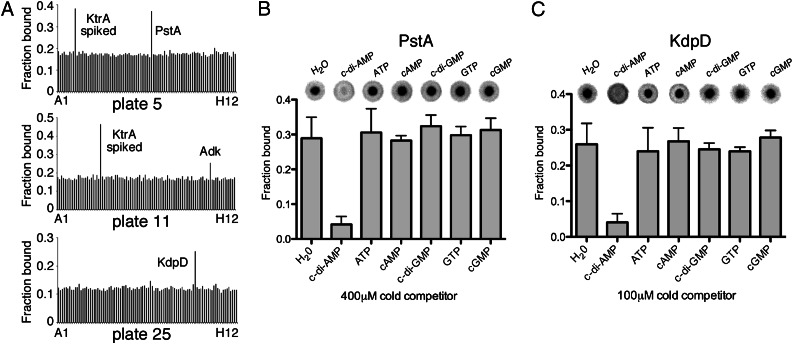
The DRaCALA method can be used to identify nucleotide/protein interactions using crude whole-cell E. coli lysates. This makes this assay ideally suited to perform a genome-wide protein/nucleotide interaction screen. An S. aureus strain COL ORFeome Gateway library is available, and we reasoned that this library together with the DRaCALA method should allow us to identify additional S. aureus c-di-AMP binding proteins. The library contains 2,343 S. aureus ORFs (86% of all S. aureus COL genes) within the Gateway entry vector pDONR221. These ORFs were recombined into the pDEST17 protein expression vector, placing each ORF under the control of the Isopropyl β-D-1-thiogalactopyranoside (IPTG)-inducible T7 promoter. With the exception of eight reactions that failed, all other resulting plasmids were recovered in the E. coli protein expression strain T7IQ. Four percent of the library strains were subsequently analyzed by PCR and all found to contain an insert of the expected size. Next, protein expression was induced and whole-cell E. coli extracts prepared. Eight percent of these extracts were analyzed by SDS/PAGE and Coomassie staining, and visible protein overproduction was observed for ∼70% of the lysates. Finally, these extracts, arrayed in 25 96-well plates, were used in DRaCALAs and the fraction of bound radiolabeled c-di-AMP determined for each spot. An average fraction bound value was determined for each plate, and the cutoff value for positive interactions was set at 1.4 times this average fraction bound background value. Extracts derived from strains expressing four different proteins gave c-di-AMP fraction bound values above background using these criteria, one of which was KtrA, thereby validating the DRaCALA ORFeome screen. The other positive clones, SACOL0525, SACOL2070, and SACOL2218, were confirmed by sequencing and renamed PstA (PII-like signal transduction protein A), KdpD (a sensor histidine kinase and annotated as KdpD in other S. aureus strains), and Adk (adenylate kinase), respectively. To determine whether these proteins are indeed bona fide c-di-AMP binding proteins, the corresponding genes were reamplified from S. aureus LAC* chromosomal DNA and cloned into the E. coli expression vector pET28b for overproduction as His-tag fusion proteins. Subsequently extracts were prepared and used in DRaCALAs (Fig. S7). Of note, whereas the fraction bound values for PstA and KdpD were twice as high as the background value in the initial whole-genome screen, the c-di-AMP fraction bound value obtained for Adk was only 1.45 times above background and so only just made the cutoff (Fig. 5A and Fig. S7). When no interaction was observed with Adk after recloning, this protein was no longer regarded as a c-di-AMP receptor protein (Fig. S7). On the other hand, c-di-AMP binding to PstA and KdpD was confirmed after recloning (Fig. S7), and both proteins interacted specifically with c-di-AMP because only the addition of an excess of cold c-di-AMP and not other cold nucleotides prevented the binding of radiolabeled c-di-AMP (Fig. 5 B and C). CpaA was not identified in this screen because the gene encoding for this protein is not present in the S. aureus COL genome. Taken together, the genome-wide DRaCALA screen identified two additional S. aureus proteins, PstA and KdpD, as bona fide c-di-AMP receptor proteins.
Fig. 5.
Identification of PstA and KdpD as specific c-di-AMP target proteins. (A) For the whole-genome DRaCALA screen, 32P-labeled c-di-AMP was dispensed into 96-well plates containing E. coli lysates, and aliquots were subsequently spotted in duplicate onto nitrocellulose membrane. The fraction of bound c-di-AMP was calculated for each well as described by Roelofs et al. (27) and the average values from the duplicate spots plotted. Plates 5, 11, and 25 with positive interactions are shown. The average fraction bound value for plate 5 was 0.178 ± 0.029. Well A10 was spiked with a KtrA lysate, and well E3 contained the PstA lysate, which had a fraction bound value of 0.370 (2× background). The average fraction bound value for plate 11 was 0.174 ± 0.032. Well B12 was spiked with a KtrA lysate, and well G11 contained the Adk lysate, which had a fraction bound value of 0.253 (1.45× background). The average fraction bound value for plate 25 was 0.122 ± 0.015. Well G2 contained the KdpD lysate, with a fraction bound value of 0.252 (2× background). (B and C) DRaCALAs were preformed with E. coli extracts prepared from strains overproducing His-PstA (B) or KdpD-His (C) and 32P-labeled c-di-AMP and an excess of cold competitor nucleotide as indicated above each spot. Fraction of bound nucleotide was determined as described by Roelofs et al. (27), and values from three independent experiments were plotted with SDs.
Discussion
Since the discovery of c-di-AMP, it has been speculated that this nucleotide binds to proteins to regulate their function. In this study we identified four c-di-AMP receptor proteins, namely KtrA, CpaA, KdpD, and PstA, by using an affinity pull-down assay, bioinformatics analysis, and a genome-wide protein nucleotide interaction screen (Figs. 1, 4, and 5). With the identification of three proteins (KtrA, CpaA, and KdpD) that have been implicated in potassium transport in other bacteria, we have linked c-di-AMP signaling to potassium transport in S. aureus. Interestingly, this distinguishes c-di-AMP from c-di-GMP, which regulates multiple cellular processes that help bacteria to transition between different lifestyles, such as extracellular carbohydrate and adhesion production, motility, and biofilm formation. The link between c-di-AMP and the ion transport may explain why c-di-AMP, in contrast to other related signaling nucleotides, is essential for growth in bacterial species. Individually ktrA, cpaA, pstA, and kdpD are not essential (28–30); however, it is plausible that combined mutations may be lethal. Alternatively the existence of an as yet unidentified essential c-di-AMP receptor is also entirely possible.
The c-di-AMP binding region in S. aureus KtrA and CpaA was narrowed down to the RCK_C domain (Figs. 2 and 4). This domain is present in a large number of bacterial and archaeal proteins, and there is a good correlation between the distribution of the c-di-AMP cyclase domain DisA_N and the presence of RCK_C domains. Most bacteria and archaea that potentially synthesize c-di-AMP also contain one or more proteins with an RCK_C domain. This raises the possibility that c-di-AMP may contribute to the regulation of ion transport in a large number of bacteria and archaea. The number of RCK_C domains per organism usually exceeds the number of cyclases, perhaps suggesting that c-di-AMP regulates the function of multiple proteins, which is similar to what we found in S. aureus. However, the RCK_C domain is phylogenetically more widely distributed than the c-di-AMP cyclase domain and is also found in some eukaryotes, such as green algae, in additional archaeal species, and most notably in a large number of Gram-negative proteobacteria where the c-di-AMP cyclase domain is absent. We would predict that in those organisms other small molecules interact with this domain to regulate transport processes. The RCK_C domain is associated as a soluble domain with potassium transporters, or in some cases directly linked to ion antiporters, such as in CpaA. However, this domain is also associated with predicted amino acid antiporters, citrate transporters, and voltage-gated channels. This suggests that c-di-AMP or other small molecules might regulate a range of different transport processes, which have not been previously associated with signaling networks.
Potassium is a major and essential intracellular ion, and therefore bacteria have evolved several different types of uptake systems. The third c-di-AMP binding protein identified in this study was KdpD, which is a widely distributed membrane-embedded sensor histidine kinase that in many bacteria controls, together with its cognate response regulator KdpE, the expression of a second type of potassium uptake system. This ATP-dependent potassium uptake system has been best characterized in E. coli and consists of four membrane components KdpABCF and the two-component system KdpDE, which is required for KdpABCF expression at a very low potassium concentration when the other uptake systems are no longer sufficient to allow the cell to acquire the necessary amount of ion (31). However, a recent study on the S. aureus KdpDE system suggested that this two-component system has a different function in this organism (32). The S. aureus KdpDE two-component system, which still responds to the extracellular potassium concentration, was found to be no longer required for bacterial survival under low potassium conditions, but instead to control the expression of several well-characterized S. aureus virulence factors (32). However, additional work is needed to fully understand the function of this two-component system in S. aureus and other Gram-positive bacteria and on the basis of this study its interplay with cellular c-di-AMP levels.
The least characterized c-di-AMP receptor protein identified in this study is the DUF970 domain-containing PII-like signal transduction protein, which belongs to the GlnB superfamily of proteins and was renamed PstA. PII-type proteins are one of the most widely distributed signal transduction proteins in nature that are present in bacteria as well as archea and plants. DUF970 domain-containing PII-like proteins are not only present in Staphylococcus species but widely distributed among Firmicutes. Characterized proteins belonging to this GlnB superfamily are the copper tolerance protein CutA1 (33) and the ATP phosphoribosyltransferase HisG, the first enzyme of the histidine pathway (34). However, the best-characterized proteins belonging to the GlnB superfamily are PII nitrogen regulatory proteins, which are key signal transduction protein that report on the nitrogen and carbon status of cells by sensing glutamine and 2-ketoglutarate levels (35). Because proteins belonging to this superfamily are known to bind diverse ligands and function by protein–protein interaction to control the activity of enzymes, transcription factor, or transport proteins, we would assume that upon c-di-AMP binding or release the S. aureus PstA protein interacts with other cellular proteins. However, these still need to be discovered.
This work demonstrates the feasibility of a DRaCALA-based ORFeome screen as a high-throughput platform for identifying c-di-AMP receptor proteins. Although the DRaCALA ORFeome screen will identify receptors whose binding site does not require additional proteins, biochemical pull-down assays will only yield receptors that are expressed in the assayed growth conditions. Together the combination of biochemical pull-down assays, bioinformatic analysis, and systematic screening of a whole genome protein expression library by DRaCALA provides a powerful synergistic approach for the systematic elucidation of protein–metabolite interaction networks (36). The discovery of the four different and widely distributed c-di-AMP receptor proteins allows future research to determine the molecular mechanisms underlying c-di-AMP dependent processes in prokaryotes.
Methods
Bacterial Strains and Culture Conditions.
E. coli strains were grown in LB or LB-M9 (37), B. subtilis strains in LB, and S. aureus strains in tryptic soy broth (TSB) or CDM at 37 °C with aeration. CDM was prepared as previously reported (38), with the following modifications: KH2PO4 was substituted with Na phosphate buffer, and KCl was added at concentrations stated in the text. In addition, Gly 50 mg/L; l-Ser 30 mg/L; l-Asp 90 mg/L; l-Lys 50 mg/L; l-Ala 60 mg/L; l-Trp 10 mg/L; l-Met 10 mg/L; l-His 20 mg/L; l-Ile 30 mg/L; l-Tyr 50 mg/L; and thymine 20 mg/L were added. Information on strain construction is provided in SI Methods. Strains and primers used are listed in Tables S1 and S2, and the methicillin-resistant S. aureus (MRSA), Strain COL Gateway Clone Set, Recombinant in E. coli, Plates 1–25, NR-19277 were obtained through BEI Resources, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH).
Affinity Pull-Down Assay.
Twenty milliliters of an S. aureus LAC* culture with an OD600 of 1 was harvested and suspended in 1 mL 10 mM Tris·HCl (pH 7.5), 50 mM NaCl buffer containing EDTA-free complete protease inhibitor (Roche). Cells were mixed with 0.1-mm glass beads and lysed in a Fast-Prep machine twice for 45 s at setting 6 (MP Biomedicals). Samples were centrifuged for 5 min at 17,000 × g and subsequently for 1 h at 100,000 × g to obtain cytoplasmic protein extracts. Forty microliters streptavidin dynabeads (Invitrogen) coupled with 2.4 μM biotinylated c-di-AMP (BioLog) were incubated with 1.2 mg cytoplasmic proteins in 1.5 mL 10% (vol/vol) glycerol, 1 mM MgCl2, 5 mM Tris (pH 7.5), 230 mM NaCl, 0.5 mM DTT, and 4 mM EDTA containing 50 μg/mL BSA for 30 min at room temperature. Samples were washed four times with the same buffer lacking BSA and suspended in 50 μL protein sample buffer. Samples were boiled for 5 min, beads removed, and 18 μL run on 12% (wt/vol) SDS/PAGE gels. Gels were stained using the SilverQuest kit (Invitrogen). Mass spectrometry was performed at the Taplin Mass Spectrometry Facility (Harvard Medical School).
Protein Purifications.
Proteins were purified from 0.5 to 4 L E. coli cultures. Cultures were grown to an OD600 of 0.5–0.7, protein expression induced with 0.5 mM IPTG, and incubated overnight at 16 °C. Protein purifications were performed by nickel affinity and size exclusion chromatography as previously described (11, 39). Protein concentrations were determined by A280 readings.
Minimum Inhibitory Concentrations.
Overnight cultures of S. aureus strains in CDM containing 2.5 mM KCl were adjusted to 5 × 105 bacteria/mL in CDM supplemented with either 2.5 or 250 mM KCl. One hundred microliters of these suspensions were incubated in 96-well plates with twofold dilutions of nigericin starting at 6.25 μM. Plates were incubated at 37 °C with shaking for 24 h. Minimum inhibitory concentrations (MICs) were determined as the antimicrobial concentration at which growth was inhibited by >75% compared with growth without antimicrobial. Five independent experiments were performed, and one representative graph is shown.
Bacterial Stress Testing.
Overnight cultures of S. aureus strains in CDM containing 2.5 mM KCl were washed three times in CDM lacking K+. Cultures were adjusted to an OD600 of 0.05, serially diluted, and 5 μL spotted onto CDM agar plates containing an extra 0.75 M NaCl. Plates were incubated at 37 °C for 24–36 h.
Construction of the S. aureus ORFeome Expression Library.
A total of 2,343 E. coli strains containing pDONR221 vectors with S. aureus strain COL ORFs (BEI Resources, NIAID, NIH) were grown in 1.5 mL LB-M9 in 2-mL 96-well deep dishes (Greiner) selecting for kanamycin resistance. The cultures were centrifuged and the plasmids extracted using 96-well MultiScreenHTS PLASMID plates (Millipore). The S. aureus gateway ORFeome library was shuttled from the pDONR221 entry plasmids into the protein overexpression destination vector pDEST17 using LR clonase enzyme II as per the manufacturer’s guidelines (Invitrogen). Subsequently, the destination plasmid library was introduced into E. coli strain T7IQ (NEB) selecting for carbenicillin resistance.
Preparation of E. coli Whole-Cell Lysates.
BL21(DE3) pET28b-containing strains or T7IQ pDEST17 containing library expression strains were grown in LB-M9 medium overnight at 30 °C and subsequently induced for 6 h with 1 mM IPTG for protein induction. Bacteria were collected by centrifugation and suspended in 1/10 of their original volume in 40 mM Tris (pH 7.5), 100 mM NaCl, 10 mM MgCl2 binding buffer containing 2 mM PMSF, 20 μg/mL DNase, and 0.5 mg/mL lysozyme. Cells were lysed by thre freeze/thaw cycles. Lysates were directly used in binding assays or stored at −20 °C.
Differential Radial Capillary Action of Ligand Assay.
The principle of the DRaCALA is described by Roelofs et al. (27). Briefly, E. coli whole-cell lysates, 20 μM purified protein (for standard assays), or 12.5 μM protein (for competition assays) in binding buffer were mixed with ∼1 nM 32P-labeled c-di-AMP, synthesized as described in SI Methods, or 5.5 nM 32P-labeled ATP and incubated at room temperature for 5 min. For the whole-genome screen the 32P-labeled c-di-AMP was dispensed into lysate-containing 96-well plates using a Multiflo Microplate Dispenser (BioTek) and the mixture spotted onto nitrocellulose membrane using a 96-well pin tool (V&P Scientific). For competition assays, 100 or 400 μM cold nucleotides [ATP, GTP, cAMP, cGMP, NAD, NADH, NADP, NADPH (Sigma); c-di-AMP, c-di-GMP (BioLog)] were added to the initial mixture and 2.5 μL of reactions were spotted onto nitrocellulose membranes (Amersham Hybond-ECL; GE Healthcare), air-dried, and radioactivity signals detected as described above. The fraction of ligand bound and Kd values were calculated as previously described (27).
Supplementary Material
Acknowledgments
This research was supported by European Research Council Grant 260371 and Wellcome Trust Grant 100289 (to A.G.) and European Molecular Biology Organisation Short-Term Fellowship 401-2011 (to R.M.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300595110/-/DCSupplemental.
References
- 1.McDonough KA, Rodriguez A. The myriad roles of cyclic AMP in microbial pathogens: From signal to sword. Nat Rev Microbiol. 2012;10(1):27–38. doi: 10.1038/nrmicro2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marden JN, Dong Q, Roychowdhury S, Berleman JE, Bauer CE. Cyclic GMP controls Rhodospirillum centenum cyst development. Mol Microbiol. 2011;79(3):600–615. doi: 10.1111/j.1365-2958.2010.07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalebroux ZD, Swanson MS. ppGpp: Magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10(3):203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 4.Witte G, Hartung S, Büttner K, Hopfner KP. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol Cell. 2008;30(2):167–178. doi: 10.1016/j.molcel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 5.Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science. 2010;328(5986):1703–1705. doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149(2):358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488(7413):680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 11.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 2011;7(9):e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamegaya T, Kuroda K, Hayakawa Y. Identification of a Streptococcus pyogenes SF370 gene involved in production of c-di-AMP. Nagoya J Med Sci. 2011;73(1-2):49–57. [PMC free article] [PubMed] [Google Scholar]
- 13.Bai Y, et al. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS ONE. 2012;7(4):e35206. doi: 10.1371/journal.pone.0035206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Römling U. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci Signal. 2008;1(33):pe39. doi: 10.1126/scisignal.133pe39. [DOI] [PubMed] [Google Scholar]
- 15.Bejerano-Sagie M, et al. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell. 2006;125(4):679–690. doi: 10.1016/j.cell.2006.03.039. [DOI] [PubMed] [Google Scholar]
- 16.Rao F, et al. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J Biol Chem. 2010;285(1):473–482. doi: 10.1074/jbc.M109.040238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 2011;12(6):594–601. doi: 10.1038/embor.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo Y, Helmann JD. Analysis of the role of Bacillus subtilis σ(M) in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol Microbiol. 2012;83(3):623–639. doi: 10.1111/j.1365-2958.2011.07953.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rallu F, Gruss A, Ehrlich SD, Maguin E. Acid- and multistress-resistant mutants of Lactococcus lactis: Identification of intracellular stress signals. Mol Microbiol. 2000;35(3):517–528. doi: 10.1046/j.1365-2958.2000.01711.x. [DOI] [PubMed] [Google Scholar]
- 20.Pozzi C, et al. Methicillin resistance alters the biofilm phenotype and attenuates virulence in Staphylococcus aureus device-associated infections. PLoS Pathog. 2012;8(4):e1002626. doi: 10.1371/journal.ppat.1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L, Li W, He ZG. DarR, a TetR-like transcriptional factor, is a cyclic di-AMP-responsive repressor in Mycobacterium smegmatis. J Biol Chem. 2013;288(5):3085–3096. doi: 10.1074/jbc.M112.428110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holtmann G, Bakker EP, Uozumi N, Bremer E. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J Bacteriol. 2003;185(4):1289–1298. doi: 10.1128/JB.185.4.1289-1298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albright RA, Ibar JL, Kim CU, Gruner SM, Morais-Cabral JH. The RCK domain of the KtrAB K+ transporter: Multiple conformations of an octameric ring. Cell. 2006;126(6):1147–1159. doi: 10.1016/j.cell.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 24.Hänelt I, et al. KtrB, a member of the superfamily of K+ transporters. Eur J Cell Biol. 2011;90(9):696–704. doi: 10.1016/j.ejcb.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417(6888):515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 26.Dong J, Shi N, Berke I, Chen L, Jiang Y. Structures of the MthK RCK domain and the effect of Ca2+ on gating ring stability. J Biol Chem. 2005;280(50):41716–41724. doi: 10.1074/jbc.M508144200. [DOI] [PubMed] [Google Scholar]
- 27.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc Natl Acad Sci USA. 2011;108(37):15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaudhuri RR, et al. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH) BMC Genomics. 2009;10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fey PD, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4(1):e00537–e12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bae T, et al. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci USA. 2004;101(33):12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epstein W. The roles and regulation of potassium in bacteria. Prog Nucleic Acid Res Mol Biol. 2003;75:293–320. doi: 10.1016/s0079-6603(03)75008-9. [DOI] [PubMed] [Google Scholar]
- 32.Xue T, You Y, Hong D, Sun H, Sun B. The Staphylococcus aureus KdpDE two-component system couples extracellular K+ sensing and Agr signaling to infection programming. Infect Immun. 2011;79(6):2154–2167. doi: 10.1128/IAI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnesano F, et al. The evolutionarily conserved trimeric structure of CutA1 proteins suggests a role in signal transduction. J Biol Chem. 2003;278(46):45999–46006. doi: 10.1074/jbc.M304398200. [DOI] [PubMed] [Google Scholar]
- 34.Lohkamp B, McDermott G, Campbell SA, Coggins JR, Lapthorn AJ. The structure of Escherichia coli ATP-phosphoribosyltransferase: Identification of substrate binding sites and mode of AMP inhibition. J Mol Biol. 2004;336(1):131–144. doi: 10.1016/j.jmb.2003.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Ninfa AJ, Atkinson MR. PII signal transduction proteins. Trends Microbiol. 2000;8(4):172–179. doi: 10.1016/s0966-842x(00)01709-1. [DOI] [PubMed] [Google Scholar]
- 36.Zhu H, et al. Global analysis of protein activities using proteome chips. Science. 2001;293(5537):2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- 37.Gründling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104(20):8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudin L, Sjöström JE, Lindberg M, Philipson L. Factors affecting competence for transformation in Staphylococcus aureus. J Bacteriol. 1974;118(1):155–164. doi: 10.1128/jb.118.1.155-164.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu D, et al. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc Natl Acad Sci USA. 2009;106(5):1584–1589. doi: 10.1073/pnas.0809020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.