Abstract
The McMurdo Dry Valleys are the largest ice-free region in Antarctica and are critically at risk from climate change. The terrestrial landscape is dominated by oligotrophic mineral soils and extensive exposed rocky surfaces where biota are largely restricted to microbial communities, although their ability to perform the majority of geobiological processes has remained largely uncharacterized. Here, we identified functional traits that drive microbial survival and community assembly, using a metagenomic approach with GeoChip-based functional gene arrays to establish metabolic capabilities in communities inhabiting soil and rock surface niches in McKelvey Valley. Major pathways in primary metabolism were identified, indicating significant plasticity in autotrophic, heterotrophic, and diazotrophic strategies supporting microbial communities. This represents a major advance beyond biodiversity surveys in that we have now identified how putative functional ecology drives microbial community assembly. Significant differences were apparent between open soil, hypolithic, chasmoendolithic, and cryptoendolithic communities. A suite of previously unappreciated Antarctic microbial stress response pathways, thermal, osmotic, and nutrient limitation responses were identified and related to environmental stressors, offering tangible clues to the mechanisms behind the enduring success of microorganisms in this seemingly inhospitable terrain. Rocky substrates exposed to larger fluctuations in environmental stress supported greater functional diversity in stress-response pathways than soils. Soils comprised a unique reservoir of genes involved in transformation of organic hydrocarbons and lignin-like degradative pathways. This has major implications for the evolutionary origin of the organisms, turnover of recalcitrant substrates in Antarctic soils, and predicting future responses to anthropogenic pollution.
The largest ice-free regions on the Antarctic continent are the McMurdo Dry Valleys, designated by international treaty as an Antarctic Special Managed Area (1) to reflect their environmental significance. The Dry Valleys are among the most threatened environments from climate change due to their polar location and unique ecology (2, 3). The terrestrial landscape is dominated by oligotrophic mineral soils (4) and extensive rocky outcrops with life restricted mainly to microbial communities due to the extreme environmental stress (5).
Extensive recent research has focused on elucidating the biodiversity of Dry Valleys landscapes (5–16). Classical microbiological studies identified edaphic Antarctic taxa by morphology and revealed a general recalcitrance to cultivation (17, 18). Molecular interrogations greatly expanded understanding of taxonomic community structure (5–16, 19) and speculated on the origin of inocula such as from streams that freeze dry and subsequently are dispersed by wind in winter (5).
These and other studies have provided major insight into the biodiversity of soil and rock niches largely from the rRNA gene perspective, although a caveat to any solely molecular study concerning biodiversity and functionality is that DNA can be isolated from nonviable propagules (20) or from inactive material blown in from elsewhere such as dessicated microbial mats on streams, dry lake beds, glaciers, coastal ice, and wetlands or brought in by snowfall (21). A limited number of in situ respirometry studies also indicated microbial contributions to carbon and nitrogen transformations may be significant (22, 23). A picture of the extent of bacterial colonization has emerged, but also soil and rock niches support algae (24), fungi (20), lichen (25), mosses (26), and invertebrates (27). A high degree of niche specialization in terms of rRNA gene-defined community assembly has been recorded between open soil, hypolith (colonized ventral surface of quartz), chasmoendolith (colonized cracks and fissures in sandstone and granite), and cryptoendolith (colonized pore spaces in weathered sandstone) (5).
Despite this, almost nothing is known about the contribution of Dry Valleys microbial communities in soils and rocks toward metabolic processes essential to biological mineral transformation and the stress tolerance mechanisms that allow them to flourish in such harsh extremes. The less extreme sub-Antarctic and maritime peninsula Antarctic locations received relatively greater attention, and the GeoChip-based functional gene arrays were successfully applied to indicate pathways and taxonomic identity of soil microbial carbon and nitrogen utilization (28–31) but they represent a fundamentally different biome compared with the extreme polar desert of the Dry Valleys ecosystem (32).
Here, we present findings from a comprehensive study using GeoChip to address key issues related to microbial contributions in geobiological processes, including carbon cycling genes, i.e., involved in autotrophy, acetogenesis, and methanogenesis; nitrogen cycling genes, i.e., nitrification and assimilatory and dissimilatory nitrogen reductions; and also, importantly, the stress tolerance strategies available to microbes to survive in these harsh environmental conditions. The data were compared with PCR-based diversity of 16S rRNA genes and biomass estimates and demonstrated autotrophic, heterotrophic, and diazotrophic pathways in Antarctic Dry Valleys microorganisms. Strikingly, we identified unique stress response pathways that can be directly related to environmental stressors in this Antarctic environment. Finally, we highlighted soil microbial pathways that indicate a potential ability to transform lignin-like molecules and anthropogenic pollutants and discussed this in view of the evolutionary origin of the microorganisms, increasing human exposure, and potential future contamination in the Dry Valleys system.
Results
Overall Differences of Microbial Communities.
Communities that displayed niche-specific metabolic potential output from the array data were grouped into functional categories related to major metabolic processes. The level of redundancy in large number of pathway-specific GeoChip oligonucleotides allowed a high degree of confidence in signal recovery inferring occurrence of any given pathway (33). GeoChip 4 contains about 84,000 50-mer oligonucleotide probes covering 152,000 gene variants (i.e., individual sequences from a gene) from 401 distinct functional gene categories involved in major biogeochemical, ecological, and other processes. Among these probes, 35,858 probes were derived from genes involved in carbon, nitrogen cycling, and stress responses. Hybridization of DNA from the four niches was achieved with an average of 45.1% of the 84,000 probes, covering an average of 91.8% of the targeted genes of interest on GeoChip 4 (Tables S1 and S2). For determining diversity, GeoChip utilized the highly specific probes targeting DNA gyrase subunit B (gyrB), which are functional genes with higher evolutional rates than 16S rRNA genes. GyrB can achieve taxonomic resolution at the species-strain level (34–37), which is higher than that of 16S rRNA gene at the genus-family level. GeoChip hybridization revealed higher phylogenetic diversity in the samples than the 16S rRNA gene cloned library (Table S3). The results differ between the two techniques due to probe specificity.
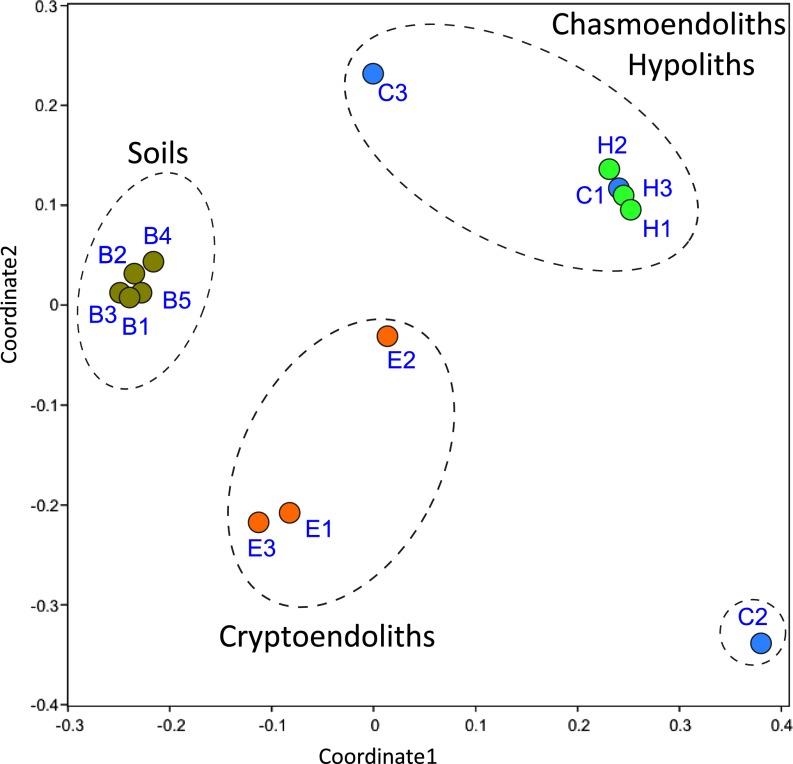
The microarray analyses revealed that all colonized niches (open soil, hypoliths, chasmoendoliths, and cryptoendoliths), encountered in McKelvey Valley, supported a functional diversity that included photoautotrophic, heterotrophic, diazotrophic, and stress response pathways. Ordination and cluster analyses of the overall functional gene profiles indicated significant variations between the communities from each of the edaphic niches analysis of similarity (ANOSIM) global R = 0.784, P = 0.0001, n = 14, permutation = 9,999] (Fig. 1). The two most exposed niches (hypolithic and chasmoendolithic) shared greatest similarity. Pairwise ANOSIM comparisons indicated the most significant differences occurred between soil and the three lithic niches (P = 0.01). A permutational multivariate analysis of variance (PERMANOVA) test also supported the same conclusion that overall soil metabolic potential was statistically different from all three lithic niches (P < 0.05). A similarity percentage (SIMPER) analysis was performed to assess which genes were primarily responsible for the observed difference between soils and the lithic niches. Overall, the most striking differences (59% of variation) between soil and lithic samples were due to the presence in soil communities of pathways involved in transformation of complex aromatic compounds (Tables S1 and S2 and Fig. S1), and variation in bacteriophage, metal resistance, and stress response gene categories each contributed around 7–10% of the total difference (Tables S1 and S2 and Fig. S1).
Fig. 1.
Nonmetric multidimensional scaling (NMDS) plot of Bray–Curtis similarities for the overall functional gene profiles (2D stress value = 0.1132). The low stress value implies little loss of information for the reduction of data into two dimensions. Other ordination analyses (de-trended correspondence analysis, correspondence analysis, and principal components analysis) also inform the same statistically significant clustering of samples.
Because many probes on GeoChip 4 are designed on the basis of the gene sequences from pure cultures of known phylogeny or from environmental sequences of known taxonomic groups based on the National Center for Biotechnology Information database, the hybridization data can be used to assess phylogenetic composition and structure of microbial communities. For example, for nitrogenase reductase gene (nifH), there are 764 sequence-specific probes and 460 group-specific probes, which covered 2,223 coding sequences. About 89.1% of the probes were from 4,332 bacterial strains (27 phyla), 3.1% from 188 archaeal strains (4 phyla), 6.1% from 420 eukaryotic strains (15 fungal phyla), and 1.3% from 273 bacteriophages (five orders). Thus, the phylogenetic nature of the data acquisition also enabled confident assignment of metabolic capabilities to specific bacterial, archaeal, or eukaryal phyla in each niche and biodiversity estimates that were compared with sequence-based assessments from extensive clone libraries of the same samples (5). Although many different types of genes were detected, in this study, we are most interested in microbial diversity important to carbon, nitrogen cycling, and survival strategies.
Carbon Utilization.
In an oligotrophic environment such as the Dry Valleys (samples from McKelvey Valley had total organic carbon mean ∼0.15 g/100 g soil and negligible carbon in rock interspace; Table S4), carbon sequestration is a major challenge and hence autotrophy is important. The key enzymes in autotrophic carbon fixation pathways used to indicate autotrophy include the ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCo) in the Calvin–Benson–Bassham cycle, the propionyl-CoA/acetyl-CoA carboxylase (pcc) in the 3-hydroxypropionate/malyl-CoA cycle, the ATP citrate lyase (aclB), and the carbon-monoxide dehydrogenase (CODH) in the reductive acetyl-CoA pathway; these genes were detected in all of the samples (Fig. S2). We assigned possession of Functional Form I RubisCo as indicative of photoautotrophic potential, which was largely indicated as cyanobacteria (Fig. 2A). Functional Forms II and III RubisCo were indicated as mainly in Archaea, Actinobacteria, and Proteobacteria (Fig. 2A) and suggested a significant capability in chemoautotrophy.
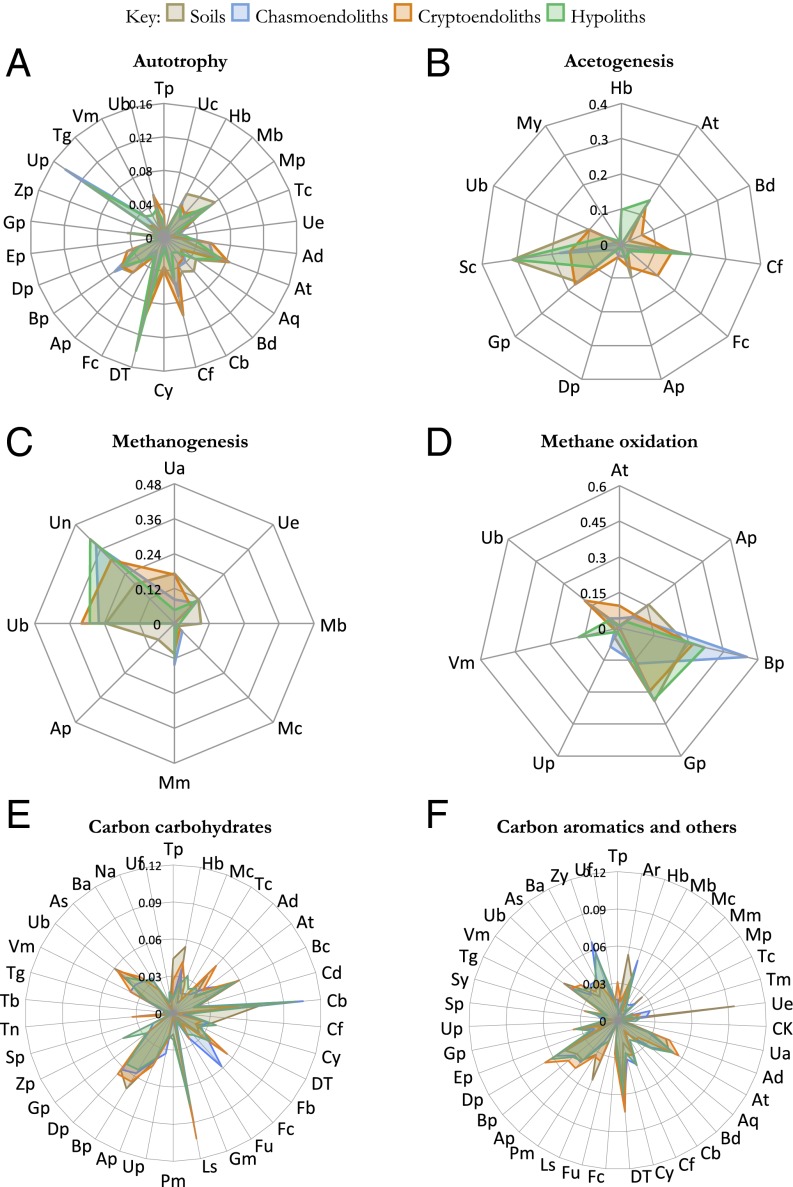
Fig. 2.
Taxa–function relationships for carbon-cycling genes. Relative signal intensity was normalized for the number of probes per taxon. Mean values of samples from respective niches (soils, hypoliths, chasmoendoliths, and cryptoendoliths) were plotted for the carbon-cycling genes involved in (A) autotrophy, (B) acetogenesis, (C) methanogenesis, (D) methane oxidation, (E) carbon carbohydrates, and (F) carbon aromatics and others. Table S5 gives two-character codes for the taxa.
We identified the presence of genes indicating acetogenesis particularly in Actinobacteria, Chloroflexi, Gammaproteobacteria, and Spirochaetes (Fig. 2B). In addition, we identified C1 pathways involving methanogenesis (Methanococci, Methanomicrobia, and unidentified archaeal taxa, Fig. 2C) and proteobacterial methane oxidation (Fig. 2D). The ability to transform various organic polymers including starch, pectin, wood polymers, and complex aromatic substrates was indicated and the ability to carry out simple and complex carbohydrate catabolism was present in 34 phyla across all domains (Fig. 2E), whereas the ability to catabolize complex aromatic compounds was indicated mainly by Actinobacteria, Deinococci, proteobacterial phyla, and both ascomycete and basidiomycete fungi (Fig. 2F). The range of detected pathway-specific genes indicated the potential to transform lignin-like compounds and other naturally occurring polyaromatics and also a range of xenobiotic aromatics including halogenated compounds, a capability demonstrated by white-rot basidiomycetes (38).
Nitrogen Utilization.
Soil-combined nitrogen levels for McKelvey Valley samples were in the order of 0.05 g/100 g soil and undetectable in rock interspaces (Table S4). Our functional analysis detected genes critical to all of the major pathways of the nitrogen cycle (Fig. 3 and Fig. S3). The data indicated diazotrophic ability and, therefore, nitrogen input to this Dry Valleys system largely by Actinobacteria, Cyanobacteria, and Alpha- and Epsilonproteobacteria (Fig. 3A). Mineralization ability (introduction of nitrate via decomposition) was indicated by basidiomycete fungi as well as Actinobacteria, Deinococci, and Alpha- and Betaproteobacteria (Fig. 3B).
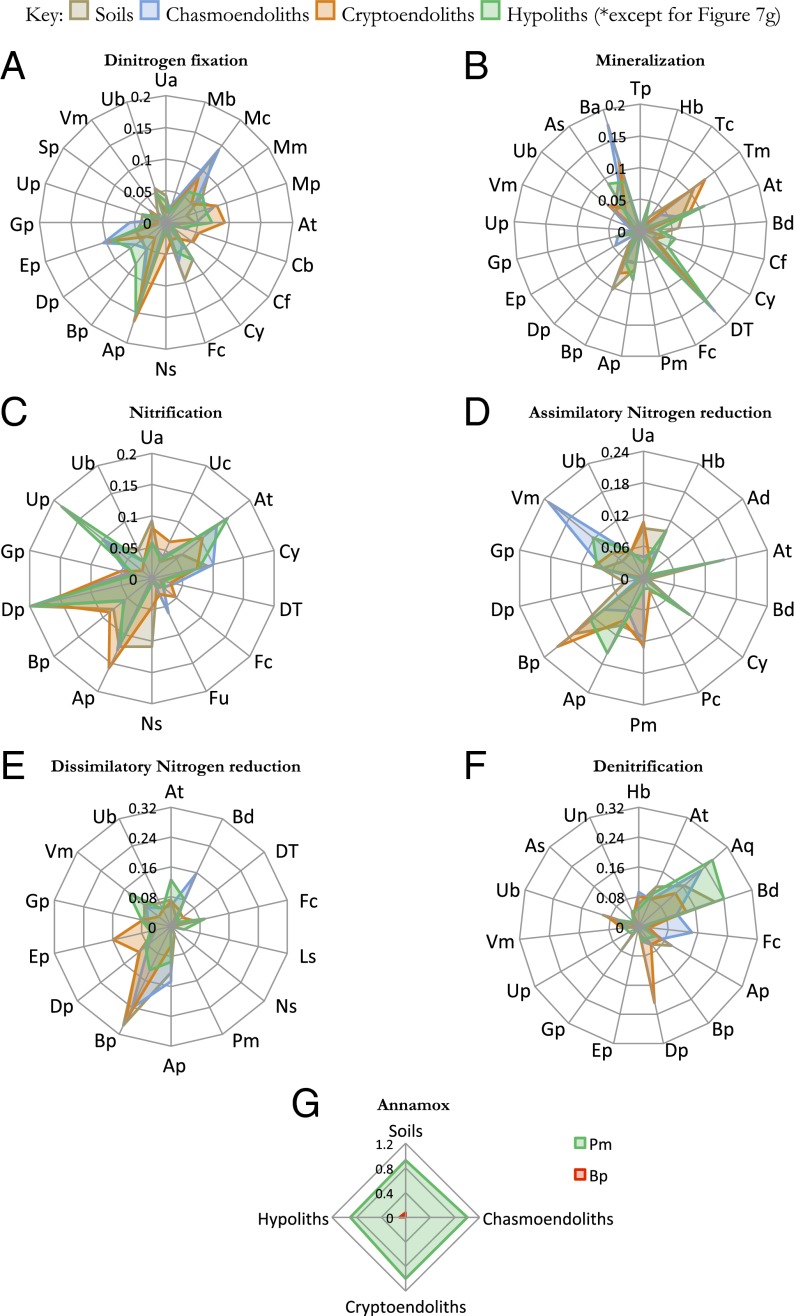
Fig. 3.
Taxa–function relationships for nitrogen-cycling genes. Relative signal intensity was normalized by the number of probes per taxon. Mean values of samples from respective niches (soils, hypoliths, chasmoendoliths, and cryptoendoliths) were plotted for the nitrogen-cycling genes involved in (A) dinitrogen fixation, (B) mineralization, (C) nitrification, (D) assimilatory nitrogen reduction, (E) dissimilatory nitrogen reduction, (F) denitrification, and (G) anaerobic ammonium oxidation (annamox). Table S5 gives two-character codes for the taxa.
The most abundant nitrifiers (oxidation of ammonium ions from nitrogen fixation and mineralization into nitrate) were Actinobacteria, Cyanobacteria, and Alpha- and Deltaproteobacteria (Fig. 3C). Anoxic pathways that lead to nitrate loss from soils were also detected, as well as anoxic nitrogen assimilation (Fig. 3D). Dissimilatory nitrate reduction was indicated largely by Betaproteobacteria (Fig. 3E). The potential for soil nitrate removal via denitrification and anaerobic ammonium oxidation (annamox) pathways was also indicated, although these were very phylum-specific to Bacteriodetes and Deltaproteobacteria (denitrifiers, Fig. 3F) and Betaproteobacteria and Planctomycetes (annamox, Fig. 3G).
Stress Response.
Almost nothing is known of stress response in edaphic Antarctic microbial communities. This study provides unique evidence that across niches a range of pathways are present in colonists, which include pathway-specific genes and σ-factors for response to osmotic, radiation (desiccation), cold-shock, heat-shock, and nutrient stress (Fig. 4 and Fig. S4). Osmotic shock pathways were largely attributed to Actinobacteria and the Proteobacteria and were abundant in rocky substrates (Fig. 4A). In soils and hypoliths in soil contact, a broader range of phyla possessed these genes. Radiation stress genes were used as a proxy for desiccation-tolerance pathway genes, and these generally occurred in phyla that were common in all niches.
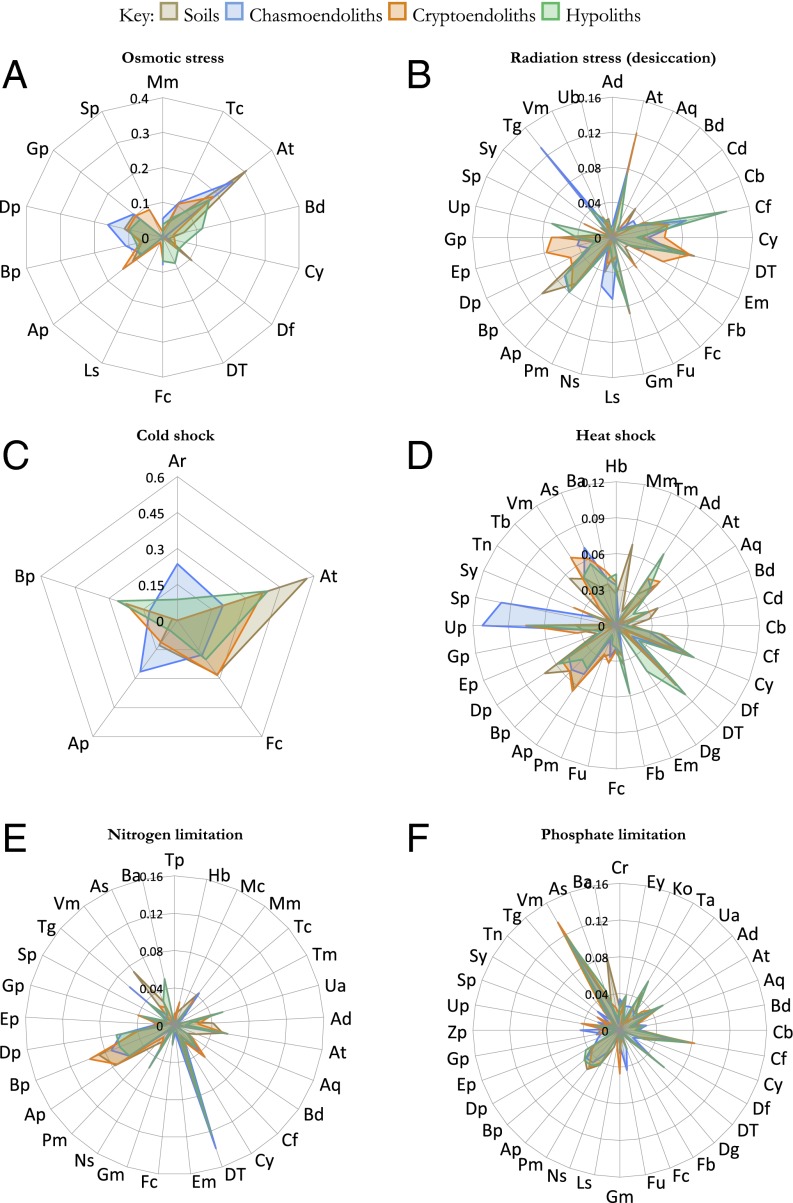
Fig. 4.
Taxa–function relationships for stress response genes. Relative signal intensity was normalized by the number of probes per taxon. Mean values of samples from respective niches (soils, hypoliths, chasmoendoliths, and cryptoendoliths) were plotted for the genes responsive to (A) osmotic stress, (B) radiation (desiccation) stress, (C) cold shock, (D) heat shock, (E) nitrogen limitation, and (F) phosphate limitation. Table S5 gives two-character codes for the taxa.
Hypolithic communities supported the most diverse range of taxa with desiccation-tolerance genes (Fig. 4B). Interestingly, cold-shock genes in these Antarctic communities were detected exclusively among Actinobacteria, Firmicutes, and the Alpha- and Betaproteobacteria (Fig. 4C). Heat-shock pathway genes were more common among phyla, including bacteria and fungi (Fig. 4D).
Genes indicative of pathways that respond to nitrogen limitation were relatively widespread, but particularly abundant among the Alpha- and Betaproteobacteria and Deinococci (Fig. 4E). Conversely, pathways related to phosphate limitation were more common in the Chloroflexi and Verrucomicrobia (Fig. 4F). The microarray also identified a large number of loci indicative of pathways encoding resistance to antibiotics and metals.
Discussion
In this study, we have revealed the autotrophic and heterotrophic strategies that support microbial colonization in soil and rock niches of the coldest and driest desert on Earth. This represents a major advance beyond biodiversity surveys in that we have identified how putative functional ecology drives microbial community assembly in this unique and threatened biome. The linkage of biogeochemically important metabolic pathways/genes with their phylogenetic identity revealed how these unique polar desert communities assemble due to functional adaptation in soil and rocky niches. The latter is of particular relevance to desert systems, where endolithic, and hypolithic colonization is extensive and often accounts for equal or greater productivity than that of soils (39). This greatly improves our understanding of the factors shaping microbial community assembly.
Functional metagenomic approaches have been successfully applied to understand temperate and tundra soils (40–42) but the present evidence is unique for functional ecology among the full range of edaphic niches in polar desert. Given that we have also previously shown that some phyla in these niches experience very limited gene flow at the rRNA level over extended timescales (43) and consequently a high degree of stochastic demography (44), the source of variation in functional genes and pathways should be considered. It is not possible to quantify the level of lateral gene transfer in these communities on the basis of our data, although the comparison between rRNA-defined taxonomy and functional diversity indicates that a high degree of predictive power is possible in terms of using taxonomic markers as indicators of functionality within a community, as recently observed in other ecosystems (45, 46). The level of exposure to environmental extremes and fluctuation appears to be a strong driver of functional ecology.
We have clearly shown that stress response pathways are more abundant and diverse in above-ground exposed niches (hypoliths and chasmoendoliths) compared with sheltered soils and cryptoendolithic substrates, and this can be related to the measurements of thermal and moisture stress and UV exposure (47). Diverse mechanisms of adaptation were also demonstrated in hyporheic cyanobacterial mats in the Arctic and Antarctic, with the former showing dominance of copper homeostasis genes and the latter showing more sigma B genes, as we observed, which were considered to be due to the severe osmotic stress during freeze-up of Antarctic ponds (48). This indicates that recruitment to exposed niches is strongly based on selection for stress tolerance traits and that these communities have a more diverse “arsenal” of responses with which to counter environmental extremes than more sheltered niches. These offer tangible clues to the mechanism behind the enduring success of microorganisms in this seemingly inhospitable terrain.
The level of similarity in core primary carbon and nitrogen metabolism between all niches supports the notion of a “blanket” of microbial productivity covering desert surfaces (39). We conclude that the Cyanobacteria, although ubiquitous, are likely also accompanied by proteobacterial and algal photoautotrophy in these systems. High abundance of a member of the Alphaproteobacteria was previously identified in cyptoendolithic cyanobacteria from Beacon sandstone in the McMurdo Dry Valleys (19). Our analysis revealed that several common edaphic phyla also possessed pathways for chemoautotrophy, expanding the knowledge of autotrophic carbon input sources in this oligotrophic system. This observation has parallels with observations of other oligotrophic microbial habitats, e.g., in oligotrophic oxygen-minimum zones of the ocean (49). Clearly, the most abundant phyla in the Dry Valleys comprise both heterotrophic and autotrophic strategies to mitigate low organic availability in this system.
A further limitation to colonization is nitrogen transformation, because this is a limiting nutrient in these systems’ input attributed to cyanobacterial diazotrophy (5, 23, 50) and also snow, as demonstrated in the East Antarctic snowpack (51). We identified that a “complete” pathway for nitrogen metabolism occurs, which indicates that during environmental conditions favorable to annamox and denitrification, net microbially mediated nitrogen loss from the system may occur. These may be envisaged to occur during the austral summer in waterlogged soils (from snowmelt). Cyanobacterial diazotrophy has traditionally been viewed to occur as a “dark” reaction due to oxygen inhibition during photosynthesis (52). However, this clearly cannot occur during a 24-h daylight cycle of the austral summer, and so interesting regulations on this pathway are likely to emerge. Ongoing research into circadian systems and their influence on this may shed additional light (53). Annamox pathways have received significant attention in other habitats, and it is of note that Antarctic soils also have potential for this via the same restricted group of Betaproteobacteria and Planctomycetes (and Archaea) as in other temperate and tropical habitats (54, 55).
The range of stress response pathways revealed by our study suggests how the Antarctic soil microflora are adapted to environmental extremes. We highlight not only stress responses directly related to moisture, radiation, and thermal shock but also others that may offer clues to community regulation. For example, the identification of antibiotic resistance genes indicates indirectly a level of competitive interactions between taxa. Similarly, shotgun metagenomic sequencing analyses of Antarctic soil metagenomes also demonstrated the presence of antibiotic resistance genes (16).
We also identified a large frequency of phage signals that suggests viral controls on population numbers may be important in this system where grazers (and hence controls via higher trophic levels) are largely absent, a role previously indicated by metaproteogenomic analysis of a hypersaline Antarctic lake (56). This notion has also been suggested as a regulatory process in hydrothermal systems, where microbial communities also dominate in the near absence of higher plants or animals (57) and so may emerge as a key community regulator in extreme systems generally. The identification of stress response signals at relatively high frequency among those taxa that were most numerically abundant in communities suggests a direct correlation may exist between the ability to withstand stress and fitness in these niches. It is also interesting to speculate the extent to which these possible keystone taxa also benefit and facilitate colonization and survival by other taxa because some stress responses (e.g., secretion of extracellular polymers for water retention) may have broader benefit to the community (58, 59).
Of interest was the observation of pathways for transformation of lignin-like, complex organic aromatic substrates, because these are among the more recalcitrant of soil substrates (4, 60) and can be envisaged to be widely available in mineral soils with low turnover and exogenous input. That these were common to soils but not to rocky niches offers further evidence that they have been selected for on this basis, because rocky niches are not reservoirs for the “legacy” carbon that is known to endure in Antarctic soils (4). Our results of lignin-like degradative pathways are supported by the findings of wood-degrading fungi in a wide variety of Antarctic soils (20, 61, 62). A link has been suggested between these saprophytic fungi and those found in the fossil record of the Triassic and Jurassic forests of Antarctica (61). Fungi are notorious contaminants especially on imported materials and around sites of human activity and the difficulty of discerning transient/introduced vs. indigenous and endemic organisms and their input to the environment is acknowledged, although indigenous and not exotic fungi dominate on imported wood (62). The structural similarities between lignins and other naturally occurring complex organics and many xenobiotic compounds led us to propose that because of the presence of these fungi, Antarctic soils may possess an innate ability to respond to future pollution threats from these compounds (38). We hypothesize that due to their physiological robustness, fungi may emerge as highly important to Antarctic geobiological processes, and ongoing study will reveal the nature of their contributions.
With this microarray approach, we have been able to identify a broad range of metagenomic “potential” with the most relevant focus on functional ecology in this system comprising the very distinct niches of an Antarctic landscape. Caveats to the study include the fact that given the endemicity of Antarctic communities, we may have underestimated stress responses that use pathways with novel evolutionary lineages. Another caveat, high variation between chasmoendoliths, indicates high levels of variation among the samples (the variation between chasmoendoliths replicates is even larger than among the niches), which may be explained by the niche characteristics, where the cracks in rocks that open to the surface have a microenvironment of steep light, moisture, and nutrient gradients, compared with the homogenous properties in the rock matrix for cryptoendoliths or under the rock for hypoliths or the soils (63). Long-term field deployments will provide a temporal perspective and, combined with expression profiles and respirometry/productivity estimates, will further advance our understanding. Nonetheless, this study provides comprehensive baseline information on functional capacity in threatened Dry Valleys niches, in a more focused manner relevant to the environment than other desert soil studies have yet been able to achieve.
Conclusions
Antarctic landscapes have emerged as far more biodiverse than previously envisaged as a result of microbial surveys. We conclude that rRNA estimates identified the dominant and endemic phylotypes that are underrepresented in databases, whereas GeoChip detected greater diversity at a broader taxonomic rank. Taken together, the two estimates are complementary and together yield a more complete view of diversity in this system. Critically, this study identified the functional traits of organisms that drive community assembly and interactions. The insights also expand our knowledge of how such communities tolerate extreme environmental stress and their capacity to respond to future changes, including climate and human impacts.
Materials and Methods
Field Sampling and Abiotic Variables.
Edaphic colonization in McKelvey Valley (central valley coordinates 77°26′ S, 161°33′ E) was surveyed during Antarctica New Zealand Event K021 in January 2008. Surface soils, sandstone, and quartz substrates were surveyed as previously described (5) and frequency of colonization for chasmoendoliths, cryptoendoliths, and hypoliths was recorded (n = 400). Colonized soil and rock samples were recovered aseptically and stored in sterilized plastic containers with no headspace at −80 °C until processed. A suite of abiotic variables including moisture content (gravimetric), pH (potentiometric), soluble salts (potentiometric), total organic carbon, and total nitrogen (gas chromatography-thermal conductivity detector at 900 °C) was measured for each substrate as previously described (5).
GeoChip Analysis.
Recovery of environmental DNAs was with a protocol optimized for edaphic desert microorganisms (64). Similarity between communities was assessed using nonmetric multidimensional scaling of Bray–Curtis similarities from terminal restriction fragment length polymorphism analysis of 16S and 18S rRNA genes as previously described (5). Independent replicates from each niche that were most representative for a given niche were then selected for GeoChip (version 4.0) analysis (n = 14). GeoChip 4.0 contained 83,992 50-mer oligo probes that covered 152,414 gene variants from 401 distinct functional gene categories involved in major biogeochemical, ecological, and other processes (12 categories: biogeochemical cycling of carbon, nitrogen, phosphorus, and sulfur; resistance to metal and antibiotics; energy process; organic compound remediation; stress response; bacteriophage related; virulence related; and others) (65, 66). About 20 ng community DNAs from each sample was amplified (67) and 1 μg of the amplified sample was used for GeoChip hybridization as previously described (65, 66). The normalized hybridization output data were then reorganized on the basis of functional categories.
Output was analyzed using the multivariate statistical software package PRIMER-E v6 (Plymouth Marine Laboratory). Alpha diversity indexes (species richness, Shannon’s index, Simpson’s diversity index, and Pielou’s evenness) were calculated using untransformed data. Nonmetric multidimensional scaling (NMDS) ordinations were used to visualize Bray–Curtis Similarities. ANOVA, AMOVA, ANOSIM, PERMANOVA, and SIMPER analyses were performed to indicate confidence in similarities/differences observed. Visualization of different phylum-level and/or class-level contributions to each metabolic pathway was achieved using spider dendrograms, where each arm of the plot was specific to a given phylum/class.
Supplementary Material
Acknowledgments
We thank Dr. Yuan Tong for GeoChip hybridization and Brett Arenz, Joel Jurgens, and Maggie C. Y. Lau for field assistance in McKelvey Valley and helpful discussions. We also thank Jackie Aislabie, Robert Blanchette, Megan Balks, and Louis Schipper for helpful discussions. We acknowledge financial support from the Vice Chancellor’s Fund of the University of Waikato and the Institute for Applied Ecology at Auckland University of Technology. Logistical and field support was provided by Antarctica New Zealand. The development of the GeoChips and associated computational pipelines used in this study was supported by Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA) through the US Department of Energy (DE-AC02-05CH11231). J. Zhou and J. D. Van Nostrand's efforts were supported by the US Department of Energy (DE-SC0004601) and the US National Science Foundation (EF-1065844).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ490210–FJ490344 and FJ895042–FJ895089). The GeoChip dataset reported in this paper is publicly available at http://ieg.ou.edu/4download/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300643110/-/DCSupplemental.
References
- 1.Scientific Committee on Antarctic Research 2004. SCAR Bulletin #155. Available at www.scar.org.
- 2.Hogg ID, Wall DH. Global change and Antarctic terrestrial biodiversity. Polar Biol. 2011;34(11):1625–1627. [Google Scholar]
- 3.Green TGA, Sancho LG, Pintado A, Schroeter B. Functional and spatial pressures on terrestrial vegetation in Antarctica forced by global warming. Polar Biol. 2011;34(11):1643–1656. [Google Scholar]
- 4.Hopkins DW, et al. Controls on the distribution of productivity and organic resources in Antarctic Dry Valley soils. Proc Biol Sci. 2006a;273(1602):2687–2695. doi: 10.1098/rspb.2006.3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pointing SB, et al. Highly specialized microbial diversity in hyper-arid polar desert. Proc Natl Acad Sci USA. 2009;106(47):19964–19969. doi: 10.1073/pnas.0908274106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wall DH, Virginia RA. Controls on soil biodiversity: insights from extreme environments. Appl Soil Ecol. 1999;13(2):137–150. [Google Scholar]
- 7.Aislabie JM, et al. Dominant bacteria in soils of Marble Point and Wright Valley, Victoria Land, Antarctica. Soil Biol Biochem. 2006;38:3041–3056. [Google Scholar]
- 8.Barrett JE, et al. Covariation in soil biodiversity and biogeochemistry in Northern and Southern Victoria Land, Antarctica. Antarct Sci. 2006;18:535–548. [Google Scholar]
- 9.Convey P, Stevens MI. Ecology. Antarctic biodiversity. Science. 2007;317(5846):1877–1878. doi: 10.1126/science.1147261. [DOI] [PubMed] [Google Scholar]
- 10.Niederberger TD, et al. Microbial community composition in soils of Northern Victoria Land, Antarctica. Environ Microbiol. 2008;10(7):1713–1724. doi: 10.1111/j.1462-2920.2008.01593.x. [DOI] [PubMed] [Google Scholar]
- 11.Wood SA, Rueckert A, Cowan DA, Cary SC. Sources of edaphic cyanobacterial diversity in the Dry Valleys of Eastern Antarctica. ISME J. 2008;2(3):308–320. doi: 10.1038/ismej.2007.104. [DOI] [PubMed] [Google Scholar]
- 12.Cary SC, McDonald IR, Barrett JE, Cowan DA. On the rocks: The microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol. 2010;8(2):129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 13.Rao S, et al. Low-diversity fungal assemblage in an Antarctic Dry Valleys soil. Polar Biol. 2011;35(4):567–574. [Google Scholar]
- 14.Lee CK, Barbier BA, Bottos EM, McDonald IR, Cary SC. The Inter-Valley Soil Comparative Survey: The ecology of Dry Valley edaphic microbial communities. ISME J. 2012;6(5):1046–1057. doi: 10.1038/ismej.2011.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magalhães C, et al. At limits of life: Multidisciplinary insights reveal environmental constraints on biotic diversity in continental Antarctica. PLoS ONE. 2012;7(9):e44578. doi: 10.1371/journal.pone.0044578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fierer N, et al. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc Natl Acad Sci USA. 2012;109(52):21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedmann EI, Hua M, Ocampo-Friedmann R. Cryptoendolithic lichen and cyanobacterial communities of the Ross Desert, Antarctica. Polarforschung. 1988;58(2–3):251–259. [PubMed] [Google Scholar]
- 18.Nienow JA, Friedmann EI. 1993. Terrestrial lithophytic rock communities. Antarctic Microbiology, ed Friedmann EI (Wiley-Liss, New York), pp 343–412.
- 19.de la Torre JR, Goebel BM, Friedmann EI, Pace NR. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl Environ Microbiol. 2003;69(7):3858–3867. doi: 10.1128/AEM.69.7.3858-3867.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arenz BE, Held BW, Jurgens JA, Farrell RL, Blanchette RA. Fungal diversity in soils and historic wood from the Ross Sea region of Antarctica. Soil Biol Biochem. 2006;38:3057–3064. [Google Scholar]
- 21.Harding T, Jungblut AD, Lovejoy C, Vincent WF. Microbes in high arctic snow and implications for the cold biosphere. Appl Environ Microbiol. 2011;77(10):3234–3243. doi: 10.1128/AEM.02611-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopkins DW, et al. Isotopic evidence for the provenance and turnover of organic carbon by soil microorganisms in the Antarctic dry valleys. Environ Microbiol. 2009;11(3):597–608. doi: 10.1111/j.1462-2920.2008.01830.x. [DOI] [PubMed] [Google Scholar]
- 23.Cowan DA, et al. Hypolithic communities: Important nitrogen sources in Antarctic desert soils. Envir Microbiol Rep. 2011;3:581–586. doi: 10.1111/j.1758-2229.2011.00266.x. [DOI] [PubMed] [Google Scholar]
- 24.Broady PA. The distribution of terrestrial and hydro-terrestrial algal associations at three contrasting locations in southern Victoria Land, Antarctica. Algol Stud. 2005;118(1):95–112. [Google Scholar]
- 25.Green TGA, Brabyn L, Beard C, Sancho LG. Extremely low lichen growth rates in Taylor Valley, Dry Valleys continental Antarctica. Polar Biol. 2011;35(4):535–541. [Google Scholar]
- 26.Colesie C, Gommeaux M, Green TGA, Büdel B. 2013. Biological Soil Crust in continental Antarctica: Garwood Valley (Southern Victoria Land) and Diamond Hill (Darwin Mountains region). Antarct Sci, in press.
- 27.Smith TE, et al. Thawing permafrost alters nematode populations and soil habitat characteristics in an Antarctic polar desert ecosystem. Pedobiologia. 2012;55(2):75–81. [Google Scholar]
- 28.Yergeau E, Kang S, He Z, Zhou J, Kowalchuk GA. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 2007;1(2):163–179. doi: 10.1038/ismej.2007.24. [DOI] [PubMed] [Google Scholar]
- 29.Yergeau E, et al. Shifts in soil microorganisms in response to warming are consistent across a range of Antarctic environments. ISME J. 2012;6(3):692–702. doi: 10.1038/ismej.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J-Z, et al. Microbial mediation of carbon cycle feedbacks to climate warming. Nat Clim Change. 2012;2:106–110. [Google Scholar]
- 31.He ZH, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010;13(5):564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- 32.Wynn-Williams DD. Ecological aspects of Antarctic microbiology. In: Marshall K, editor. Advances in Microbial Ecology. Vol 2. New York: Plenum Press; 1990. pp. 71–146. [Google Scholar]
- 33.He ZL, Van Nostrand JD, Zhou J. Applications of functional gene microarrays for profiling microbial communities. Curr Opin Biotechnol. 2012;23(3):460–466. doi: 10.1016/j.copbio.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 34.Wu LY, et al. Development and evaluation of functional gene arrays for detection of selected genes in the environment. Appl Environ Microbiol. 2001;67(12):5780–5790. doi: 10.1128/AEM.67.12.5780-5790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee SK, et al. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl Environ Microbiol. 2004;70(7):4303–4317. doi: 10.1128/AEM.70.7.4303-4317.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He Z, Wu LY, Li X, Fields MW, Zhou J-Z. Empirical establishment of oligonucleotide probe design criteria. Appl Environ Microbiol. 2005;71(7):3753–3760. doi: 10.1128/AEM.71.7.3753-3760.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Z, et al. GeoChip: A comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISME J. 2007;1(1):67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- 38.Thwaites JM, Farrell RL, Duncan SD, Lamar RL, White RB. 2006. Environmental Bioremediation Technologies, ed Singh SN (Springer, Heidelberg), pp 465–479.
- 39.Pointing SB, Belnap J. Microbial colonization and controls in dryland systems. Nat Rev Microbiol. 2012;10(8):551–562. doi: 10.1038/nrmicro2831. [DOI] [PubMed] [Google Scholar]
- 40.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308(5721):554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 41.Delmont TO, Robe P, Clark I, Simonet P, Vogel TM. Metagenomic comparison of direct and indirect soil DNA extraction approaches. J Microbiol Methods. 2011;86(3):397–400. doi: 10.1016/j.mimet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Mackelprang R, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480(7377):368–371. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 43.Bahl J, et al. Ancient origins determine global biogeography of hot and cold desert cyanobacteria. Nat Commun. 2011;2:163. doi: 10.1038/ncomms1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caruso T, et al. Stochastic and deterministic processes interact in the assembly of desert microbial communities on a global scale. ISME J. 2011;5(9):1406–1413. doi: 10.1038/ismej.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raes J, Letunic I, Yamada T, Jensen LJ, Bork P. Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol Syst Biol. 2011;7:473. doi: 10.1038/msb.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cowan DA, et al. Distribution and abiotic influences on hypolithic microbial communities in an Antarctic dry Valley. Polar Biol. 2010;34:307–311. [Google Scholar]
- 48.Varin T, et al. Metagenomic analysis of stress genes in microbial mat communities from extreme Arctic and Antarctic environments. Appl Environ Microbiol. 2012;78:549–559. doi: 10.1128/AEM.06354-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward BB, Kilpatrick KA, Renger E, Eppley RW. Biological nitrogen cycling in the nitracline. Limnol Oceanogr. 1989;34:493–513. [Google Scholar]
- 50.Niederberger TD, et al. Diverse and highly active diazotrophic assemblages inhabit ephemerally wetted soils of the Antarctic Dry Valleys. FEMS Microbiol Ecol. 2012;82(2):376–390. doi: 10.1111/j.1574-6941.2012.01390.x. [DOI] [PubMed] [Google Scholar]
- 51.France JL, King MD, Frey MM. Snow optical properties at Dome C (Concordia), Antarctica; implications for snow emissions and snow chemistry of reactive nitrogen. Atmos Chem Phys. 2011;11(18):9787–9801. [Google Scholar]
- 52.Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev. 1992;56(2):340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dvornyk V, Jahan AS. Extreme conservation and non-neutral evolution of the cpmA Circadian locus in a globally distributed Chroococcidiopsis sp. from naturally stressful habitats. Mol Biol Evol. 2012;29(12):3899–3907. doi: 10.1093/molbev/mss191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Francis CA, Beman JM, Kuypers MM. New processes and players in the nitrogen cycle: The microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J. 2007;1(1):19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]
- 55.Jetten MS, et al. Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol Biol. 2009;44(2–3):65–84. doi: 10.1080/10409230902722783. [DOI] [PubMed] [Google Scholar]
- 56.Yau S, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci USA. 2011;108(15):6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Breitbart M, Wegley L, Leeds S, Schoenfeld T, Rohwer F. Phage community dynamics in hot springs. Appl Environ Microbiol. 2004;70(3):1633–1640. doi: 10.1128/AEM.70.3.1633-1640.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong FK, et al. Hypolithic microbial community of quartz pavement in the high-altitude tundra of central Tibet. Microb Ecol. 2010;60(4):730–739. doi: 10.1007/s00248-010-9653-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chan Y, et al. Hypolithic microbial communities: Between a rock and a hard place. Environ Microbiol. 2012;14(9):2272–2282. doi: 10.1111/j.1462-2920.2012.02821.x. [DOI] [PubMed] [Google Scholar]
- 60.Hopkins DW, et al. Carbon, nitrogen and temperature controls on microbial activity in soils from an Antarctic dry valley. Soil Biol Biochem. 2006b;38:3130–3140. [Google Scholar]
- 61.Farrell RL, et al. Introduced and indigenous fungi of the Ross Island historic huts and pristine areas of Antarctica. Polar Biol. 2011;34(11):1669–1677. [Google Scholar]
- 62.Blanchette RA, et al. 2010. An Antarctic hot spot for fungi at Shackleton’s historic hut on Cape Royds. Microb Ecol 60:29–38.
- 63.Omelon CR. Endolithic microbial communities in polar desert habitats. Geomicrobiol J. 2008;25:404–414. [Google Scholar]
- 64.Pointing SB, Warren-Rhodes KA, Lacap DC, Rhodes KL, McKay CP. Hypolithic community shifts occur as a result of liquid water availability along environmental gradients in China’s hot and cold hyperarid deserts. Environ Microbiol. 2007;9(2):414–424. doi: 10.1111/j.1462-2920.2006.01153.x. [DOI] [PubMed] [Google Scholar]
- 65.Lu ZM, et al. Microbial gene functions enriched in the Deepwater Horizon deep-sea oil plume. ISME J. 2012;6(2):451–460. doi: 10.1038/ismej.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hazen TC, et al. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science. 2010;330(6001):204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 67.Wu LY, Liu X, Schadt CW, Zhou J-Z. Microarray-based analysis of subnanogram quantities of microbial community DNAs by using whole-community genome amplification. Appl Environ Microbiol. 2006;72(7):4931–4941. doi: 10.1128/AEM.02738-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.