All negative-sense, single-stranded RNA (−ssRNA) viruses encode a nucleoprotein (NP), the major function of which is to bind viral RNAs and encapsidate them as ribonucleoprotein complexes (RNPs) (1, 2). Consequently, the genomes of −ssRNA viruses do not exist as naked RNA but rather as protein–RNA complexes with high-order structures. In PNAS, Li et al. (3), Niu et al. (4), and Reguera et al. (5) report the crystal structures of NP-RNA complexes from three orthobunyaviruses, namely the Bunyawera virus (BUNV), Leanyer virus (LEAV), and La Crosse virus (LACV). Meanwhile, the apo-NP structure from another orthobunyavirus, the Schmallenberg virus (SBV), has been reported by Dong et al. (6). The structures of the BUNV NP-RNA complex and SBV apo-NP have also been reported by Ariza et al. (7). Altogether, these findings make a significant impact on our understanding of Orthobunyavirus genome encapsidation and condensation.
Bunyaviridae is a large family of viruses encompassing more than 350 species that are organized into five genera, including Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus (8, 9). Orthobunyavirus, which is the focus of study by these five research teams listed above, includes more than 170 species and constitutes the largest genus within the family (9). Except for Tospovirus, members from the other four bunyavirus genera infect animals, and many of them are considered as serious human pathogens (e.g., the Crimean-Congo hemorrhagic fever virus, La Crosse virus, Rift valley fever virus, and Sin Nombre virus) (8, 9). For instance, the Yosemite hantavirus outbreak last summer caused ten confirmed infections with three deaths (10).
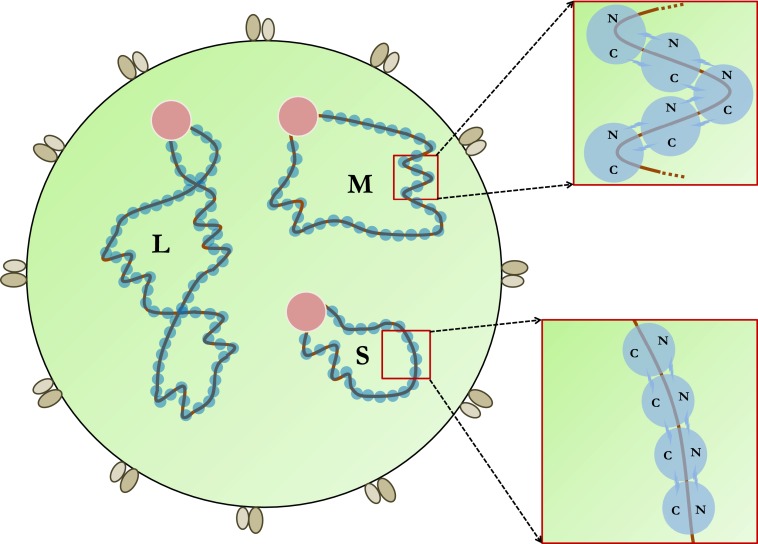
Bunyaviruses are enveloped viruses that are generally spherical in shape, with a diameter of 80–120 nm (8) (Fig. 1). Their genome consists of three −ssRNA segments that encode four structural proteins (i.e., L, Gn, Gc, and N) and in some viruses also two nonstructural proteins (i.e., NSm and NSs) (8). Like in other −ssRNA viruses, the genomic RNAs of bunyaviruses are extensively coated by NP and associated with the viral RNA polymerase or L protein to form RNPs, which are responsible for viral RNA synthesis as well as viral genome packaging (1, 8, 9). Electron microscopy (EM) images show that bunyavirus RNPs form closed circular structures that are often loosely coiled without apparent symmetry (3, 4, 8).
Fig. 1.
Schematic drawing of a bunyavirus. The three gene segments are designated as L, M, and S. The two glycoproteins Gn and Gc are associated with the viral envelope. Each RNP is associated with a viral polymerase (pink) and multiple copies of NP (blue). The two enlarged areas highlight a linear and a helical NP-RNA structure, respectively.
These five recent research papers (3–7) reveal that Orthobunyavirus NPs adopt a unique structural fold not previously observed in NPs from other −ssRNA viruses or even those from other genera within the same Bunyaviridae family (1). Each Orthobunyavirus NP folds into a compact structure that can be divided into four parts: the N-arm, N-terminal domain, C-terminal domain, and C-arm. The N-/C-terminal domains, formed by the N-/C-terminal halves of the polypeptide chain, comprise the NP structure core (3–7). The N-/C-arms are short sequences from the N/C termini, and they play important roles in mediating NP–NP interaction. In between the N- and C-terminal domains lies a positively charged RNA-binding groove. Although the core structure of NP is rigid, the N-/C-arms can adopt different conformations due to flexible hinge connections.
The Orthobunyavirus NP–RNA complex structures by Li et al. (3), Niu et al. (4), Reguera et al. (5), and Ariza et al. (7) provide valuable insights into the mechanism of NP-mediated viral genome encapsidation. The bound RNAs in these complexes were either derived from the Escherichia coli expression host or reconstituted using synthetic oligos. All three Orthobunyavirus NP–RNA complexes were crystallized as tetramers, in which each NP interacts with two other NP molecules in a head-to-tail fashion through the N-/C-arms. The positive-charged grooves from neighboring protomers line up sequentially, thereby forming a continuous RNA-binding channel along the inner perimeter of the NP tetrameric ring. Each NP subunit binds 11 nucleotides of RNA according to Niu et al., Reguera et al., and Ariza et al. (4, 5, 7). Bound RNA is largely inaccessible, consistent with the observation that Orthobunyavirus NP–RNA complexes are generally resistant to RNase treatment (3–5, 7).
The apo-NP structures by Ariza et al., Dong et al., and Reguera et al. show few structural differences compared with the RNA-bound form in terms of the NP core (5–7). This observation indicates that local structural rearrangement should be sufficient to release the RNA template from RNP during viral RNA synthesis. Furthermore, the N-/C-arms of apo-NP were found to make contacts that are sometimes not the same as in the NP–RNA complexes. NP regions other than the N-/C-arms were also found at the NP–NP interface in an SBV apo-NP structure (6). It is proposed that the different NP–NP contacts observed in apo-NP may reflect structural changes in RNP upon RNA dissociation induced by the viral polymerase (6).
Concrete RNP models have been proposed by Ariza et al., Li et al., Niu et al., and Reguera et al. to explain various RNP structures observed in EM (3–5, 7). In particular, the flexible N-/C-arms of NP should allow Orthobunyavirus RNPs to form both linear, 4-nm-wide NP-RNA polymers as well as helical, 10-nm-wide filaments (3–5, 7). Further supercoiling of circularized RNPs would result in highly compact, rod-shaped structures. These three RNP structures condense the associated RNA to different extents, which may be required at different stages of the virus replication cycle. For instance, highly condensed RNPs may facilitate viral RNA packaging into infectious particles whereas extended RNPs may be required for viral RNA synthesis.
Studies by these five research teams (3–7) and others (1) indicate that NP proteins from −ssRNA viruses with segmented genomes exhibit substantial differences in size, sequence, structure, and even function. Considering that NP proteins encoded by bunyaviruses from different genera have nonhomologous sequences (8), it is not unexpected that Orthobunyavirus NP possesses a different structural fold than the NPs of Rift Valley fever virus (RVFV, genus Phlebovirus) (11–13) and Crimean-Congo hemorrhagic fever virus (CCHFV, genus Nairovirus) (14–16). RVFV NP is similar in size to Orthobunyavirus NP, but CCHFV NP is much larger with an additional endonuclease activity (14).
NP proteins from \x{2212}ssRNA viruses with segmented genomes exhibit substantial differences in size, sequence, structure, and even function.
CCHFV NP is structurally related to the NP of Lassa fever virus (LASV), a member of the Arenaviridae family with a bisegmented −ssRNA genome (15, 17). LASV NP is associated with multiple activities, including RNA binding, cap snatching, and an exonuclease activity related to immunosuppression (17–19). Lastly, the structure of the influenza virus NP is different from any of these NPs described above but is conserved within the Orthomyxoviridae family (1, 20–22). By comparison, NPs from −ssRNA viruses with nonsegmented genomes are usually related in both structure and function (1).
Now, with the Orthobunyavirus RNP structure established, many questions regarding RNP assembly and function start to emerge. For example, how does Orthobunyavirus keep its NP in a nonaggregated form that is competent for RNP encapsidation? Does Orthobunyavirus selectively package its genomic RNA, and, if so, what mechanism is used to ensure specific packaging of the three RNA segments? Furthermore, how does Orthobunyavirus RNP change its structure during RNA synthesis to allow the readout of nucleotide sequences? Such studies are crucial for our understanding of Orthobunyavirus biology, and the results will likely lead to better prevention and treatment of Orthobunyavirus infection.
Acknowledgments
We are supported by Robert A. Welch Foundation Grant C-1565.
Footnotes
References
- 1.Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14(4):504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Zheng W, Tao YJ. Structure and assembly of the influenza A virus ribonucleoprotein complex. FEBS Lett. 2013;587(8):1206–1214. doi: 10.1016/j.febslet.2013.02.048. [DOI] [PubMed] [Google Scholar]
- 3.Li B, et al. Bunyamwera virus possesses a distinct nucleocapsid protein to facilitate genome encapsidation. Proc Natl Acad Sci USA. 2013;110:9048–9053. doi: 10.1073/pnas.1222552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu F, et al. Structure of the Leanyer orthobunyavirus nucleoprotein–RNA complex reveals unique architecture for RNA encapsidation. Proc Natl Acad Sci USA. 2013;110:9054–9059. doi: 10.1073/pnas.1300035110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reguera J, Malet H, Weber F, Cusack S. Structural basis for encapsidation of genomic RNA by La Crosse Orthobunyavirus nucleoprotein. Proc Natl Acad Sci USA. 2013;110(18):7246–7251. doi: 10.1073/pnas.1302298110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong H, Li P, Elliott RM, Dong C. Structure of schmallenberg orthobunyavirus nucleoprotein suggests a novel mechanism of genome encapsidation. J Virol. 2013;87(10):5593–5601. doi: 10.1128/JVI.00223-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ariza A, et al. Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guu TS, Zheng W, Tao YJ. Bunyavirus: Structure and replication. Adv Exp Med Biol. 2012;726:245–266. doi: 10.1007/978-1-4614-0980-9_11. [DOI] [PubMed] [Google Scholar]
- 9.Schmaljohn CS, Nichol ST. 2007. Bunyaviridae. Fields Virology, eds Knipe DM, et al. (Lippincott Williams & Wilkins, Philadelphia), 5th Ed, pp 1741–1789.
- 10.Centers for Disease Control and Prevention (CDC) Hantavirus pulmonary syndrome in visitors to a national park—Yosemite Valley, California, 2012. MMWR Morb Mortal Wkly Rep. 2012;61(46):952. [PubMed] [Google Scholar]
- 11.Raymond DD, Piper ME, Gerrard SR, Smith JL. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci USA. 2010;107(26):11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferron F, et al. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7(5):e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci USA. 2012;109(47):19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo Y, et al. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci USA. 2012;109(13):5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carter SD, et al. Structure, function, and evolution of the Crimean-Congo hemorrhagic fever virus nucleocapsid protein. J Virol. 2012;86(20):10914–10923. doi: 10.1128/JVI.01555-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, et al. Structure of Crimean-Congo hemorrhagic fever virus nucleoprotein: Superhelical homo-oligomers and the role of caspase-3 cleavage. J Virol. 2012;86(22):12294–12303. doi: 10.1128/JVI.01627-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hastie KM, et al. Crystal structure of the Lassa virus nucleoprotein-RNA complex reveals a gating mechanism for RNA binding. Proc Natl Acad Sci USA. 2011;108(48):19365–19370. doi: 10.1073/pnas.1108515108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastie KM, Kimberlin CR, Zandonatti MA, MacRae IJ, Saphire EO. Structure of the Lassa virus nucleoprotein reveals a dsRNA-specific 3′ to 5′ exonuclease activity essential for immune suppression. Proc Natl Acad Sci USA. 2011;108(6):2396–2401. doi: 10.1073/pnas.1016404108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi X, et al. Cap binding and immune evasion revealed by Lassa nucleoprotein structure. Nature. 2010;468(7325):779–783. doi: 10.1038/nature09605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444(7122):1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 21.Ng AK, et al. Structure of the influenza virus A H5N1 nucleoprotein: Implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008;22(10):3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng AK, et al. Structural basis for RNA binding and homo-oligomer formation by influenza B virus nucleoprotein. J Virol. 2012;86(12):6758–6767. doi: 10.1128/JVI.00073-12. [DOI] [PMC free article] [PubMed] [Google Scholar]