Abstract
Bunyamwera virus (BUNV), which belongs to the genus Orthobunyavirus, is the prototypical virus of the Bunyaviridae family. Similar to other negative-sense single-stranded RNA viruses, bunyaviruses possess a nucleocapsid protein (NP) to facilitate genomic RNA encapsidation and virus replication. The structures of two NPs of members of different genera within the Bunyaviridae family have been reported. However, their structures, RNA-binding features, and functions beyond RNA binding significantly differ from one another. Here, we report the crystal structure of the BUNV NP–RNA complex. The polypeptide of the BUNV NP was found to possess a distinct fold among viral NPs. An N-terminal arm and a C-terminal tail were found to interact with neighboring NP protomers to form a tetrameric ring-shaped organization. Each protomer bound a 10-nt RNA molecule, which was acquired from the expression host, in the positively charged crevice between the N and C lobes. Inhomogeneous oligomerization was observed for the recombinant BUNV NP–RNA complex, which was similar to the Rift Valley fever virus NP–RNA complex. This result suggested that the flexibility of one NP protomer with adjacent protomers underlies the BUNV ribonucleoprotein complex (RNP) formation. Electron microscopy revealed that the monomer-sized NP–RNA complex was the building block of the natural BUNV RNP. Combined with previous results indicating that mutagenesis of the interprotomer or protein–RNA interface affects BUNV replication, our structure provides a great potential for understanding the mechanism underlying negative-sense single-stranded RNA RNP formation and enables the development of antiviral therapies targeting BUNV RNP formation.
Keywords: assembly, packaging
Bunyaviruses constitute the largest segmented negative-sense single-stranded RNA (–ssRNA) virus family, which is subdivided into Orthobunyavirus, Hantavirus, Nairovirus, Phlebovirus, and Tospovirus genera (1). The genomes of Bunyaviridae members are featured by three segments, the large (L), middle (M), and small (S) segments (2, 3). The L segment encodes an RNA-dependent RNA polymerase (RdRp), the M segment encodes a precursor of glycoproteins (Gn and Gc), and the S segment encodes a nucleocapsid protein (NP). Moreover, a few Bunyaviridae members possess a nonstructural protein (NSs and/or NSm) using an ambisense coding strategy by the S and M segments (4).
Similar to other –ssRNA viruses, all members of the Bunyaviridae family contain a ribonucleoprotein (RNP) complex composed of genomic RNA enwrapped by the NP (4). After entry into the cytoplasm through membrane fusion mediated by glycoproteins, the RNP is released from the virion and serves as the template with which the copackaged RdRp transcribes mRNAs from the viral genome in the RNP. In the later stage of virus replication, complementary positive-strand RNA (cRNA) is produced in the form of an RNP. The RNP serves as the template for replication that generates the viral genomic RNA in the form of an RNP ready to be packaged in the virion. Throughout the entire virus replication cycle of a –ssRNA virus, the genome-length viral RNA (cRNA or viral genomic RNA) is only present in the form of an RNP that either serves as a template for RNA synthesis or is packaged in the virion. Therefore, RNP assembly is a critical step in –ssRNA virus replication.
All bunyaviruses encode NPs to facilitate genome encapsidation. However, the structure, function, and molecular weight of these NPs show distinct variations (Fig. S1). The crystal structure of the NP from the Crimean–Congo hemorrhagic fever virus (CCHFV), which is a member of the Nairovirus genus, revealed an unusual metal-dependent DNA-specific endonuclease activity but shared high structural similarity to the N-terminal domain of the Lassa fever virus (LAFV) NP (1). Moreover, recent studies have revealed that the NP of the Rift Valley fever virus (RVFV; genus Phlebovirus) oligomerizes with distinct molecular architectures to facilitate RNA encapsidation (5–7). A monomeric structure of the RVFV NP that was purified using a denaturation/renaturation method was first reported, and a distinct architecture for RNA encapsidation in –ssRNA viruses was found (6). Subsequently, the structure of the RVFV NP purified under native conditions was determined, and the formation of a hexameric ring-shaped structure was revealed (5). The most significant difference between these two results appears in the conformations of the N terminus of the RVFV NP. In the monomeric RVFV NP, the N-terminal arm (the first 30 amino acids) folds backward to the core region, which allows the formation of a compact architecture for the entire molecule, whereas this N-terminal arm was found to extend to the adjacent molecule to form the ring-shaped architecture of the hexameric structure. The structures of the RVFV NP complexed with RNA of variable lengths have been determined recently, demonstrating that the flexibility of the N-terminal arm of the RVFV NP underlies higher-order RNP formation (7). Another interesting observation from crystallographic studies on all of the bunyavirus-encoded NPs is that all of them cannot properly protect the packaged RNA against exogenous RNases. This result is consistent with the finding that bound RNA can be well protected by NPs from nonsegmented –ssRNA viruses but not by NPs from segmented –ssRNA viruses (8, 9).
Bunyamwera virus (BUNV), which can cause severe hemorrhagic fever and other diseases in livestock and humans (10), is the prototypic member of both the Orthobunyavirus genus and the entire Bunyaviridae family. BUNV is also known to serve as an excellent model for studying the biology of other Bunyaviridae family members (11, 12). BUNV possesses a small NP with a molecular weight of 25 kDa (2). Numerous investigations have revealed its biochemical features. However, its potential oligomerization state, effect on virus replication, specificity and stoichiometry of RNA binding (12–15), detailed structure of the BUNV NP–RNA complex, and the underlying mechanism for RNP formation remain unclear. To further understand the NPs encoded by Bunyaviridae members, particularly the prototypic member of each genus belonging to the Bunyaviridae family, we determined the crystal structure of the BUNV NP–RNA complex. Our findings reveal a distinct virally encoded NP and provide insight into how NP oligomerization contributes to the regulation of RNP assembly.
Results
Recombinant BUNV NP Oligomerizes and Binds Cellular RNA.
Previous studies have suggested that bunyavirus NPs cannot well protect the bound RNA against exogenous RNases (1, 5). These observations are consistent with the finding that the NPs of nonsegmented –ssRNA viruses can protect the bound RNA, in contrast to the NPs of segmented –ssRNA viruses, which cannot (9).
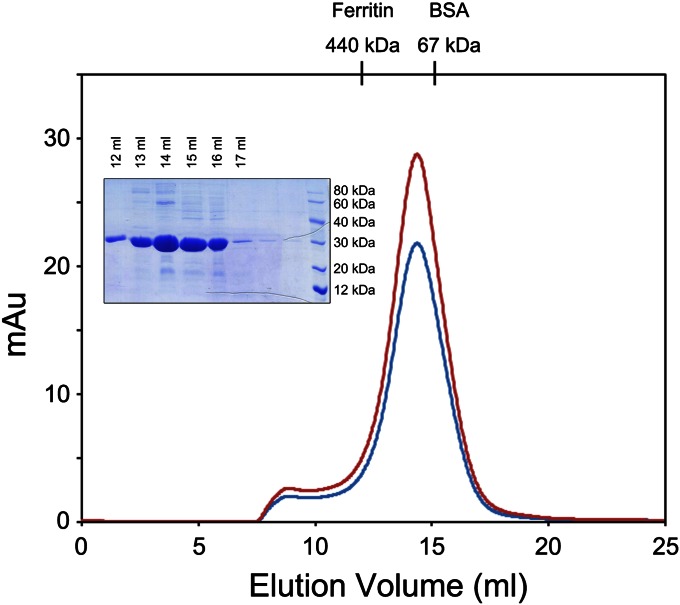
We first analyzed the RNA-binding and solution properties of the recombinant BUNV NP. We expressed the full-length BUNV NP (residues M1–M233) in Escherichia coli and purified it under physiological conditions. The retention volume of the target protein in size-exclusion chromatography peaks at 14.5 mL (Fig. 1), corresponding to a molecular weight greater than 100 kDa. An SDS/PAGE analysis indicated that the major peaks contained a protein that was the size expected for the BUNV NP (25 kDa), suggesting that no protein contamination occurred during purification. These results revealed that the recombinant BUNV NP existed as a higher-order oligomer in solution. Moreover, the ratio of the OD at 260 nm to that at 280 nm (OD260/OD280) for the major peak was 1.35, which indicated that the recombinant BUNV NP oligomers possessed encapsidated nucleic acids.
Fig. 1.
Size-exclusion chromatography (SEC) of the BUNV NP–RNA complex with RNase pretreatment. The E. coli-expressed BUNV NP (0.2 mg) pretreated with RNase (0.2 mg/mL at 16 °C for 12 h) was injected onto a Superdex 200 HR 10/30 column. The absorbance at 260 and 280 nm are indicated in red and blue, respectively. The retention volume for the major peak was 14.5 mL. The retention volumes for the molecular-weight standards are shown above. SDS/PAGE analysis of the SEC elution fractions corresponding to the peaks is shown with standard protein markers.
As aforementioned, the NPs of segmented –ssRNA viruses, for example, bunyavirus, cannot well protect bound RNA against exogenous RNases. Therefore, we determined whether the BUNV NP possessed a similar property. Surprisingly, although we incubated the preliminarily purified BUNV NP–RNA mixture with RNase at a final concentration of 0.2 mg/mL for 12 h at 16 °C, we observed that the bound RNA was not digested. An identical peak at an identical retention volume was observed from size-exclusion chromatography, and an identical OD260/OD280 ratio was obtained from BUNV NP–RNA mixtures with or without RNase treatment. These results demonstrated that the BUNV NP can form a higher-order oligomer and protect the bound RNA against exogenous RNase. Therefore, we used this BUNV NP–RNA complex in subsequent analyses.
EM of the Recombinant BUNV NP–RNA Complex.
Previous studies on the RVFV NP or NP–RNA complex indicated that the oligomerization state of the RVFV NP was inhomogeneous in solution, that is, pentamers and hexamers were found to be the most prevalent species of the digested endogenous viral RNP and recombinant NP–RNA complex (5–7). The recombinant BUNV NP–RNA complex was found to exist as a multimer; thus, we first analyzed the RNase-treated NP–RNA sample using negative-stain EM to assess the homogeneity of the oligomerization state of the BUNV NP–RNA complex in solution.
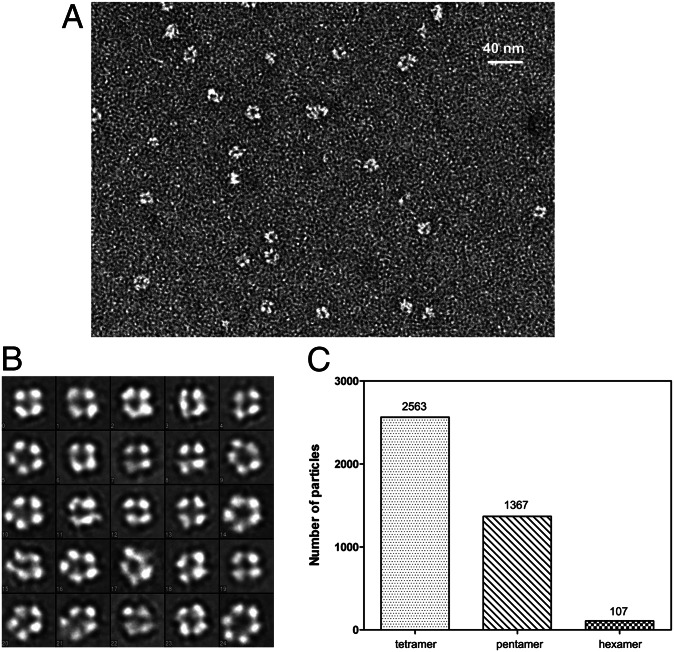
From a total of 300 images recorded by EM, we selected 4,037 particles that were categorized into 24 classes by a reference-free classification. The results indicated that the BUNV NP–RNA complex formed three types of higher-order ring-shaped oligomers with diameters ranging from 80 Å to 120 Å (Fig. 2 A and B). These particles consisted of predominantly tetramers (2,363 particles, 59%), followed by pentamers (1,367 particles, 33%) and very few hexamers (107 particles, 3%) (Fig. 2C). This observation differed from that for the RVFV NP–RNA complex, for which the amount of pentameric and hexameric structures were found to be equivalent followed by a small amount of tetramers and other higher-order multimers (7).
Fig. 2.
EM analysis of the recombinant BUNV NP–RNA complex. (A) A typical EM version of the negatively stained BUNV NP–RNA complex. (B) The reference-free classifications of 4,037 BUNV NP–RNA particles in 24 classes (the length of the side of one square is 150 Å). The distribution of multimeric states is summarized in (C).
The inhomogeneous oligomerization of the RVFV NP insignificantly affected the crystallization of the most abundant species (5, 7). Given that the tetramer was the major form of the BUNV NP–RNA complex in solution, we supposed that the contaminants, namely, the small amount of pentamers and hexamers, would not influence the crystallization of the BUNV NP–RNA complex. Thus, we used this purified sample for crystallographic study.
Overall Structure of the BUNV NP–RNA Complex.
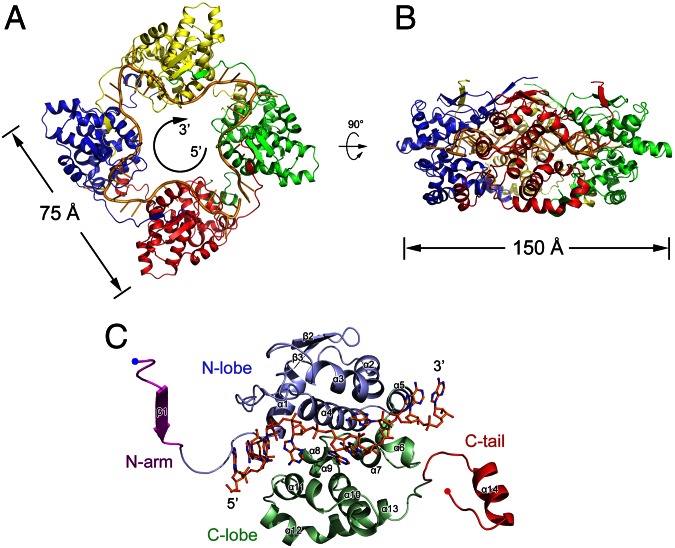
The BUNV NP–RNA complex was successfully crystallized, and the crystal structure was subsequently determined using the single-wavelength anomalous dispersion (SAD) method and refined to 3.2 Å resolution with a final Rwork of 22.0% (Rfree = 27.6%) (Table S1). Each protomer of the BUNV NP bound one 10-nt RNA, and four NP–RNA complexes formed a ring-shaped architecture with an inner diameters of 30 Å and outer diameters of 75 Å through the fourfold crystallographic axis. This finding was consistent with the results of EM analysis (Fig. 3 and Fig. S2).
Fig. 3.
Structure of the BUNV NP–RNA complex. The tetrameric ring of the BUNV NP–RNA complex is shown as a ribbon diagram from the top (A) and side (B) views. Each protomer is presented as a colored cartoon, and the bound RNA is shown as a yellow cartoon. The bound RNA presents a 5′–3′ clockwise orientation. The molecular dimensions are also indicated. (C) The structure of the BUNV NP monomer. The N arm, N lobe, C lobe, and C tail are shown in magenta, blue, green, and red, respectively. The N and C termini are indicated. The bound RNA is shown as colored sticks.
The polypeptide of the BUNV NP possessed a compact body part with an additional N-terminal extension and a C-terminal tail and featured four key regions: an N-terminal arm (termed N arm, hereafter) (M1–A10), an N lobe (A11–S120), a C lobe (K121–K214), and a C-terminal tail (termed C tail, hereafter) (K215–M233) (Fig. 3C and Fig. S3). Topology comparison using DALI (16) and secondary-structure matching (17) did not generate evident structural homologs, suggesting a unique protein fold structure for virally encoded NPs. The compact body of the BUNV NP protomer consisted of the N and C lobes. The C lobe was predominantly composed of α-helices, whereas the N lobe consisted of four α-helices and two β-strands. The one-stranded N arm and the helical C tail winged the main body and participate in intermolecular interactions with the adjacent two protomers. The RNA-binding site was identified as a large positively charged crevice located at the interface of the N and C lobes with a 10-nt RNA molecule bound within the inner side of the tetrameric ring. Regarding the coexistence of tetramers, pentamers, and hexamers of the BUNV NP–RNA complex in solution, the length of the bound RNA ranged from 40 nt to 60 nt, which is consistent with previous results (12). The body parts of four molecules in one asymmetric unit are nearly identical with rmsd values for the Cα atoms of less than 0.7 Å, whereas the rmsd values for the Cα atoms of N arms and C tails are approximate 2.0 Å, suggesting the flexibility of the N- and C-terminal extensions in the formation of NP–RNA oligomers.
NP–RNA Interaction.
Consistent with the OD260/OD280 ratio during purification, the continuous experimental electron density map indicated the presence of an RNA molecule bound to the BUNV NP (Fig. S4). Each BUNV NP protomer clamped a 10-nt RNA molecule in the deep positively charged RNA-binding cavity located at the interface between the N and C lobes, in contrast to previous biochemical results (12, 15). Given that the bound RNA molecule in the NP–RNA complex was randomly acquired from the expression host, we used adenine to build the final model (Fig. 4).
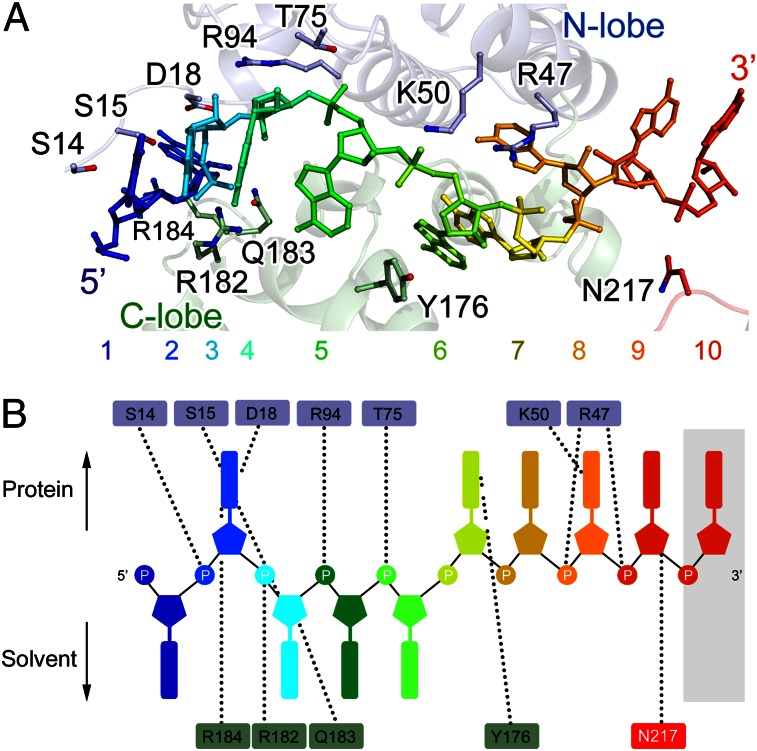
Fig. 4.
The NP-RNA interaction. (A) A magnified side view of the interaction between the BUNV NP and bound RNA. RNA is shown as a colored stick in rainbow colors from the 5′- to 3′-end. The NP polypeptide is shown as a semitransparent cartoon representation with an identical color scheme as that in Fig. 3C. The residues contacting the RNA are shown as colored sticks. (B) Schematic top view of the NP–RNA interactions. The last nucleotide located in the cleft exposed to the solvent between two NP protomers is shadowed. The dashed lines denote the contacts between the RNA and residues.
The electron densities for the bases indicated that the RNA molecule in the tetrameric complex structure bound in a 5′–3′ direction in a clockwise orientation (Fig. 3A). The RNA strand was twisted clockwise along the inner perimeter of the tetrameric ring. The RNA-binding cleft formed the N lobe (top) and C lobe (bottom) interface. The first nucleotide pointed toward the solvent and stacked with the last base of the preceding NP protomer (N−1) to form a ring-shaped structure. Nucleotides 1, 3, 4, and 5 were exposed to the solvent; nucleotides 2, 6, 7, 8, and 9 faced the protein, whereas nucleotide 10 was located in the cleft, that is, was exposed to the solvent between two NP protomers and did not contact the residues from the polypeptides of the NP (Fig. 4 and Fig. S5). The residues of the BUNV NP that interact with the RNA were primarily charged residues, including S14, S15, D18, R47, K50, T75, R94, Y176, R182, Q183, R184, and N217. These residues not only interacted with the ribose–phosphate backbone but also interacted with the bases to stabilize the bound RNA molecule.
Oligomerization of the BUNV NP Protomer.
The assembled RNP comprised a large number of copies of the NP protein bound to genomic RNA (13). In the RNP, one NP protomer interacted with adjacent NP molecules to maintain the oligomeric architecture.
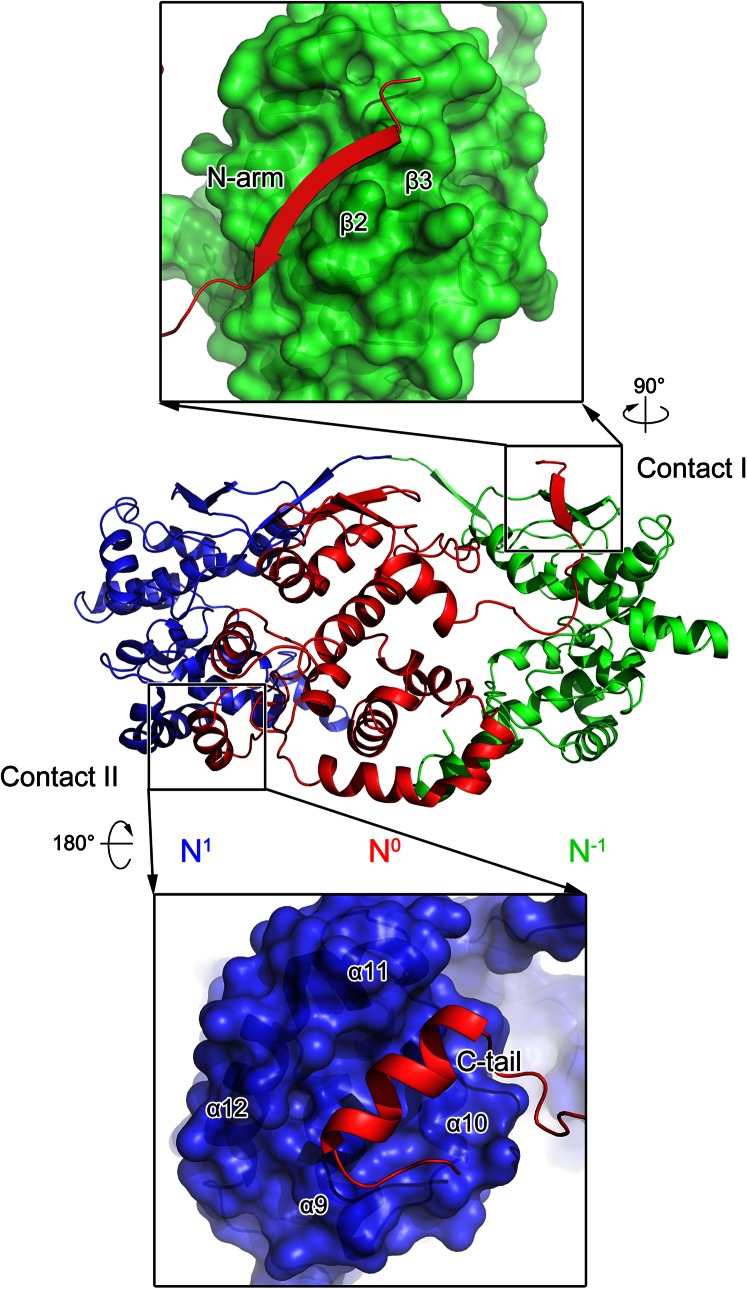
The interactions between the adjacent molecules within the tetrameric BUNV NP–RNA complex buried a total surface area of 1,300 Å2 of the 14,000-Å2 total surface area of one protomer, as calculated by PISA (17), which suggested strong interactions for the assembly of the higher-order oligomer. The most substantial interactions were contributed by the N arm (contact I) and C tail (contact II) with neighboring NP protomers (Fig. 5).
Fig. 5.
Interprotomer interaction within the BUNV NP–RNA complex. Three adjacent molecules are shown as cartoons in blue, red, and green, respectively. Contacts I and II are labeled and detailed in the top and bottom panels. In the enlarged panels, the N0 protomer is shown as a colored cartoon, whereas the N1 and N−1 protomers are depicted as molecular surfaces. Two indicators show how the two magnified views are related in orientation to the original diagram in the center.
The one-stranded N arm of one protomer (N0) interacted with the two β-strands in the N lobe of the protomer on the right (N−1) to form a three-stranded β-sheet, which constituted contact I. Within the contact-I region, a subset of hydrogen bonds was formed by residues E3–V9 of the N0 protomer with V61–L65 of the β2-strand of the N−1 protomer to stabilize this intermolecular three-stranded β-sheet. Contact I contributed ∼40% of the interaction surface of the total 1,300-Å2 interprotomer interface, whereas the remaining interaction was provided by contact II. Contact II was formed by the C tails of N0 with the C lobe of a neighboring protomer (N1) on the left. Residues A222, F225, L226, F229, and I231 of the C tails of the N0 protomer, particularly their aliphatic side chains and aromatic rings, occupied the hydrophobic groove formed by the hydrophobic side chains of F158, L161, I165, L177, M181, W193, L198, V201, and L205 of the C lobe of the N1 protomer. Similar to the interactions observed in the ring-shaped viral NP–RNA complex, contacts I and II were repeated in a directional manner such that the protomer was linked by contact I in a clockwise direction and by contact II in a counterclockwise direction, constituting the tetrameric ring of the BUNV NP–RNA complex. This structural information concerning the BUNV NP oligomerization was consistent with the results of mutagenesis analysis that mapped the homotypic interaction of the BUNV NP (14) and the finding that deletions of both the N arm and the C tail completely eliminated the ring-shaped oligomer in EM analysis.
Visualization of the Authentic BUNV RNP.
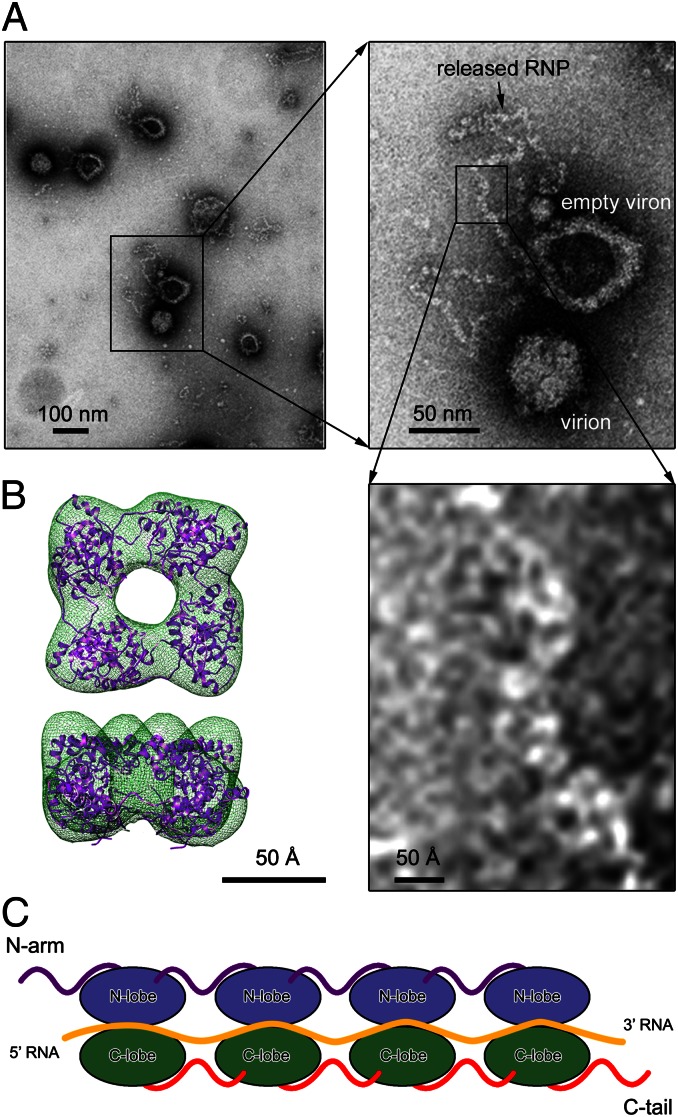
The oligomerization state of the recombinant BUNV NP–RNA complex was found to be inhomogeneous in solution. Thus, we extracted natural RNP from BUNV virions and visualized it using negative-stain EM to verify the precise oligomerization state of the BUNV NP in vivo (Fig. 6A). BUNV virions released by infected BHK-21 cells were purified and treated with 1 M sucrose to open the virus particles and release the natural RNPs. These RNPs were not disrupted or modified by these isolation procedures. The RNPs extracted from BUNV virions demonstrated a relaxed structure without remarkable helical symmetry, which was similar to observations of the RVFV RNPs but clearly differed from those of the influenza virus or nonsegmented –ssRNA viruses, in which the RNP is supercoiled in structure (18). The width of the virion-extracted BUNV RNP is ∼40 Å and was consistent with the size of an NP–RNA monomer in either the crystal structure or the 3D reconstruction that was obtained using cryonegative-staining EM (Fig. 6B and Fig. S6), suggesting that the monomer-sized NP–RNA complex was likely the building block of the relaxed BUNV RNP.
Fig. 6.
The monomer-sized NP–RNA is the building block of natural BUNV RNPs. (A) Negative-stained EM image of the natural BUNV RNP extracted from bunyamwera viruses. The inset frames represents the enlarged regions. (B) The 3D reconstruction of the recombinant BUNV NP–RNA tetramer obtained using cryonegatively stained samples with docking of the crystal structure into the EM density map. (C) A model of the linear BUNV RNP formed using the monomer-sized NP–RNA as the building block. The color scheme for each structural element is identical to that used in Fig. 3C.
Discussion
BUNV is the prototypic virus of the Orthobunyavirus genus and the entire Bunyaviridae family. The structure of the BUNV NP–RNA complex allows a better understanding of the molecular mechanism underlying segmented –ssRNA RNP formation and the significant difference between NPs encoded by the members of each genus belonging to the Bunyaviridae family.
The crystal structure of the tetrameric BUNV NP–RNA complex reveals a distinct fold for virally encoded NPs and presents the predominant oligomeric state of the recombinant NP–RNA in solution. Although BUNV is a segmented –ssRNA virus, it possesses a distinct NP among segmented –ssRNA viruses but demonstrates structural similarity to NPs encoded by nonsegmented –ssRNA viruses. To date, the structures of four NPs of nonsegmented –ssRNA viruses are available: the tetrameric Borna disease virus NP (19) and the oligomeric NP–RNA rings of the rabies virus (20), vesicular stomatitis virus (21), and respiratory syncytial virus (22) that contain 10 or 11 NP protomers. Although the nucleotides bound by a single protomer and the position of RNA binding differ, these four nonsegmented –ssRNA-encoded NPs demonstrate clear structural homology. All of these NPs possess an N- and a C-terminal extension for interprotomer interaction, in addition to conserved N and C lobes for RNA binding (Fig. S7, upper row). In contrast, the reported structures of segmented –ssRNA virus NPs differ, and the N- and C-terminal extensions for interprotomer interaction are not conserved (Fig. S7, lower row). The influenza A virus NP possesses a C-terminal tail loop, whereas the RVFV NP extends an N-terminal arm for oligomerization. Moreover, the LAFV NP and CCHFV NP do not demonstrate a clear structural element for multimerization. The BUNV NP clearly presents both an N- and a C-terminal extension, which play a central role in RNP formation in nonsegmented –ssRNA viruses. Another interesting structural observation is the presence of a large cleft at the interface of two protomers, exposing the last nucleotide to the solvent (Figs. S5 and S8). However, this cleft was not observed in other reported NP–RNA complex structures. We speculate that this cleft may represent the flexibility between two protomers during RNP formation.
The RNA binding of the BUNV NP was found to be similar to that of nonsegmented –ssRNA virus NPs. The BUNV NP possesses two clear N and C lobes in the central core that face each other to form a positively charged crevice for RNA binding. In the BUNV NP–RNA complex, the RNA is twisted around the inner perimeter of the tetrameric ring with individual bases facing either the solvent or the protein. In contrast, the RVFV NP sequestered the bases of only four nucleotides in a narrow hydrophobic pocket and interacted through polar contacts only with the ribose–phosphate backbone, which faced the solvent (7).
Furthermore, the presence of pentamers and hexamers in addition to the predominate tetramers that crystallized and the relaxed structure of the endogenous RNP as visualized by EM suggests that the multimer may not be the building block for the BUNV RNP (Fig. 6C). This feature was similar to that observed in the RVFV NP–RNA complex structures (7) but differed from that observed in the influenza virus and nonsegmented –ssRNA viruses (18). The monomeric BUNV NP–RNA complexes twisted to form a linear RNP with a flexible conformation between two protomers. We propose that flexibility within the hinge regions linking the N arm and C tail to the central core was responsible for the formation of this linear RNP during replication and transcription. Such a role for these regions would account for the essential nature of the residues in the hinge regions between the N arm and C tail and the central core, for example, F17 and W213, for BUNV replication (2).
Previous functional studies have revealed that the deletion of the last 17 residues of the BUNV NP (NP1–216) or the first 10 residues (NP11–233) can attenuate the oligomerization of the BUNV NP in a mammalian cell expression system, whereas the deletion of both N and C termini can significantly abolish the multimerization to monomer (14). This work also demonstrated that destroying the assembly of the NP complex can evidently abolish viral replication using a minireplicon system (14). Another systematic analysis of BUNV NP residues revealed that a subset of residues were critical for BUNV replication (2). Among them, the substitutions of F26, R94, I118, P125, G131, W134, Y141, F144, Y158, L160, Y176, L177, K179, Y185, W193, F225, L226, and I231, together with F17 and W213, can completely abolish virus rescue (2). Moreover, the deletions of the N- and C-terminal residues destroyed the interprotomer interactions, thereby attenuating RNP formation and BUNV replication. Consistent with the ratio of the surface area buried by the interaction of the N arm and C tail with neighboring protomers, deletion of the N or C termini alone cannot completely eliminate the intermolecular interaction, whereas the double deletion completely abolished oligomerization to monomer (14). Furthermore, F225, L226, and I231 of the N0 protomer projected their side chains into the hydrophobic pocket formed by Y158, L161, Y176, L177, and other residues from the N1 protomer, thereby contributing to the interaction in contact II. Surprisingly, the single substitution of the residue from the N arm retained >65% minireplicon activity, and all mutated viruses were rescued (2), indicating that the hydrogen bond networks between the three-stranded β-sheet formed by the N arm and β2 and β3 of an adjacent protomer were essential to BUNV RNP formation. In addition, R94, P125, K179, Y185, and W193 all interacted with the bound RNA or contributed to the formation of the structural motif to stabilize the bound RNA, thereby significantly affecting BUNV RNP formation and viral replication. Notably, most of the residues essential to BUNV replication by RNP formation are conserved among the members of different serotypes of the Orthobunyavirus genus (Fig. S9). During the preparation of this manuscript, another group determined the crystal structure of the NP–RNA complex from Leanyer virus (LEAV), which is a member of the Orthobunyavirus genus and was isolated from arthropods in Australia (23). In addition to the high primary sequence conservation (42% sequence identity), the monomer fold of the BUNV NP and LEAV NP are highly homologous with an rmsd value of 1.4 Å for Cα atoms of 204 residues (Fig. S10A). Moreover, the architectures of the ring-shaped tetramers of the recombinant BUNV and LEAV NP–RNA complex in addition to the key residues for BUNV NP and LEAV NP oligomerization, RNA binding, and viral replication were also very similar (Fig. S10 B and C). Interestingly, one BUNV NP protomer binds a 10-nt RNA molecule, which is in contrast with the 11-nt RNA molecule bound to LEAV NP protomer. This difference suggests that the numbers of nucleotides bound to different orthobunyaviral NP protomers may be variable. All of these results suggest a conserved network for RNP formation in this genus, which suggests its potential role in discovering wide-spectrum anti-Orthobunyavirus therapeutics.
In summary, the crystal structure and EM visualization of the BUNV NP–RNA complex clearly present how orthobunyaviruses encapsidate their genomic RNA and form the RNP. This information also aids in understanding the structural and functional differences among NPs encoded by bunyavirus or segmented –ssRNA viruses, which can benefit the development of antiviral therapies targeting RNP formation.
Materials and Methods
The gene of the full-length BUNV NP (residues 1–233) was cloned into the pQE-30 expression vector with a 6× His tag fused at the N terminus following a general protocol. The target protein was expressed and purified as previously described (13) with a few modifications (SI Materials and Methods). The purified BUNV NP–RNA complex was concentrated to 10 mg/mL and stored at 193 K. The crystals for the native BUNV NP were obtained in a reservoir solution containing 100 mM sodium malonate (pH 7.0) and 10% (wt/vol) PEG 3350. The selenomethionine derivatives of the BUNV NP were purified following a general procedure (24) and then crystallized under conditions similar to those used for the native protein. The selenomethionine SAD data set of the BUNV NP–RNA complex was collected to 4.0 Å at the wavelength corresponding to the Se peak at the Photon Factory beamline BL17A. All datasets were processed using the HKL2000 package (25). The SAD data phases were calculated and substantially improved by solvent flattening using the PHENIX program (26). The coordinates and structure factors were deposited in the Research Collaboratory for Structural Bioinformatics under the Protein Data Bank ID code 4IJS. For visualization of the recombinant BUNV NP–RNA complex using EM, the samples were prepared by negative staining with uranyl acetate and imaged using an FEI Tecnai 20 electron microscope with a 2 k × 2 k Gatan CCD camera at a magnification of 50,000× using a defocus value of –1.5 μm. Two-dimensional class averages and 3D reconstructions were calculated using the Electron Micrograph Analysis (EMAN) software package (27). For the EM analysis of natural BUNV RNPs, Bunyamwera virions released by infected BHK-21 cells were produced and purified as previously described (28). The purified virions were adsorbed to EM grids and incubated for 3 min with 1 M sucrose in TEN buffer [0.01 M Tris⋅HCl (pH 7.4) with 0.1 M NaCl and 1 mM EDTA] before negative staining with 2% uranyl acetate for 30 s. The samples were subsequently imaged in a JEOL JEM 1011 electron microscope operating at 100 kV.
Supplementary Material
Acknowledgments
We thank Dr. Richard M. Elliott for generous help in providing the BUNV NP expression plasmid and the staff of the Photon Factory for their assistance with the diffraction data collection. We also thank Dr. Gang Ji (Institute of Biophysics, Chinese Academy of Sciences) for his assistance with EM data collection. This work was supported by Ministry of Science and Technology of China 973 Project Grants 2013CB911103 and 2010CB833600; National Natural Science Foundation of China Grants 31000332, 31170678, and 31230016; and Key Projects in the National Science and Technology Pillar Program Grants 10ZCKFSY07200 and 10ZCKFSY08700.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 4IJS).
See Commentary on page 8769.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222552110/-/DCSupplemental.
References
- 1.Guo Y, et al. Crimean-Congo hemorrhagic fever virus nucleoprotein reveals endonuclease activity in bunyaviruses. Proc Natl Acad Sci USA. 2012;109(13):5046–5051. doi: 10.1073/pnas.1200808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eifan SA, Elliott RM. Mutational analysis of the Bunyamwera orthobunyavirus nucleocapsid protein gene. J Virol. 2009;83(21):11307–11317. doi: 10.1128/JVI.01460-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elliott RM. Molecular biology of the Bunyaviridae. J Gen Virol. 1990;71(Pt 3):501–522. doi: 10.1099/0022-1317-71-3-501. [DOI] [PubMed] [Google Scholar]
- 4.Jiao Y, et al. Preparation and evaluation of recombinant severe fever with thrombocytopenia syndrome virus nucleocapsid protein for detection of total antibodies in human and animal sera by double-antigen sandwich enzyme-linked immunosorbent assay. J Clin Microbiol. 2012;50(2):372–377. doi: 10.1128/JCM.01319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferron F, et al. The hexamer structure of Rift Valley fever virus nucleoprotein suggests a mechanism for its assembly into ribonucleoprotein complexes. PLoS Pathog. 2011;7(5):e1002030. doi: 10.1371/journal.ppat.1002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raymond DD, Piper ME, Gerrard SR, Smith JL. Structure of the Rift Valley fever virus nucleocapsid protein reveals another architecture for RNA encapsidation. Proc Natl Acad Sci USA. 2010;107(26):11769–11774. doi: 10.1073/pnas.1001760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raymond DD, Piper ME, Gerrard SR, Skiniotis G, Smith JL. Phleboviruses encapsidate their genomes by sequestering RNA bases. Proc Natl Acad Sci USA. 2012;109(47):19208–19213. doi: 10.1073/pnas.1213553109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Y, Guo Y, Lou Z. 2012. A versatile building block: The structures and functions of negative-sense single-stranded RNA virus nucleocapsid proteins. Protein Cell 3(12):893–902. [DOI] [PubMed]
- 9.Kranzusch PJ, Whelan SP. Architecture and regulation of negative-strand viral enzymatic machinery. RNA Biol. 2012;9(7):941–948. doi: 10.4161/rna.20345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calisher CH, Sever JL. Are North American Bunyamwera serogroup viruses etiologic agents of human congenital defects of the central nervous system? Emerg Infect Dis. 1995;1(4):147–151. doi: 10.3201/eid0104.950409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tauro LB, Diaz LA, Almirón WR, Contigiani MS. Infection by Bunyamwera virus (Orthobunyavirus) in free ranging birds of Cordoba city (Argentina) Vet Microbiol. 2009;139(1-2):153–155. doi: 10.1016/j.vetmic.2009.04.032. [DOI] [PubMed] [Google Scholar]
- 12.Mohl BP, Barr JN. Investigating the specificity and stoichiometry of RNA binding by the nucleocapsid protein of Bunyamwera virus. RNA. 2009;15(3):391–399. doi: 10.1261/rna.1367209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodgers JW, Zhou Q, Green TJ, Barr JN, Luo M. Purification, crystallization and preliminary X-ray crystallographic analysis of the nucleocapsid protein of Bunyamwera virus. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62(Pt 4):361–364. doi: 10.1107/S1744309106006397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leonard VH, Kohl A, Osborne JC, McLees A, Elliott RM. Homotypic interaction of Bunyamwera virus nucleocapsid protein. J Virol. 2005;79(20):13166–13172. doi: 10.1128/JVI.79.20.13166-13172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walter CT, Bento DF, Alonso AG, Barr JN. Amino acid changes within the Bunyamwera virus nucleocapsid protein differentially affect the mRNA transcription and RNA replication activities of assembled ribonucleoprotein templates. J Gen Virol. 2011;92(Pt 1):80–84. doi: 10.1099/vir.0.024240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm L, Rosenstrom P. 2010. Dali server: Conservation mapping in 3D. Nucleic Acids Res 38(Web Server issue):W545–549.
- 17.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1):2256–2268.
- 18.Ruigrok RW, Crépin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14(4):504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Rudolph MG, et al. Crystal structure of the borna disease virus nucleoprotein. Structure. 2003;11(10):1219–1226. doi: 10.1016/j.str.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Albertini AA, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313(5785):360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 21.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313(5785):357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 22.Tawar RG, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326(5957):1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 23.Savji N, et al. Genomic and phylogenetic characterization of Leanyer virus, a novel orthobunyavirus isolated in northern Australia. J Gen Virol. 2011;92(Pt 7):1676–1687. doi: 10.1099/vir.0.028308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ren L, et al. Structural insight into substrate specificity of human intestinal maltase-glucoamylase. Protein Cell. 2011;2(10):827–836. doi: 10.1007/s13238-011-1105-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Part A, Methods in Enzymology, eds Carter, Jr CW, Sweet RM (Academic, New York), Vol 276, pp 307–326.
- 26.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58(Pt 11):1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 27.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 28.Novoa RR, Calderita G, Cabezas P, Elliott RM, Risco C. Key Golgi factors for structural and functional maturation of bunyamwera virus. J Virol. 2005;79(17):10852–10863. doi: 10.1128/JVI.79.17.10852-10863.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.