Abstract
To what extent are evolutionary outcomes determined by a population's recent environment, and to what extent do they depend on historical contingency and random chance? Here we apply a unique experimental system to investigate evolutionary reproducibility and path dependence at the protein level. We combined phage-assisted continuous evolution with high-throughput sequencing to analyze evolving protein populations as they adapted to divergent and then convergent selection pressures over hundreds of generations. Independent populations of T7 RNA polymerase genes were subjected to one of two selection histories (“pathways”) demanding recognition of distinct intermediate promoters followed by a common final promoter. We observed distinct classes of solutions with unequal phenotypic activity and evolutionary potential evolve from the two pathways, as well as from replicate populations exposed to identical selection conditions. Mutational analysis revealed specific epistatic interactions that explained the observed path dependence and irreproducibility. Our results reveal in molecular detail how protein adaptation to different environments, as well as stochasticity among populations evolved in the same environment, can both generate evolutionary outcomes that preclude subsequent convergence.
Keywords: directed evolution, evolutionary biology, tape of life
Stephen J. Gould famously hypothesized that if the “tape of life”— the long evolutionary trajectory that has led to present life on earth—were rewound and played again, the outcomes would be very different (1). Different evolutionary outcomes could arise from mutational stochasticity (random chance) or from differences in past selection environments (selection history). Adaptation to a common environment can theoretically restore the similarity of evolutionary outcomes by consistently enriching for a subset of these mutations, resulting in evolutionary convergence.
Several studies have investigated the reproducibility of evolution by evolving parallel populations from an identical ancestral state. Although phenotypic outcomes are often similar, the underlying genetic changes frequently differ across populations (2–12). Identical genetic outcomes occur more frequently when fitness hinges on the performance of very few genes, as with the evolution of small phages with only a handful of genes (13, 14), cellular traits determined by a single gene (15, 16), and especially for single proteins evolved in vitro (11). Indeed, biochemical explorations of all hypothetical evolutionary trajectories from a single starting sequence to a known evolutionary endpoint (11, 12, 17–20) have demonstrated that there are many more accessible paths to genotypes involving mutations scattered across the genome (21) than those with a similar number of mutations concentrated in a single gene (8). These results have led to suggestions that replaying the tape of life for protein-encoding genes might be surprisingly repetitive (8).
If parallel protein evolution frequently yields similar or identical outcomes, what conditions are sufficient to cause distinct ancestral populations to converge on similar solutions? Despite the importance of this question for the predictability of evolutionary outcomes in common environments, only a handful of experiments have directly or indirectly examined the ability of adaptation to overcome historical differences. These studies have observed high phenotypic convergence for Escherichia coli populations initially separated by genetic drift (2), limited genetic convergence among two ribozyme populations initially evolved with or without a denaturant (10), and no genetic convergence among moderately related phages adapted to high temperature (14). No experiments have directly examined the extent to which closely related protein-encoding genes can undergo convergent genetic and phenotypic evolution. Systematically investigating evolutionary convergence will require a method capable of generating protein populations with a desired level of divergence, then subjecting them to convergent selection pressures over hundreds of generations.
We hypothesized that our recently developed phage-assisted continuous evolution (PACE) system (22) could serve as an experimental platform for the investigation of protein evolutionary convergence and reproducibility in a continuous format, without concern for secondary fitness effects caused by host genome mutations. During PACE, host E. coli cells continuously dilute an evolving population of ∼1010 filamentous bacteriophages in a fixed-volume vessel (a “lagoon”). Dilution occurs faster than cell division but slower than phage replication, ensuring that only the phage can accumulate mutations. Each phage carries a protein-encoding gene to be evolved instead of a phage gene (gene III) that is required for infection. Phage encoding active variants trigger host-cell expression of gene III in proportion to the desired activity and consequently produce infectious progeny, but phage encoding less-active variants produce less infectious progeny that are diluted out of the lagoon.
Because PACE allows protein populations to be evolved in parallel over hundreds of generations under controlled mutation and selection conditions, it can be used to systematically investigate the evolutionary convergence and reproducibility of protein-encoding genes in a manner previously restricted to entire genomes. We recently demonstrated that PACE can experimentally explore the effects of mutation rate and selection stringency on evolutionary outcomes (23). In this work, we used PACE to experimentally address the following questions: (i) If initially identical enzyme populations are subjected to distinct selection pressures before converging toward a common evolutionary goal (Fig. 1A), will they evolve a common set of amino acid changes? (ii) How reproducible are the evolved similarities or differences?
Fig. 1.
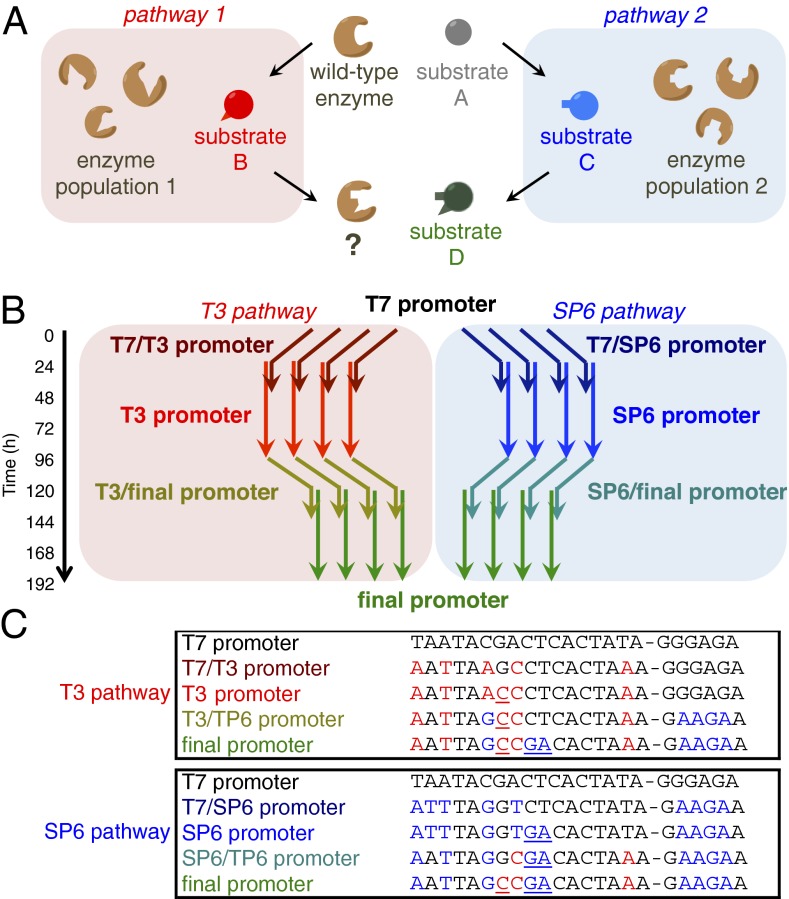
Design of two evolutionary pathways. (A) Schematic overview of this study. An enzyme is guided through two separate evolutionary pathways before undergoing convergent evolution towards the same final target. (B) Independent populations of phage-encoded T7 RNAP were continuously evolved over 192 h (∼200 generations) to recognize one of two distinct intermediate promoters (T3 and SP6) followed by a common “final” promoter. Each pathway included two hybrid “stepping stone” promoters (T7/T3, T7/SP6, T3/final, and SP6/final) preceding and following each intermediate. Arrows represent times during which phage populations were fed host cells bearing the indicated promoter. Overlapping arrows represent mixtures of host cell cultures. Evolution was simultaneously performed in four replicate populations for each pathway (eight populations total). (C) Promoter sequences for each target and intermediate promoter, with changed positions from the T3 (red) or SP6 (blue) promoters colored and the transcriptional start site indicated by a dash. Critical contacts at position −11 for T3 and position −8 and −9 for SP6 promoter recognition are underlined.
Results
Design of Two Selection Pathways.
To study the incidence of evolutionary convergence during protein evolution, we designed two selection pathways that subject T7 RNA polymerase (T7 RNAP) to selection pressure schedules with a common beginning and ending but that are otherwise distinct. Both pathways begin with phage encoding the wild-type T7 RNAP gene, which recognizes the T7 promoter with a high degree of sequence specificity (24), and diverge by demanding recognition of either the T3 or SP6 promoter, both of which are orthogonal to one another and to the T7 promoter in nature (25). Each pathway proceeds through a series of evolutionary “stepping stones” that introduce several nucleotide changes at a time (Fig. 1 B and C). The two initial stepping-stone promoters were designed such that wild-type T7 RNAP has enough activity on the T7/T3 and the T7/SP6 promoters to support robust plaque formation. We found that wild-type T7 RNAP retained ∼20% activity and supported phage propagation on each of two hybrid promoters in which all of bases were changed to the T3 promoter except −11G, or to the SP6 promoter except for −8C and −9T (SI Appendix, Fig. S1). These three nucleotides have previously been identified as the primary determinants of T7 vs. T3 (25–28) and T7 vs. SP6 promoter specificity (25, 29–31). The selection schedule for both pathways begins with 24 h of selection using host cells that demand recognition of the first hybrid promoter, 24 h of selection on a 1:1 mixture of host cells containing the first hybrid promoter and host cells containing the intermediate target (either T3 or SP6), and 48 h of selection on the first intermediate target (Fig. 1B).
After 96 h of evolution (∼100 phage generations), the two separate pathways then converge by requiring recognition of the same “final” promoter, a hybrid of the T3 and SP6 promoters in which 12 of 23 positions are altered relative to the starting T7 promoter. Starting with a sample from each of the 96-h populations, we performed PACE for 24 h on host cells that require recognition of the second hybrid promoter (either T3/final or SP6/final, which again contain all changes except those at either the −11 or the −8/−9 positions), then for 24 h on a mixture of host cells harboring the second hybrid promoter and the final promoter, and finally for 48 h on host cells that contain only the final promoter. To evaluate evolutionary stochasticity and reproducibility, we performed evolution in four replicate populations for each pathway. Replicate populations were housed in separate lagoon vessels that were diluted with the same host cell culture at a flow rate of 2 volumes per hour, ensuring that the selection histories of sibling populations were as similar as possible. Throughout the experiment, all eight populations were subjected to a mutation rate that is ∼100-fold higher than the basal E. coli mutation rate by inducing a mutagenesis plasmid with arabinose (22). In total, each of the eight populations was continuously evolved for 192 h, representing ∼200 phage generations for the average surviving phage in each population (22).
Evolved Population Phenotypes.
At the end of each step of the evolution, lagoons contained phage populations of 108 to 109 pfu/mL. To measure the phenotypic fitness of evolved clones at the middle and end of each experiment, we subcloned T7 RNAP genes from ∼20 randomly chosen phage from each population into an E. coli expression vector after 96 and 192 h of evolution and quantified transcriptional activity on different promoters using a luciferase assay. Evolved RNAP populations at each time point possessed 13–250% average activity on their respective target promoters relative to that of wild-type T7 RNAP on its native T7 promoter, which is 100% by definition (Fig. 2 A–C). Wild-type T7 RNAP has no detectable activity (< ∼1%) on the T3, SP6, or final promoters in this assay.
Fig. 2.
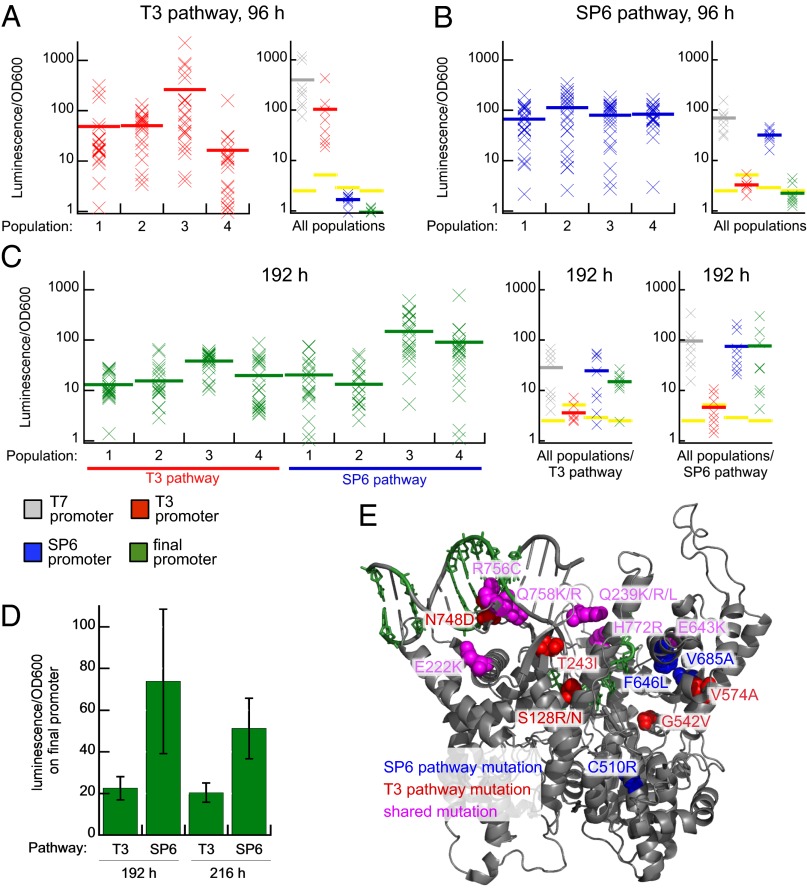
Phenotypes of evolved RNA polymerases. (A) T3 promoter activity of the four populations on the T3 pathway after 96 h of continuous evolution. Each “X” represents the luciferase activity of a single randomly chosen clone on the T3 promoter luciferase reporter in E. coli cells, normalized to wild-type T7 RNAP on the T7 promoter (100%). Gray, red, blue and green bars represent the average activity of all of the assayed clones from one population and yellow bars represent the background signal with no exogenous RNA polymerase present. The activity of a subset of clones from all T3 pathway populations on the full panel of promoters is also shown (SI Appendix, Fig. S3A). (B) SP6 promoter activity of the four populations on the SP6 pathway after 96 h of continuous evolution. The activity of a subset of clones from all SP6 pathway populations on the full panel of promoters is also shown (SI Appendix, Fig. S3B). (C) Final promoter activity of all eight populations after 192 h of continuous evolution. The activity of a subset of clones from all populations on the full panel of promoters is also shown (SI Appendix, Fig. S3 C and D). (D) Average final promoter activity of each pathway at 192 and 216 h. The average final promoter activity of the four populations from each pathway is shown. Error bars represent the SE of the four populations. (E) Crystal structure of the initiation complex of T7 RNAP (25) highlighting some of the key mutations identified by HTS and reversion analysis. The green nucleotides denote positions changed in the final promoter. Red and blue residues show sites of T3-pathway and SP6-pathway mutations, respectively. PDB Structure: 1QLN.
We selected a subset of individual clones from 96 h and 192 h that span the range of observed activities to assay on the T7, T3, SP6, and final promoters. At 96 h, clones from each pathway were very active on their respective T3 promoter (16–260% average activity) (Fig. 2A) or SP6 promoter (66–113% average activity) (Fig. 2B), but exhibited minimal or no detectable activity on the promoter of the other pathway (Fig. 2 A and B, and SI Appendix, Fig. S2 A and B), demonstrating strongly divergent evolved phenotypes at 96 h.
By 192 h, clones from both pathways exhibit > 10% average activity on the T7, SP6, and final promoters, but lower activity on the T3 promoter (Fig. 2C, and SI Appendix, Fig. S2 C and D), suggesting that recognition of the T3 promoter and the final promoter are mutually exclusive. Variants from the T3 pathway lost most of their ability to recognize the T3 promoter as they evolved activity on the final promoter, and six of the eight assayed 192-h T3 pathway variants also gained significant SP6 promoter activity even though these clones were never explicitly selected to recognize the SP6 promoter (Fig. 2C). The two variants from the T3 pathway assayed that did not gain SP6 activity also lost T7 activity and exhibited robust activity only on the final promoter (SI Appendix, Fig. S2C, variants T3-192-2-3 and T3-192-3-14). Variants from the SP6 pathway maintained their ability to recognize the SP6 and T7 promoters while acquiring final promoter activity (Fig. 2C).
Significantly, the 192-h evolved clones from the T3 pathway exhibited an average of ∼three- to fourfold lower average activity on the final promoter than those from the SP6 pathway (Fig. 2D). This activity difference suggests that the two pathways differed in their ability to evolve high levels of final promoter activity. Moreover, the average activity of assayed clones evolved within sibling populations that experienced identical selection histories also varied by up to 11-fold (e.g., SP6 population 2 vs. SP6 population 3) (Fig. 2C), indicating that even within the same pathway, evolutionary stochasticity was a strong determinant of phenotypic outcome.
Additional Evolution Does Not Resolve Differences in Evolved Activity Levels.
To test whether these activity differences between pathways and within each pathway reflected populations that were still evolving, we subjected all eight populations to an additional 24 h of PACE on the final promoter under increased selection pressure at a high flow rate of ∼3.5 volumes per hour, corresponding to ∼40 additional generations per population. This additional evolution allowed SP6 population 1 to evolve final promoter activity levels comparable to SP6 populations 3 and 4, but did not significantly alter the average final promoter activities of the T3 pathway populations (Fig. 2D and SI Appendix, Fig. S3). These results suggest that some populations reached local fitness maxima by 192 h and indicate that pathway-specific differences in phenotypic outcome persist even after many generations of convergent selection pressure.
Evolved Population Genotypes.
We sequenced five to eight complete clones from each of the eight populations at both the 96-h and 192-h time points, including those that had been assayed on the full panel of promoters (SI Appendix, Figs. S4–S8). Additionally, we submitted each population to high-throughput sequencing (HTS) at a coverage level sufficient to identify mutations at frequencies of 2.5% or greater to obtain a more comprehensive picture of sequence variation during evolution (SI Appendix, Fig. S9 and Dataset S1). We used the HTS data to calculate the average diversity of the populations and the number of unique and average mutational compositions within each pathway (SI Appendix, Fig. S10 A and B), all of which tended to increase over the course of the evolution. We also analyzed the HTS data using FST, a widely applied measure of population differentiation that estimates the variation between populations (32) (SI Appendix, Fig. S10C). Finally, we constructed phylogenetic trees using the single clone data (SI Appendix, Figs. S11 and S12).
At 96 h, the four populations from the T3 pathway evolved a variety of mutations that we previously observed (22) to confer T3 promoter-recognition activity, including E222K, G542V, V574A, and N748D. The SP6 pathway at 96 h evolved a different set of predominant mutations, including V685A and Q758K/R, in addition to E222K.
At 192 h, the T3 pathway populations evolved a variety of additional mutations not observed at 96 h, including E643K (two of four populations), R756C (three of four populations), Q758K/R (four of four populations), and H772R (three of four populations), and all four SP6 pathway populations enriched R756C (four of four populations). Although N748D, a mutation known to facilitate recognition of the −11 nucleotide in the T3 promoter (26), was highly enriched during the T3 pathway evolution, this mutation did not significantly enrich in the four populations from the SP6 pathway at 192 h, even though the final promoter contains the same nucleotide at position −11, strongly suggesting that N748D is preferred during the T3 pathway. R756 and Q758 interact directly with the template DNA strand at the −8 and −9 positions and are thought to account for the inability of T7 RNAP to recognize the SP6 promoter (25). Collectively, these results reveal that despite many generations of evolution on the same final promoter, each pathway by 192 h evolved sets of mutations that were distinct from those that evolved in the other pathway (Fig. 2E).
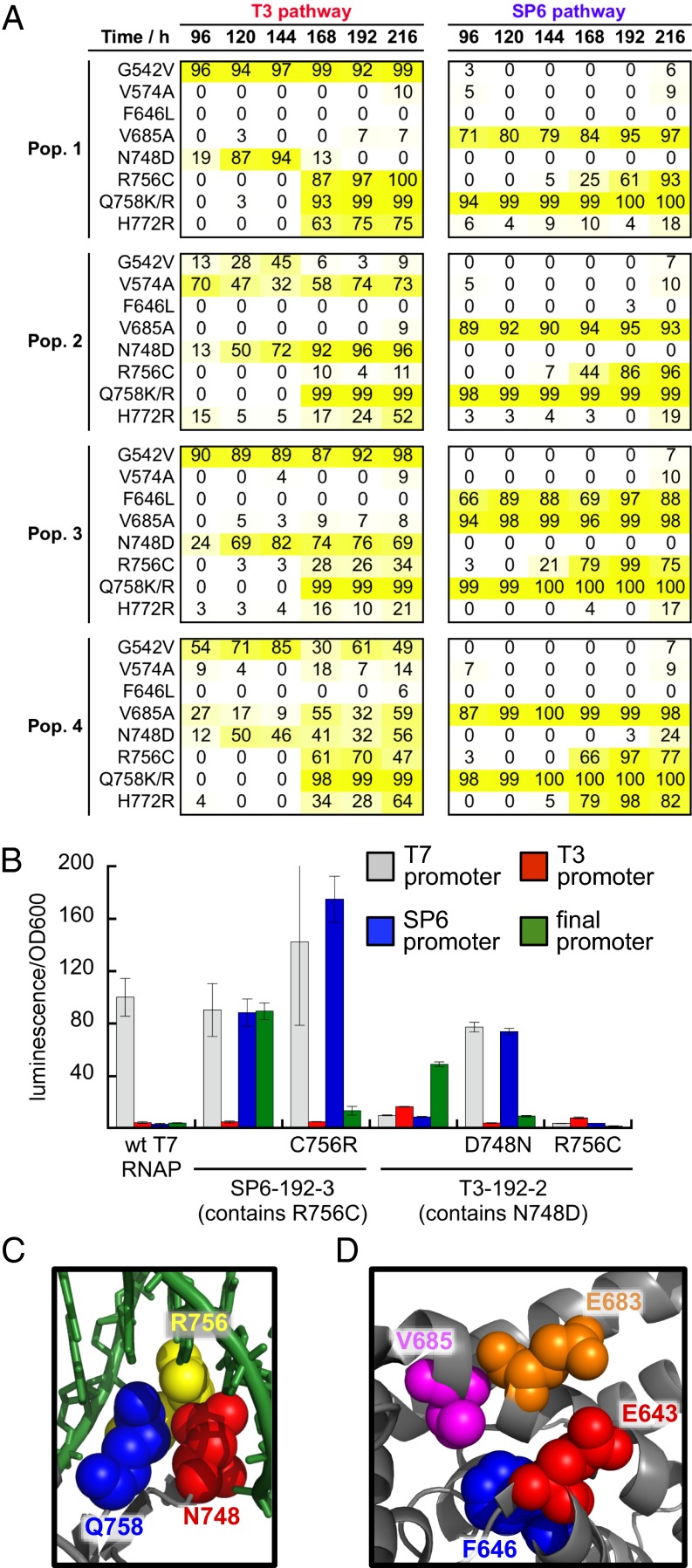
We also observed striking genotypic differences between sibling lagoons in the same pathway. For example, F646L was present at an abundance of 97% of SP6 pathway population 3 by 192 h, but was found in ≤ 3% abundance in the other three SP6 pathway populations. Similarly, V574A was present in 74% of T3 pathway population 2 at 192 h, but virtually absent from the other three T3 pathway populations at 192 h (Fig. 3A). These observations establish that stochasticity can strongly limit evolutionary reproducibility, even among populations surviving many generations of evolution under identical selection histories.
Fig. 3.
Epistasis and stochasticity drive evolutionary outcomes. (A) The abundance of a subset of key mutations in each population (pop.) is shown during the course of the evolution from 96 to 216 h as determined by HTS. (B) Normalized luciferase activity of wild-type T7 RNAP, an evolved RNAP clone with R756C (SP6-192-3-9) (SI Appendix, Fig. S7), SP6-192-3-9 with R756C reverted, an evolved RNAP clone with N748D (T3-192-2-3) (SI Appendix, Fig. S6), T3-192-2-3 with N748D reverted, and T3-192–2-3 with the addition of R756C on luciferase reporter vectors driven by each promoter. Error bars in B reflect SE (n = 3). (C) X-ray diffraction structure of T7 RNAP bound to the T7 promoter (25) showing the proximity of N748, R756, and Q758 at the DNA-binding interface. (D) X-ray diffraction structure of T7 RNAP bound to the T7 promoter (25) showing the proximity of E643, F646, E683, and V685. PDB Structure: 1QLN.
Functional Dissection of Key Mutations.
To understand the functional significance of some of the most highly enriched mutations from the two pathways, we chose four representative clones from the 192-h time point (two from each pathway) and analyzed the role of the most abundant mutations. First, we incorporated each of the mutations (E222K, V685A, F646L, N748D, R756C, and Q758K) into wild-type T7 RNAP. No single mutation significantly altered the promoter specificity profile of T7 RNAP, which exhibits no significant activity on the T3, SP6, or final promoters, although some mutations resulted in a loss of T7 promoter activity (SI Appendix, Fig. S13). We then reverted each mutation in the four evolved clones back to the wild-type amino acid and assayed the resulting clone’s complete promoter activity profile (SI Appendix, Fig. S14).
Reversion of E222K, present in all four clones from both pathways, results in a global loss of activity across all promoters assayed, regardless of the genetic background, demonstrating that this pathway-independent mutation is required to maintain high activity levels. Consistent with its high enrichment at the end of the evolution in both pathways, reversion of R756C results in decreased activity on the final promoter, but either negligible or increased activities on the T7, SP6, and T3 promoters. Reversion of N748D, the T3 pathway-preferred mutation that contacts the −11 position of the promoter, in a T3 variant that lacks R756C results in a loss of final promoter activity and a gain of SP6 promoter activity. N748D largely excludes recognition of the SP6 promoter, but is critical for final promoter activity in the absence of R756C. Surprisingly, reversion of Q758K/R in either T3 or SP6 pathway variants results in a complete loss of both SP6 and final promoter activities, as well as a gain in T3 promoter activity. This result is intriguing because it not only demonstrates that Q758K/R is the mutation responsible for the loss of T3 activity in the T3 pathway, but it shows that the variants from the SP6 pathway are only one mutation away from robust T3 activity.
That different selection pathways and populations within each pathway gave rise to genetic and phenotypic differences following convergent selection suggests that epistatic interactions may have precluded certain populations from achieving high final promoter activity. We therefore examined the phenotypic effects of individual mutations for evidence of epistasis.
Pathway Dependence Arising from Negative Epistasis Between R756C and N748D.
The four T3 pathway populations stochastically enriched either N748D or R756C by 192 h. In contrast, the four SP6 pathway populations predominantly enriched R756C, and little or no N748D, by 192 h. We sought to understand the molecular basis of this striking path-dependent outcome.
The abundance of R756C was strongly anticorrelated with the abundance of N748D (Fig. 3A). Previous biochemical studies on R756, which contacts N748 in the crystal structure (25) (Fig. 3C), suggested that mutations at this residue might reposition N748 (33). The epistasis of R756C and N748D is strongly supported by HTS data. If randomly distributed, these mutations should be present in the same clone at a frequency of 20% in T3 population 3 based on the individual abundance of each mutation. Instead, both mutations occurred together in only 6% of 6,661 individual T3 population 3 sequencing reads covering both positions.
This epistatic interaction between N748D and R756C has important phenotypic consequences, as the 192-h clones that lost T7 promoter recognition activity and do not recognize the SP6 promoter are the clones that contain N748D instead of R756C (SI Appendix, Fig. S2C, clones T3-192-2-3 and T3-192-3-14), whereas all of the 14 assayed clones containing the R756C maintain T7 promoter activity and obtain SP6 promoter activity as well. Reversion of R756C in a clone from the SP6 pathway that contains this mutation results in a sevenfold loss of final promoter activity (Fig. 3B). Reversion of N748D in a clone from the T3 pathway also results in a loss of final promoter activity (fourfold), as well as a 15-fold increase in SP6 activity (Fig. 3B). Introduction of R756C into a clone containing N748D indeed results in an almost complete loss of enzyme activity on all promoters, confirming the strong negative epistasis between these two mutations (Fig. 3B).
These results collectively provide a detailed explanation for the observed path-dependent “choice” between N748D and R756C. Although N748D and R756C both substantially increase final promoter activity, N748D strongly decreases SP6 promoter recognition. Therefore, SP6 pathway clones evolved R756C and not N748D because their histories necessarily avoided N748D. Even after selection pressure shifts entirely to transcription of the final promoter, epistasis enforces a fitness valley between these two mutations that prevented SP6 pathway clones from acquiring the N748D mutation.
Evolutionary Irreproducibility Arising from Sign Epistasis at F646L.
We next sought to address how SP6 population 3 was able to achieve the highest activity on the final promoter. The most notable genetic differences between sibling SP6 populations is the predominance of F646L in SP6 pathway population 3, compared with its virtual absence in any other population (Fig. 3A). We speculated that specific epistatic interactions prevented evolutionary parallelism at F646L by enabling high final promoter activity only in specific genetic contexts. Because the biochemical reversion analysis revealed that final promoter recognition requires the synergistic effects of E222K and Q758K/R with either N748D or R756C, and combinations of these core mutations are present in all eight populations from both selection pathways (Fig. 3A), the unusually high final promoter activity of SP6 population 3 must arise from additional mutations.
Reverting F646L in a clone from SP6 pathway population 3 resulted in a 10-fold decrease in final promoter recognition activity (SI Appendix, Fig. S14D), indicating that this mutation is important for strong final promoter activity in the genetic background of SP6 population 3. V685A, which is also required for robust final promoter activity in SP6 populations (SI Appendix, Fig. S14 C and D) and is highly enriched in all SP6 pathway populations (>70% at 96 h, and >94% at 192 h), is predicted to contact F646L (25). E643K, enriched in SP6 population 1, and E683D, enriched in SP6 population 4, are predicted to closely pack next to F646L and V685A, respectively (Fig. 3D).
The existence of this cluster of highly enriched mutations around the SP6 pathway-specific mutation V685A reveals that this region of the enzyme is an important mutational hotspot for evolving SP6 promoter recognition activity. We note that SP6 pathway population 2, which was unable to achieve high levels of final promoter activity even after ∼40 additional rounds of evolution, does not contain a highly enriched mutation in the direct vicinity of V685A. We therefore hypothesized that F646L may be unusually well suited for final promoter activity in a genetic context that appeared only in that population.
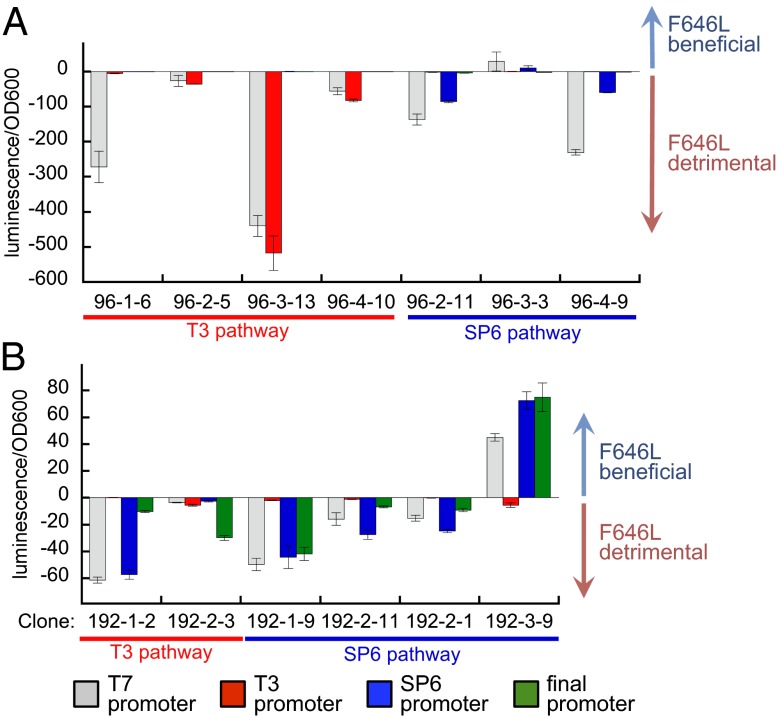
To test whether F646L can enhance the activity of clones with other genetic backgrounds, we introduced the F646L mutation into a variety of other 96- and 192-h clones, isolated from both pathways, which lack additional mutations near residue 685. Although F646L was beneficial to clones isolated from SP6 population 3, this mutation is detrimental when added to clones isolated from all other populations from both the 96- and 192-h time points (Fig. 4). These results indicate that the ability to benefit from F646L has already been lost by 96 h from all populations other than SP6 population 3, and therefore represents an example of “sign epistasis” (17), because F646L results in either increased or decreased activity, depending on genetic context. All four SP6 populations had equivalent SP6 activity levels at 96 h and all shared a common set of core mutations, but clones in SP6 population 3 were uniquely able to use F646L for future productive evolution. Interestingly, F646L increased the activity of wild-type T7 RNAP on the T7 promoter (SI Appendix, Fig. S13), indicating that seven of the eight populations first acquired mutations that exhibit negative epistasis with F646L. These findings collectively provide a molecular explanation for the basis of the nonreproducibility of evolutionary outcomes even among sibling populations subjected to identical selection histories.
Fig. 4.
Normalized difference in luciferase activity of evolved RNAP clones from 96 h (A) and 192 h (B) on luciferase reporter vectors driven by each promoter upon introduction of F646L. Clones are identified as timepoint-population#-clone#.
Discussion
Our experimental investigation of evolutionary convergence points to the existence of at least two clusters of local fitness peaks, accessed by the different selection pathways, on the final transcriptional activity fitness landscape (SI Appendix, Fig. S15). Populations subjected to the T3 pathway were unable to access the higher-activity cluster discovered by populations exposed to the SP6 pathway, despite being subjected to many generations of evolution on the same final promoter, establishing the importance of selection history in this system and the limitations of convergent selection pressure. Similarly, populations occupying lower fitness peaks within one cluster were unable to colonize higher peaks in the same cluster, directly demonstrating the limits of evolutionary reproducibility, which in the cases studied here was a result of epistasis.
Although we observed key mutations that consistently arise and contribute to overall activity, a result consistent with previous single-gene protein studies (8), we also observed stochastic outcomes between sibling populations subjected to identical selection histories, both phenotypically (such as SP6 populations 2 vs. 3, which evolved 13% vs. 150% average activity on the final promoter, respectively) and genotypically (such as T3 population 2 that enriched N748D vs. T3 population 1, which enriched R756C instead). These observations more closely parallel previous results in RNA-based evolutionary systems, which featured population sizes and total generations (34–37) more similar to those explored by PACE than did previous protein evolution experiments.
Taken together, our results demonstrate how the evolution of large protein populations over long evolutionary trajectories, even when subjected to many generations under the same selection pressure, can be strongly influenced both by stochastic occurrences and prior selection history. Surprisingly, epistasis-driven stochasticity within a pathway was a primary determinant of both genotypic outcome and phenotypic evolutionary potential. These results contrast with many previous fixed-endpoint and limited-duration single-gene evolution studies, indicating that parallel protein evolution experiments performed over many rounds of evolution are unlikely to result in highly similar genotypic or phenotypic outcomes. At least for T7 RNAP, the protein tape of life is not highly repetitive.
Divergent populations were even less likely to converge upon the same solution. Only in the grossest phenotypic sense—the ability to recognize the final promoter at any significant activity level—did the evolutionary trajectories studied here exhibit pathway-independence, whereas both the level of final promoter activity as well as the mutations that conferred activity were found to be strongly influenced by evolutionary pathway. Populations guided through the SP6 pathway were more likely to reach a higher activity regime (three of four SP6 pathway populations reached >50% relative activity vs. zero of four for the T3 pathway). This effect may be a result of the order in which mutations must arise, but may also be influenced by the strength and order of the selection pressures along each pathway. The convergence observed during evolution toward T3/final promoter recognition suggests that recognition of the altered SP6 bases at promoter positions −8/−9 may have been the most difficult step of the evolution. That the two pathways were forced to cross the most difficult stages of the specificity change at different times may have been a key determinant of the differing evolutionary outcomes. Future PACE experiments involving more replicate populations, different pathways, other protein activities, or different degrees of pathway divergence may further illuminate the factors governing evolutionary convergence.
Finally, our results have implications for future laboratory evolution efforts. If independent populations and divergent pathways are important outcome determinants during evolution, it may be more effective to subdivide one large evolving population into several isolated subpopulations and to guide those populations through alternative evolutionary stepping-stones. Multiple isolated populations, especially if occasionally subjected to differing selection pressures, may be more likely to avoid local fitness peak traps than a single large population, a result consistent with Sewall Wright’s predictions in his original formulation of the fitness landscape (38).
Methods
For experimental methods see the SI Appendix.
Phage-Assisted Continuous Evolution.
PACE was performed as previously described (22). Briefly, during the single-promoter stages of the evolution the lagoon volumes were fixed at 40 mL and during mixing stages of the evolutions the lagoon volumes were raised to 80 mL to keep the dilution rate constant.
Supplementary Material
Acknowledgments
We thank Irene Chen, Jacob Carlson, David Yang, and John Guilinger for their assistance. This research was supported by Defense Advanced Research Planning Agency Grants HR0011-11-2-0003 and N66001-12-C-4207; the Howard Hughes Medical Institute; National Science Foundation Grant DMR-1121053; National Science Foundation Grant CNS-0960316; National Institutes of Health National Research Service Award Fellowship F32GM095028 (to A.M.L.); and the John Templeton Foundation (B.A.). B.C.D. is a Fellow of the Jane Coffin Childs Memorial Fund for Medical Research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220670110/-/DCSupplemental.
References
- 1.Gould SJ. Wonderful Life: The Burgess Shale and the Nature of History. New York: WW Norton & Co; 1989. [Google Scholar]
- 2.Travisano M, Mongold JA, Bennett AF, Lenski RE. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995;267(5194):87–90. doi: 10.1126/science.7809610. [DOI] [PubMed] [Google Scholar]
- 3.Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 4.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature. 2012;489(7417):513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2012;44(1):101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenaillon O, et al. The molecular diversity of adaptive convergence. Science. 2012;335(6067):457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 7.Meyer JR, et al. Repeatability and contingency in the evolution of a key innovation in phage lambda. Science. 2012;335(6067):428–432. doi: 10.1126/science.1214449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinreich DM, Delaney NF, Depristo MA, Hartl DL. Darwinian evolution can follow only very few mutational paths to fitter proteins. Science. 2006;312(5770):111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 9.Romero PA, Arnold FH. Exploring protein fitness landscapes by directed evolution. Nat Rev Mol Cell Biol. 2009;10(12):866–876. doi: 10.1038/nrm2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayden EJ, Ferrada E, Wagner A. Cryptic genetic variation promotes rapid evolutionary adaptation in an RNA enzyme. Nature. 2011;474(7349):92–95. doi: 10.1038/nature10083. [DOI] [PubMed] [Google Scholar]
- 11.Salverda ML, et al. Initial mutations direct alternative pathways of protein evolution. PLoS Genet. 2011;7(3):e1001321. doi: 10.1371/journal.pgen.1001321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gumulya Y, Sanchis J, Reetz MT. Many pathways in laboratory evolution can lead to improved enzymes: How to escape from local minima. ChemBioChem. 2012;13(7):1060–1066. doi: 10.1002/cbic.201100784. [DOI] [PubMed] [Google Scholar]
- 13.Holder KK, Bull JJ. Profiles of adaptation in two similar viruses. Genetics. 2001;159(4):1393–1404. doi: 10.1093/genetics/159.4.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bollback JP, Huelsenbeck JP. Parallel genetic evolution within and between bacteriophage species of varying degrees of divergence. Genetics. 2009;181(1):225–234. doi: 10.1534/genetics.107.085225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gresham D, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4(12):e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Couñago R, Chen S, Shamoo Y. In vivo molecular evolution reveals biophysical origins of organismal fitness. Mol Cell. 2006;22(4):441–449. doi: 10.1016/j.molcel.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445(7126):383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- 18.O’Maille PE, et al. Quantitative exploration of the catalytic landscape separating divergent plant sesquiterpene synthases. Nat Chem Biol. 2008;4(10):617–623. doi: 10.1038/nchembio.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lozovsky ER, et al. Stepwise acquisition of pyrimethamine resistance in the malaria parasite. Proc Natl Acad Sci USA. 2009;106(29):12025–12030. doi: 10.1073/pnas.0905922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobler S, Dalla S, Wagschal V, Agrawal AA. Community-wide convergent evolution in insect adaptation to toxic cardenolides by substitutions in the Na,K-ATPase. Proc Natl Acad Sci USA. 2012;109(32):13040–13045. doi: 10.1073/pnas.1202111109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quan S, et al. Adaptive evolution of the lactose utilization network in experimentally evolved populations of Escherichia coli. PLoS Genet. 2012;8(1):e1002444. doi: 10.1371/journal.pgen.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esvelt KM, Carlson JC, Liu DR. A system for the continuous directed evolution of biomolecules. Nature. 2011;472(7344):499–503. doi: 10.1038/nature09929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leconte AM, et al. A population-based experimental model for protein evolution: Effects of mutation rate and selection stringency on evolutionary outcomes. Biochemistry. 2013;52(8):1490–1499. doi: 10.1021/bi3016185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joho KE, Gross LB, McGraw NJ, Raskin C, McAllister WT. Identification of a region of the bacteriophage T3 and T7 RNA polymerases that determines promoter specificity. J Mol Biol. 1990;215(1):31–39. doi: 10.1016/S0022-2836(05)80092-0. [DOI] [PubMed] [Google Scholar]
- 25.Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286(5448):2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 26.Raskin CA, Diaz G, Joho K, McAllister WT. Substitution of a single bacteriophage T3 residue in bacteriophage T7 RNA polymerase at position 748 results in a switch in promoter specificity. J Mol Biol. 1992;228(2):506–515. doi: 10.1016/0022-2836(92)90838-b. [DOI] [PubMed] [Google Scholar]
- 27.Klement JF, et al. Discrimination between bacteriophage T3 and T7 promoters by the T3 and T7 RNA polymerases depends primarily upon a three base-pair region located 10 to 12 base-pairs upstream from the start site. J Mol Biol. 1990;215(1):21–29. doi: 10.1016/s0022-2836(05)80091-9. [DOI] [PubMed] [Google Scholar]
- 28.Rong M, He B, McAllister WT, Durbin RK. Promoter specificity determinants of T7 RNA polymerase. Proc Natl Acad Sci USA. 1998;95(2):515–519. doi: 10.1073/pnas.95.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SS, Kang C. Two base pairs at -9 and -8 distinguish between the bacteriophage T7 and SP6 promoters. J Biol Chem. 1993;268(26):19299–19304. [PubMed] [Google Scholar]
- 30.Ikeda RA, Ligman CM, Warshamana S. T7 promoter contacts essential for promoter activity in vivo. Nucleic Acids Res. 1992;20(10):2517–2524. doi: 10.1093/nar/20.10.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee SS, Kang C. A two-base-pair substitution in T7 promoter by SP6 promoter-specific base pairs alone abolishes T7 promoter activity but reveals SP6 promoter activity. Biochem Int. 1992;26(1):1–5. [PubMed] [Google Scholar]
- 32.Holsinger KE, Weir BS. Genetics in geographically structured populations: Defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10(9):639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imburgio D, Rong M, Ma K, McAllister WT. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000;39(34):10419–10430. doi: 10.1021/bi000365w. [DOI] [PubMed] [Google Scholar]
- 34.Hanczyc MM, Dorit RL. Replicability and recurrence in the experimental evolution of a group I ribozyme. Mol Biol Evol. 2000;17(7):1050–1060. doi: 10.1093/oxfordjournals.molbev.a026386. [DOI] [PubMed] [Google Scholar]
- 35.Lehman N. Assessing the likelihood of recurrence during RNA evolution in vitro. Artif Life. 2004;10(1):1–22. doi: 10.1162/106454604322875887. [DOI] [PubMed] [Google Scholar]
- 36.Spiegelman S, Haruna I, Holland IB, Beaudreau G, Mills D. The synthesis of a self-propagating and infectious nucleic acid with a purified enzyme. Proc Natl Acad Sci USA. 1965;54(3):919–927. doi: 10.1073/pnas.54.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breaker RR, Joyce GF. Emergence of a replicating species from an in vitro RNA evolution reaction. Proc Natl Acad Sci USA. 1994;91(13):6093–6097. doi: 10.1073/pnas.91.13.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wright S. 1932. The roles of mutation, inbreeding, crossbreeding, and selection in evolution. Proceedings of the Sixth International Congress on Genetics 1:355–366.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.