Abstract
As a managed pollinator, the honey bee Apis mellifera is critical to the American agricultural enterprise. Recent colony losses have thus raised concerns; possible explanations for bee decline include nutritional deficiencies and exposures to pesticides and pathogens. We determined that constituents found in honey, including p-coumaric acid, pinocembrin, and pinobanksin 5-methyl ether, specifically induce detoxification genes. These inducers are primarily found not in nectar but in pollen in the case of p-coumaric acid (a monomer of sporopollenin, the principal constituent of pollen cell walls) and propolis, a resinous material gathered and processed by bees to line wax cells. RNA-seq analysis (massively parallel RNA sequencing) revealed that p-coumaric acid specifically up-regulates all classes of detoxification genes as well as select antimicrobial peptide genes. This up-regulation has functional significance in that that adding p-coumaric acid to a diet of sucrose increases midgut metabolism of coumaphos, a widely used in-hive acaricide, by ∼60%. As a major component of pollen grains, p-coumaric acid is ubiquitous in the natural diet of honey bees and may function as a nutraceutical regulating immune and detoxification processes. The widespread apicultural use of honey substitutes, including high-fructose corn syrup, may thus compromise the ability of honey bees to cope with pesticides and pathogens and contribute to colony losses.
Keywords: abaecin, cytochrome P450
The western honey bee Apis mellifera is the most important managed pollinator species in the world; in the United States, its pollination services are estimated at contributing $14 billion annually to the economy (1). The apicultural industry in the United States, however, has been threatened in recent years by substantial colony losses. During the past 5 y, annual losses have amounted to ∼30% of managed colonies in the United States (2–5). Colony collapse disorder, a suite of symptoms associated with many of these losses, is characterized by a sudden disappearance of worker bees (2–6). Multiple factors have been investigated as potential causes of or factors contributing to colony collapse disorder, including honey bee pathogens and parasites spanning several kingdoms (7–9) as well as exposure to pesticides that compromise immune responses (10, 11), navigation ability, learning, and memory (12).
Because bees must gather nectar for honey from spring through fall in temperate regions to make sufficient honey to maintain the colony through the winter months, they use a spectacular diversity of plant species as nectar sources. Indeed, their ability to pollinate so many different plant species contributes to their status as the premier managed pollinator in agricultural systems worldwide. However, despite the potential exposure to a broad diversity of phytochemicals in the nectar of the diverse flower species visited, the honey bee genome is characterized by a paucity of genes associated with detoxification. Cytochrome P450 monooxygenases (P450s) are among the principal phase I detoxification enzymes used by organisms, including insects, to metabolize xenobiotics, including phytochemicals and insecticides (13). Whereas most other insect genomes contain 80 or more cytochrome P450 genes, A. mellifera has only 46 P450 genes (14). Honey bees metabolize phytochemicals found in honey and pollen as well as acaricides used in-hive for management of Varroa destructor, an ectoparasitic mite of honey bees, via a number of CYP6 and CYP9 family members. Quercetin, a flavonoid constituent of honey and pollen, is metabolized by three enzymes in the CYP6AS subfamily and two enzymes in the CYP9Q subfamily (15, 16), whereas the acaricides coumaphos and τ-fluvalinate are detoxified by three enzymes in the CYP9Q subfamily (16). Regulation of these detoxification genes in A. mellifera differs in some respects from P450 regulation in other insects (17) in that CYP6AS3, which metabolizes quercetin, is not inducible by its substrate or by phenobarbital, a classic experimental inducer of insect P450 transcription. Regulation of genes involved in detoxification of dietary phytochemicals may be different in bees because honey, the principal source of energy to meet the metabolic needs of the hive, is processed from diverse floral nectar sources and its phytochemical composition varies according to locality and phenology. That P450s are regulated by predictable constituents of host plants in most insect herbivores (17) suggests that there may be predictable constituents of honey that serve as specific inducers of detoxification enzymes.
Results
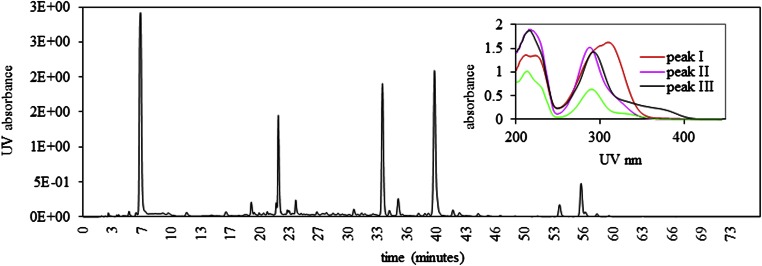
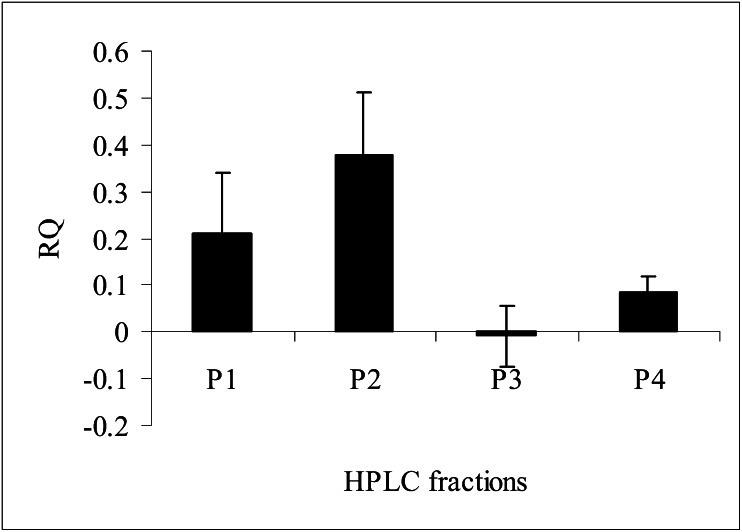
Honey extracts have previously been documented to up-regulate transcription of genes in the CYP6AS (18) and CYP9Q (16) subfamilies. We have now identified the specific constituents responsible for induction of these genes. HPLC separation of ethyl acetate extracts of honey yielded four peaks (Fig. 1), the constituents of which were checked for their ability to up-regulate detoxification genes by isolating them from 0.6 mL of ethyl acetate extract (equivalent to 60 mL honey) and bioassaying them in “bee candy” (a mixture of powdered sugar and sucrose syrup). Each fraction was evaporated to dryness, taken up in methanol, and added to 1 g of bee candy to compare its effects vs. those of bee candy prepared with an equivalent amount of methanol. Quantitative RT-PCR analyses demonstrated that three of the four peaks (peaks 1, 2, and 4) induced CYP9Q3 transcript accumulation (Fig. 2).
Fig. 1.
Isolation of CYP9Q3 inducers in honey. Reverse-phase HPLC separation of CYP9Q3 inducers in the ethyl acetate fraction of honey. (Inset) UV spectra from 200∼450 nm of peaks 1, 2, 3, and 4 (ie, I–IV).
Fig. 2.
Quantitative RT-PCR analyses of CYP9Q3 inducibility by the four peaks HPLC-fractionated in Fig.1. Data represent mean ± SEM (three technical replicates). P1, p-coumaric acid; P2, pinobanksin 5-methyl ether; P3, pinobanksin; P4, pinocembrin.
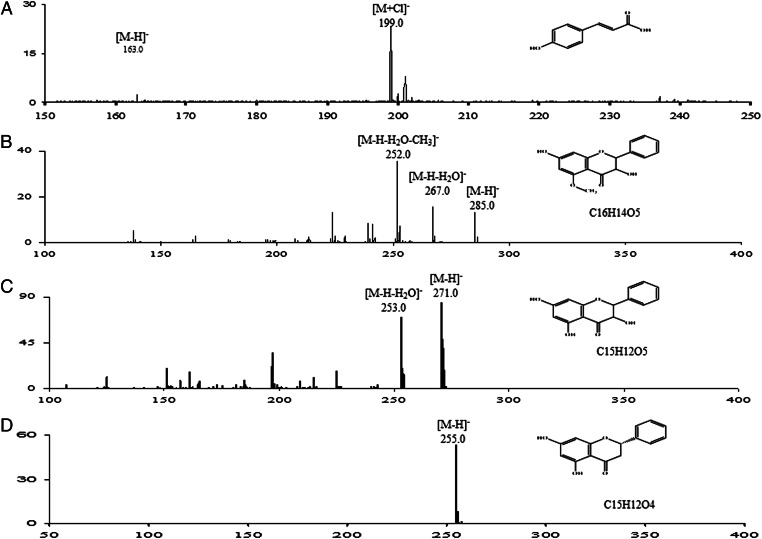
To identify the compounds represented by each of these peaks, we used several methods. The UV/visible spectra of the four peaks (Fig. 1, Inset) indicated that peak 1 is a hydroxycinnamic acid and peaks 2, 3, and 4 contain flavanones or dihydroflavonols. Peak 1, with a molar mass determined to be 164 g/mol by MS analysis (Fig. 3A), was identified as p-coumaric acid and confirmed by HPLC comparison with an authentic standard. Peak 4, with a molar mass of 256 g/mol and an element composition of C15H12O4, was determined to be pinocembrin (Fig. 3D) and confirmed by HPLC comparison with an authentic standard. Peaks 2 and 3 were determined to be dihydroflavonols with a hydroxyl group at C3 on the C-ring (Fig. 3 B and C). The identity of peak 2 as pinobanksin-5-methyl ether, with a molar mass of 287 g/mol and an element composition of C16H14O5 (Fig. 3B), is consistent with methylation of the 5-hydroxyl group on pinobanksin causing a 5- to 10-nm hypsochromic shift compared with methylation of the 7-hydroxyl group (Fig. 1). Similarly, peak 3 was determined to be pinobanksin, with a molar mass of 272 g/mol and an element composition of C15H12O5 (Fig. 3C).
Fig. 3.
Structure determination of four inducers purified from honey extract. Other than p-coumaric acid (P1 in A), whose structure was determined by electrospray MS method, the structures of all inducers were determined by quadrupole TOF MS-MS with elemental compositions predicted by using the built-in software of this system (P2 in B, pinobanksin 5-methyl ether; P3 in C, pinobanksin; P4 in D, pinocembrin).
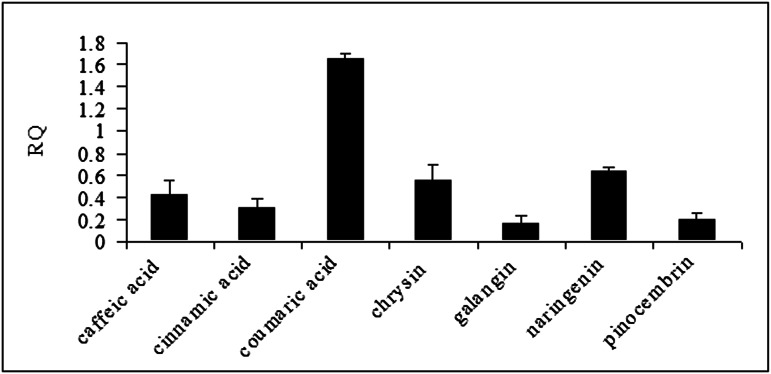
Induction experiments with p-coumaric acid and pinocembrin administered in increasing concentrations in bee candy revealed that p-coumaric acid is a stronger inducer of CYP9Q3, continuously increasing in induction capacity with concentration, whereas pinocembrin induction increased to its highest point at 23.4 µmol/g bee candy (Fig. S1). Therefore, this concentration was chosen for all potential inducers in subsequent experiments monitoring CYP9Q3 transcript levels. Among the three phenolic acids tested (Fig. S2), p-coumaric acid is the strongest inducer of CYP9Q3 (Fig. 4). Among the flavonoids, chrysin and naringenin were more effective inducers than were pinocembrin and galangin (Fig. 4). Pinobanksin (Fig. 2, peak 3) and galangin (Fig. 4) may be less effective inducers as a result of the presence of 3-hydroxyl groups on their C-rings. In contrast, pinobanksin 5-methyl ether is highly effective, suggesting that methylation of the 5-hydroxyl group on the A-ring of this compound enhances efficacy (Fig. 2, peak 2).
Fig. 4.
CYP9Q3 inducibility of representative phenolic acid and flavonoid constituents of honey as determined via quantitative RT-PCR. Data represent mean ± SEM (three biological replicates). RQ, relative quantification (i.e., fold increase vs. control, calculated as 2−ΔΔCt.
To determine the full range of genes regulated by p-coumaric acid in the honey bee midgut, an important site of pathogen and toxin entry for insects (17), three pairs of samples (control vs. treatment) were subjected to RNA-seq analysis. The collected RNA-seq data were analyzed by using Cuffdiff, DESeq, and edgeR (19). Of all genes up-regulated by p-coumaric acid, 31 were shared by the three methods (Fig. S2), and 110 genes were common in the lists of up-regulated genes from the DESeq and edgeR analyses. Remarkably, among these 31 genes (Table 1), 12 xenobiotic-metabolizing genes, including seven genes encoding phase I enzymes, four genes encoding phase II enzymes (three of which comprise one-quarter of all uridine-diphosphate-glucosyl transferases in the honey bee genome) (20), and two genes encoding phase III enzymes, were up-regulated by p-coumaric acid. In addition to CYP9Q3, known to metabolize pesticides (16), four CYP6AS enzymes known to metabolize honey flavonoids (10) and CYP6BD1 were induced 1.9- to 3.11-fold. Also up-regulated by almost twofold was the gene encoding abaecin (Dataset S1), an antimicrobial peptide mediating immunity against bacteria (21). In addition, the DESeq and edgeR analyses identified GB19392-RA, another honey bee gene encoding an antimicrobial peptide, defensin1, as more than twofold up-regulated by p-coumaric acid (Datasets S2 and S3). No genes were down-regulated by any of the compounds tested.
Table 1.
Genes up-regulated at least 1.4-fold by p-coumaric acid as identified by three independent methods of RNA-seq expression analysis
| Gene name | Fold change | Accession no. |
| Antimicrobial genes | ||
| Drug- metabolizing genes | ||
| Phase I | ||
| Abaecin | 1.90 | NP_001011617 |
| CYP6AS2 | 2.70 | XP_395085 |
| CYP6AS3 | 3.11 | XP_001122413 |
| CYP6AS4 | 2.60 | XP_395671 |
| CYP6AS5 | 2.55 | NP_001035324 |
| CYP6BD1 | 1.92 | XP_623955 |
| CYP9Q3 | 2.55 | XP_392001 |
| β-Esterase, E-class | 1.47 | XP_392696 |
| Phase II | ||
| α-Glutamyltransferase 1 | 1.50 | XP_393584 |
| UGT334C1 glucosyltransferase | 1.46 | XP_001123301 |
| UGT318A3 glucosyltransferase | 2.26 | XP_396494 |
| UGT332F1 glucosyltransferase | 1.81 | XP_392727 |
| Phase III | ||
| Multidrug resistance-associated protein 4 | 1.60 | XP_623460 |
| Multidrug resistance-associated protein 1 | 1.80 | XP_003249371 |
| Other genes | ||
| FABP-like protein | 1.58 | NP_001011636 |
| Inorganic phosphate cotransporter-like protein | 1.60 | XP_393759 |
| Similar to CG13424 (genome assembly nucleotide accession) | 1.59 | NW_003377962 |
| Protein lethal(2)essential for life-like | 1.92 | XP_393575 |
| Hypothetical protein LOC408807 | 3.69 | XP_397526 |
| Sarcoplasmic calcium-binding protein 2 | 2.43 | XP_003249222 |
| Hypothetical protein LOC552202 | 1.55 | XP_624582 |
| Venom acid phosphatase | 1.71 | NP_001013377 |
| Luciferin 4-monooxygenase-like | 2.81 | XP_001122105 |
Dataset S1 provides Cuffdiff analysis of differential gene expression in the midguts of honey bees fed with p-coumaric acid-containing sugar candy (CoA) vs “bee candy” (CK); Dataset S2 provides DESeq analysis of differential gene expression in the midguts of honey bees fed with p-coumaric acid-containing sugar candy (CoA) vs “bee candy” (CK); Dataset S3 provides edgeR analysis of differential gene expression in the midguts of honey bees fed with p-coumaric acid-containing sugar candy (CoA) vs “bee candy” (CK). See Fig. S2 for depiction of intersecting sets of genes up-regulated by p-coumaric acid as determined by RNA-seq of genes and identified by all three analytical methods.
To determine the functional significance of P450 up-regulation by p-coumaric acid, newly emerged adult bees were fed with bee candy for 3 d with or without augmentation with an ecologically realistic concentration of p-coumaric acid. Because this compound is present in honey and pollen/beebread, daily rates of p-coumaric acid ingestion will depend largely on floral source of nectar and pollen, which can vary over several orders of magnitude, and relative consumption of honey and beebread. To use an ecologically reasonable concentration and at the same time maximize the likelihood of observing a functional response, we selected a concentration of 1 mg/g, approximately double the concentrations reported in certain pollen (0.41 mg/g) (22) and beebread (0.367 mg/g) (23). Midguts were removed and assayed for rates of substrate disappearance of coumaphos, an acaricide known to be metabolized by CYP9Q3 (16), which is included among the genes up-regulated by p-coumaric acid in the RNA-Seq analysis. Activities of control and p-coumaric acid–induced midguts toward coumaphos were 2.57 and 4.10 nmol/min per midgut, respectively. A statistical comparison by t test demonstrated that the activity of p-coumaric acid–induced midguts is significantly greater than the activity of guts from bees consuming only bee candy (P = 0.047; i.e., significant at P < 0.05).
Discussion
Pollen ingestion is known to reduce honey bee susceptibility to pesticides and pathogens (reviewed in ref. 24); this effect may result in part from the up-regulation of nutrient-sensing and metabolic pathways as well as genes encoding certain antimicrobial peptides in response to pollen diets (24). To date, the specific constituents of pollen responsible for this up-regulation have not been identified. In that p-coumaric acid is a structural component of sporopollenin, the principal material comprising the outer wall, or exine, of pollen grains (25), it is likely be consumed by bees in beebread and honey (which in the hive invariably contains some quantity of pollen). Sporopollenin itself, however, is not readily digested by bees, so how much p-coumaric acid is actually consumed on a regular basis is difficult to estimate across all pollen types (although as much as 98% of pollen contents can ultimately be extracted by bees) (24).
Our analysis of honey extracts revealed that p-coumaric acid can induce detoxification genes; moreover, RNA-seq analysis demonstrates that it up-regulates a select suite of genes required for defense against pesticides and pathogens. Thus, this ubiquitous constituent of honey and beebread may act as a nutraceutical, defined as a nonnutrient food constituent contributing to health, in honey bees. The content of p-coumaric acid in pollen/beebread and honey varies with floral source, so, although it is a ubiquitous cue, it may not always be a sufficient cue for up-regulating detoxification functions. In our study of honey derived predominantly from soybeans/wildflowers, we also identified three other constituents (pinobanksin, pinobanksin 5-methyl ether, and pinocembrin) that were effective inducers of CYP9Q enzymes. These compounds are not reported from nectar but rather are abundant in bud exudates of poplars (Populus spp.) and other salicaceous plants, which are among the tree resins collected by bees to produce propolis, the resinous “bee-glue” that seals cracks and lines cells in the hive (26). The activity of these honey constituents raises the possibility that other honey compounds (including nectar-derived flavonoids; Fig. 3) and/or hive products may well interact with p-coumaric acid with additive or even synergistic consequences to regulate detoxification and/or immune status.
That honey up-regulates detoxification genes whereas sucrose and high-fructose corn syrup do not (18) suggests estimates of pesticide toxicity based on assays that used these honey substitutes may need reexamination (e.g., ref. 12). More importantly, the practice of using honey substitutes is widespread in commercial beekeeping operations as a cost-saving measure. This longstanding practice was adopted after laboratory studies demonstrated the acceptability and nutritional equivalence of substitutes (27). These studies, however, were conducted before the introduction of varroa mites in the mid-1980s; since that introduction, the pathogen load of US bees has been substantially increased because of the ability of varroa mites to act as vectors, and pesticide exposures have increased due to the use of in-hive acaricides and nontarget encounters with pesticides in agricultural fields. In view of current knowledge of contemporary levels of honey bee exposure to pesticides (28) and of increased pathogen loads caused by globalization of trade (e.g., ref.7), examining the ability of honey and honey substitutes to regulate expression of detoxification and immunity genes would seem to be a high priority. At minimum, after comprehensive testing and development, p-coumaric acid may find use as an additive to honey substitutes to allow beekeepers to maintain colonies during food shortages without compromising the ability of their bees to defend themselves against the pesticides and pathogens that currently bedevil beekeeping in the United States.
Materials and Methods
Phenolic Standards.
Galangin was obtained from Indofine Chemical. Caffeic acid, trans-cinnamic acid, p-coumaric acid, chrysin, naringenin, and pinocembrin were purchased from Sigma-Aldrich.
Isolation and Structural Determination of Phenolics from Honey.
For preparation of a honey extract to be used for isolation of active phenolics, 100 mL of honey from the University of Illinois Bee Research Facility was diluted with 900 mL warm distilled water. After extracting with three 300-mL volumes of petroleum ether, 100 g of Amberlite XAD16 (Sigma-Aldrich) was added in the water phase to absorb the compounds with phenol rings. Then, the Amberlite was drained on a 150 mL ASTM coarse (40–60 μm) Buchner funnel, washed three times with 150 mL of water, and eluted with 300 mL of methanol. After the methanol in the eluent was evaporated by rotary evaporator (Buchi Rotavapor; Brinkmann), the remaining solution was extracted with the same volume of ethyl acetate three times. The pooled ethyl acetate extracts were again dried by rotary evaporator and resuspended in 1 mL ethyl acetate.
The ethyl acetate extract was fractionated by using a 4.6 × 250 mm i.d. 5-μm Symmetry C18 column (Waters) connected to a Waters HPLC system coupled with a Waters 996 photodiode array detector at a flow rate of 1 mL/min. The separation was performed by means of a linear gradient elution (solvent A, water with 0.1% formic acid; solvent B, acetonitrile with 0.1% formic acid): 10% (vol/vol) solvent B for 5 min, 10% to 26% (vol/vol) solvent B for 5 min, 26% to 60% (vol/vol) solvent B for 60 min, 60% to 100% (vol/vol) solvent B for 1 min, 100% (vol/vol) solvent B for 5 min, and 10% (vol/vol) solvent B for 1 min. The photodiode array detector was set at 280 nm to monitor the UV/visible absorption. UV/visible spectra were recorded from 200 to 450 nm. Four peaks—I (22 min), II (33.7 min), III (39.5 min), and IV (56 min)—were collected (Fig. 1), lyophilized, and resuspended in 1 mL of methanol for bioassays, MS, and tandem MS (MS-MS) analysis.
MS analyses of the peaks were performed at the Mass Spectrometry Service Facility and VOICE NMR Lab at University of Illinois at Urbana–Champaign with a quadrupole TOF Ultima AP1 MS unit (Waters) equipped with an ESI interface. For each peak except I, an MS spectrum in the positive mode and an MS-MS spectrum in negative mode were obtained to analyze their element composition and to acquire information on their fragmentation patterns. With respect to peak I, only an MS spectrum in negative mode was obtained because its signals were too weak for MS-MS analysis in negative mode and MS analysis in positive mode.
Quantitative RT-PCR.
Approximately 15 1-d-old worker bees were placed in plastic Solo Deli cups (16 oz, 454 g) covered with cotton cheesecloth and were fed with 1 g bee candy as a control or bee candy containing a test compound as a treatment. Bee candy is made from equal parts powdered sugar and heavy sucrose syrup (two parts sucrose to one part water) and serves as a medium for delivering test materials via ingestion. Ten midguts per treatment were dissected after 3 d, frozen in liquid nitrogen, and stored at −80 °C to extract total RNA for quantitative RT-PCR analysis of CYP9Q3 expression. Whereas each treatment was independently replicated three times, the bioassays for the eluents corresponding to the three peaks were performed a single time.
RNA was extracted from the frozen midguts with the TRIzol method (Invitrogen), treated with DNase (Ambion), and subsequently used for cDNA synthesis. Quantitative RT-PCR analyses were carried out as described earlier (16).
RNA-Seq Analyses.
Approximately 15 1-d-old bees, placed as before in plastic Solo Deli cups covered with cotton cheesecloth, were fed with 1 g of bee candy as a control or bee candy containing 31.4 μmol of p-coumaric acid as a treatment. Each treatment was replicated three times. Ten midguts per treatment were dissected after 3 d, frozen in liquid nitrogen, and stored at −80 °C to extract total RNA for RNA-seq analysis. RNA was extracted from the frozen midguts with the TRIzol method and cleaned up with an RNeasy Mini Kit, and the RNase-free DNase set for on-column DNase digestion (Qiagen).
RNA sequencing and analysis of the RNA-seq data were carried out at the W. M. Keck Center for Comparative and Functional Genomics at the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana–Champaign. The RNA-seq libraries were prepared using the TruSeq RNA-seq Sample Prep kit according to the manufacturer’s instructions (Illumina). The libraries were quantified by quantitative PCR and sequenced on one lane for 100 cycles on a HiSeq2000 by using a TruSeq sequencing by synthesis kit (version 3) and analyzed with Casava1.8 (pipeline 1.9). TopHat was used to align RNA-Seq reads to the A. mellifera genome assembly 4 (amel4.fa). Three different methods (Cuffdiff, DESeq, and edgeR) were used to analyze differentially expressed genes (adjusted P ≤ 0.05) caused by p-coumaric acid treatment (19). Clean Genes.gff3 was used as the annotation file for amel4.fa.
Inducibility of Coumaphos Metabolism by p-Coumaric Acid.
Between 10 and 20 1-d-old bees, placed in plastic Solo Deli cups covered with cotton cheesecloth, were fed with 1 g of bee candy as a control or bee candy containing 1 μg of p-coumaric acid as a treatment. Each treatment was replicated three times. After 3 d, the midguts of bees in both treatment groups were dissected and cleaned on ice in ice-cold grinding buffer (0.1 M sodium phosphate buffer, pH 7.4, containing 20% glycerol, 1.1 mM EDTA, 0.1 mM DTT, 0.5 mM PMSF, and 5 µg/mL leupeptin), homogenized in 50 µL grinding buffer per midgut, and centrifuged at 2,300 × g for 5 min at 4 °C. The supernatants were frozen in liquid nitrogen and stored at −80 °C for bioassays of coumaphos metabolism (i.e., rate of substrate disappearance). Metabolism reactions were set up with 50 µL of the supernatant (for each midgut), 5 µL of 5 mM coumaphos in ethanol, 400 µL of 0.1 M phosphate buffer (pH 7.4), and 50 µL of the phosphate buffer for controls or 50 µL of the phosphate buffer with NADPH for treatments. The reactions were incubated for 45 min in a 35 °C shaking water bath. The reaction mixture was extracted with 500 mL of ethyl acetate and centrifuged at 17,000 × g for 5 min by using a benchtop centrifuge at room temperature. Twenty microliters of the ethyl acetate phase was analyzed by reverse-phase HPLC (Synergi 4 µ Fusion-RP 80A column, 250 × 4.6 mm). Gradient elution (solvent A, water containing 0.1% formic acid; solvent B, acetonitrile containing 0.1% formic acid) was performed at a flow rate of 1 mL/min with gradient conditions ranging from 70% solvent B to 90% solvent B over 10 min after 5 min at 70% solvent B.
Supplementary Material
Acknowledgments
We thank Charley Nye [University of Illinois at Urbana–Champaign (UIUC)] for assistance with honey bees, Dr. Barry Pittendrigh and Dr. Weilin Sun (UIUC) for assistance with quantitative PCR analyses, Dr. Alvaro Hernandez (UIUC) for advice on and assistance with RNA-seq analysis, Dr. Gene Robinson (UIUC) for advice and assistance, Dr. Marla Spivak (University of Minnesota) for valuable discussions, and Dr. Jay D. Evans [US Department of Agriculture (USDA)–Agricultural Research Service Bee Research Laboratory, Beltsville, MD] and Dr. Rene Feyereisen (Institut National de la Recherche Agronomique–Centre National de la Recherche Scientifique, Université de Nice Sophia-Antipolis) for their critical reviews of the manuscript. This project was funded by USDA Agriculture and Food Research Initiative 2010-03760.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information RefSeq database. For a list of accession numbers, see Table 1.
See Commentary on page 8763.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303884110/-/DCSupplemental.
References
- 1.Calderone NW. Insect pollinated crops, insect pollinators and US agriculture: trend analysis of aggregate data for the period 1992-2009. PLoS ONE. 2012;7(5):e37235. doi: 10.1371/journal.pone.0037235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.vanEngelsdorp D, Underwood R, Caron D, Hayes J., Jr An estimate of managed colony losses in the winter of 2006 - 2007: A report commissioned by the Apiary Inspectors of America. Am Bee J. 2007;147:599–603. [Google Scholar]
- 3.van Engelsdorp D, Hayes J, Jr, Underwood RM, Pettis J. A survey of honey bee colony losses in the U.S., fall 2007 to spring 2008. PLoS ONE. 2008;3(12):e4071. doi: 10.1371/journal.pone.0004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.vanEngelsdorp D, Hayes J, Jr, Underwood RM, Pettis JS. A survey of honey bee colony losses in the United States, fall 2008 to spring 2009. J Apic Res. 2010;49:7–14. [Google Scholar]
- 5.vanEngelsdorp D, et al. A national survey of managed honey bee 2010-11 winter colony losses in the USA: Results from the Bee Informed Partnership. J Apic Res. 2012;51:115–124. [Google Scholar]
- 6.Oldroyd BP. What’s killing American honey bees? PLoS Biol. 2007;5(6):e168. doi: 10.1371/journal.pbio.0050168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox-Foster DL, et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science. 2007;318(5848):283–287. doi: 10.1126/science.1146498. [DOI] [PubMed] [Google Scholar]
- 8.Evans JD, Schwarz RS. Bees brought to their knees: Microbes affecting honey bee health. Trends Microbiol. 2011;19(12):614–620. doi: 10.1016/j.tim.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 9.Cornman RS, et al. Pathogen webs in collapsing honey bee colonies. PLoS ONE. 2012;7(8):e43562. doi: 10.1371/journal.pone.0043562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alaux C, et al. Interactions between Nosema microspores and a neonicotinoid weaken honeybees (Apis mellifera) Environ Microbiol. 2010;12(3):774–782. doi: 10.1111/j.1462-2920.2009.02123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pettis JS, vanEngelsdorp D, Johnson J, Dively G. Pesticide exposure in honey bees results in increased levels of the gut pathogen Nosema. Naturwissenschaften. 2012;99(2):153–158. doi: 10.1007/s00114-011-0881-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henry M, et al. A common pesticide decreases foraging success and survival in honey bees. Science. 2012;336(6079):348–350. doi: 10.1126/science.1215039. [DOI] [PubMed] [Google Scholar]
- 13.Feyereisen R. Insect CYP genes and P450 enzymes. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Amsterdam: Elsevier/Academic Press; 2012. pp. 236–316. [Google Scholar]
- 14.Claudianos C, et al. A deficit of detoxification enzymes: Pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15(5):615–636. doi: 10.1111/j.1365-2583.2006.00672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mao W, et al. Quercetin-metabolizing CYP6AS enzymes of the pollinator Apis mellifera (Hymenoptera: Apidae) Comp Biochem Physiol B Biochem Mol Biol. 2009;154(4):427–434. doi: 10.1016/j.cbpb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Mao W, Schuler MA, Berenbaum MR. CYP9Q-mediated detoxification of acaricides in the honey bee (Apis mellifera) Proc Natl Acad Sci USA. 2011;108(31):12657–12662. doi: 10.1073/pnas.1109535108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 18.Johnson RM, et al. Ecologically appropriate xenobiotics induce cytochrome P450s in Apis mellifera. PLoS ONE. 2012;7(2):e31051. doi: 10.1371/journal.pone.0031051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11(10):R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn S-J, Vogel H, Heckel DG. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem Mol Biol. 2012;42(2):133–147. doi: 10.1016/j.ibmb.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Casteels P, et al. Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera) Eur J Biochem. 1990;187(2):381–386. doi: 10.1111/j.1432-1033.1990.tb15315.x. [DOI] [PubMed] [Google Scholar]
- 22.Kao Y-T, Lu M-J, Chen C. Preliminary analyses of phenolic compounds and antioxidant activities in tea pollen extracts. J Food Drug Analysis. 2011;19:470–477. [Google Scholar]
- 23.Isidorov VA, Isidorova AG, Sczczepaniak L, Czyzewska U. Gas chromatographic–mass spectrometric investigation of the chemical composition of beebread. Food Chem. 2009;115:1056–1063. [Google Scholar]
- 24.Alaux C, Dantec C, Parrinello H, Le Conte Y. Nutrigenomics in honey bees: digital gene expression analysis of pollen’s nutritive effects on healthy and varroa-parasitized bees. BMC Genomics. 2011;12:496–509. doi: 10.1186/1471-2164-12-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wehling K, et al. p-Coumaric acid – a monomer in the sporopollenin skeleton. Planta. 1989;179:376–380. doi: 10.1007/BF00391083. [DOI] [PubMed] [Google Scholar]
- 26.Greenaway W, Scaysbrook T, Whatley FR. The composition and plant origins of propolis: A report of work at Oxford. Bee World. 1990;71:107–118. [Google Scholar]
- 27.Barker RJ, Lehner Y. Laboratory comparison of high fructose corn syrup, grape syrup, honey, and sucrose syrup as maintenance food for caged honey bees. Apidologie (Celle) 1978;9:111–116. [Google Scholar]
- 28.Mullin CA, et al. High levels of miticides and agrochemicals in North American apiaries: Implications for honey bee health. PLoS ONE. 2010;5(3):e9754. doi: 10.1371/journal.pone.0009754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.