Abstract
Pathogenic retroviruses have evolved multiple means for evading host restriction factors such as apolipoprotein B editing complex (APOBEC3) proteins. Here, we show that murine leukemia virus (MLV) has a unique means of counteracting APOBEC3 and other cytosolic sensors of viral nucleic acid. Using virus isolated from infected WT and APOBEC3 KO mice, we demonstrate that the MLV glycosylated Gag protein (glyco-Gag) enhances viral core stability. Moreover, in vitro endogenous reverse transcription reactions of the glyco-Gag mutant virus were substantially inhibited compared with WT virus, but only in the presence of APOBEC3. Thus, glyco-Gag rendered the reverse transcription complex in the viral core resistant to APOBEC3. Glyco-Gag in the virion also rendered MLV resistant to other cytosolic sensors of viral reverse transcription products in newly infected cells. Strikingly, glyco-Gag mutant virus reverted to glyco-Gag–containing virus only in WT and not APOBEC3 KO mice, indicating that counteracting APOBEC3 is the major function of glyco-Gag. Thus, in contrast to the HIV viral infectivity factor protein, which prevents APOBEC3 packaging in the virion, the MLV glyco-Gag protein uses a unique mechanism to counteract the antiviral action of APOBEC3 in vivo—namely, protecting the reverse transcription complex in viral cores from APOBEC3. These data suggest that capsid integrity may play a critical role in virus resistance to intrinsic cellular antiviral resistance factors that act at the early stages of infection.
Keywords: intrinsic immunity, trex1, virus restriction factors
Retroviruses are enveloped single-stranded RNA viruses that replicate via a DNA intermediate synthesized by viral reverse transcriptase. All retroviruses contain the three basic viral genes, gag, pol, and env, that encode the viral core, enzymes, and envelope proteins, respectively. Many gammaretroviruses, including Moloney murine leukemia virus (M-MLV) and Friend MLV (F-MLV), as well as feline leukemia virus, encode an additional glycosylated form of the Gag protein termed gPr80gag or glyco-Gag, originating from an upstream CUG initiation codon in frame with the Gag polyprotein AUG (1–3). Glyco-Gag has 88 additional amino acids at the N terminus compared with the Gag precursor and is cleaved by a cellular protease to yield two proteins of 55 and 40 kDa (4, 5). The C-terminal protein, containing much of the viral capsid (CA) and NC sequences, is secreted, whereas the N-terminal fragment, composed of the glyco-Gag unique sequences, matrix, and pp12gag, remains associated with the cell as a type II integral membrane protein (1, 4). The N-terminal fragment is also found in MLV virions (6–8). The conservation of glyco-Gag in many gammaretroviruses suggests that it plays a significant role in their replication. Indeed, although mutations of glyco-Gag do not affect virus replication in vitro (9–11), they lead to lower in vivo infectivity (9, 12, 13). Moreover, in mice inoculated with glyco-Gag–deficient M-MLV, there is reversion to WT virus (10, 12, 13).
Different investigators have reported distinct glyco-Gag effects on MLV infection. We found that glyco-Gag is important for late stage virus budding/release from infected fibroblasts and that mutant virus-infected cells show aberrant tubelike structures at the surface (13). Moreover, glyco-Gag appears to direct virus budding through lipid rafts, which results in high cholesterol content in the virus (13, 14). Pizzato (15) reported that MLV glyco-Gag can complement an infectivity defect for nef-negative HIV-1 that is specific for lymphocytes and does not affect virus release. Kolokithas et al. (8) suggested that glyco-Gag counteracts the antiviral action of apolipoprotein B editing complex 3 (APOBEC3), because APOBEC3 KO mice were equally infected with either glyco-Gag WT or mutant F-MLV, whereas WT mice were only efficiently infected with glyco-Gag–containing virus.
APOBEC3 genes belong to a family of genes that encode DNA and RNA editing enzymes (16). The human genome contains seven genes, whereas mice have only one gene (17). APOBEC3G was initially discovered because it is counteracted by the HIV-1 viral infectivity factor (Vif). In vif-deficient–HIV-1 producer cells, APOBEC3 proteins are packaged into progeny virions via interaction with the nucleocapsid (NC) protein and viral RNA. The packaged APOBEC3 proteins inhibit infection in target cells by deaminating deoxycytidine residues on the DNA minus strand during reverse transcription, inducing hypermutation in newly synthesized HIV-1 DNA. APOBEC3 proteins also inhibit replication by cytidine deaminase–independent mechanism(s), such as blocking reverse transcription (18–21). Vif binds several APOBEC3 proteins and targets them for ubiquitinylation and degradation in the proteasome in virus producer cells, thereby blocking the antiviral activity (22). In addition to inhibiting infection when packaged into virions, cellular APOBEC3 proteins can restrict incoming virus particles when expressed in target cells. For example, APOBEC3G expressed in recipient cells functions as a postentry restriction block for HIV in resting CD4+ T cells, monocytes, and immature dendritic cells (DCs) (23–25).
Mouse APOBEC3 also restricts endemic murine viruses, including mouse mammary tumor virus (MMTV), F-MLV, M-MLV, and AKR-MLV (AKV); this has been demonstrated in vivo in APOBEC3 KO mice (26–29). Although there is only one mouse APOBEC3 gene, different inbred strains of mice carry variant alleles (30–32). The predominant transcript in C57BL/6 mice, APOBEC3BL6, lacks the fifth exon and is expressed at high levels in vivo, whereas that found in BALB/c mice, APOBECBALB, contains all nine exons and is expressed at lower levels (31). There are also amino acid sequence changes in the conserved exons of the two alleles (30–32). APOBEC3BL6 restricts infection by MMTV, F-MLV, and AKV more effectively than APOBECBALB (27, 29–31). Interestingly, mouse APOBEC3 does not hypermutate viral DNA in F-MLV–, M-MLV–, or MMTV-infected cells but instead inhibits infection at an earlier replication step (21, 26, 29, 30, 33–35). How APOBEC3-restricted viruses that lack vif-like genes, such as MLV, MMTV, and equine infectious anemia virus (36, 37), counteract APOBEC3 proteins and persist in their hosts is not known.
Here we provide insight into how glyco-Gag’s effects on virus structure and APOBEC3 sensitivity may be related. We show that both APOBEC3 alleles are packaged in WT and glyco-Gag mutant M-MLV in vivo and that both inhibit early reverse transcription of glyco-Gag mutant viruses. Glyco-Gag mutant viruses were also more susceptible to both target cell–expressed APOBEC3 and cytosolic sensors of viral reverse transcription products in newly infected cells. Moreover, we demonstrate that viruses lacking glyco-Gag have a more unstable capsid than WT M-MLV. Finally, we show that glyco-Gag’s ability to counteract APOBEC3 is critical for efficient virus infection, because glyco-Gag mutant virus reverts to WT virus shortly after in vivo infection of APOBEC3+ but not APOBEC3 KO mice.
Results
Glyco-Gag Mutant MLV Is Restricted by APOBEC3.
Previous studies showed that APOBEC3BL6 more potently restricts F-MLV than the APOBEC3BALB allele and that glyco-Gag mutant F-MLV replicates more poorly in WT mice bearing either allele than in APOBEC3 KO mice (8, 27, 30). To determine whether this was also the case for M-MLV, we infected C57BL/6, BALB/c, and APOBEC3 KO pups with WT and glyco-Gag mutant M-MLV; the APOBEC3 null allele is present on the BL/6 background (26). The mutant virus has a stop codon in the gPr80Gag reading frame (UAU to UAG) at nt 608, 12 nt 5′ of the normal Gag AUG (9). BL/6 mice infected with the glyco-Gag mutant virus showed significantly lower levels of infectious virus in spleens and thymi (Fig. S1) compared with APOBEC3 KO mice. In contrast, WT virus replicated to the same higher levels in both BL/6 and APOBEC3 KO mice. Moreover, in APOBEC3 KO mice, mutant and WT M-MLV replicated to the same levels, similar to results previously reported for F-MLV (8). Although the overall infection levels were higher in BALB/c than BL/6 with either virus, glyco-Gag mutant virus also infected BALB/c mice more poorly than did WT virus (Fig. S1).
These results were confirmed in a macrophage cell line NR-9456 expressing the APOBEC3BL6 allele (Fig. S2). Equal amounts of WT and glyco-Gag mutant M-MLV isolated from NIH 3T3 cells, which express no APOBEC3, were used to infect the murine NR-9456 cells, and at different times after infection, DNA was isolated and examined for proviral DNA. There were significantly lower levels of viral DNA present in cells infected with the glyco-Gag mutant M-MLV compared with the cells infected with the WT virus at all time points (Fig. S2).
The remaining studies addressing the role of packaged APOBEC3 proteins were largely performed with virus recovered from splenocytes of M-MLV–infected WT and APOBEC3 KO mice by short-term in vitro culture, thereby ensuring that endogenous levels of APOBEC3 were packaged. Virus levels were normalized by quantitative real-time-PCR (RT-qPCR) for viral RNA, by Western blots for the p30CA protein found in mature virions, and were titered on NIH 3T3 cells (Table S1).
Glyco-Gag Mutant and WT MLV Produced In Vivo Contains APOBEC3.
To determine whether glyco-Gag affected virus packaging of APOBEC3 expressed at physiological levels, 2-d-old BL/6, KO, and BALB/c pups were infected with WT and glyco-Gag negative virus, and at 16 d after infection (dpi), splenocytes and thymocytes were isolated and cultured. Virus released into the supernatants was recovered, and viral RNA was quantified by reverse-transcribed RT-qPCR. The presence of mature virions was demonstrated by Western blots with anti-MLV antisera; equal amounts of p30CA were found in the viruses, which were also analyzed by Western blots with anti-APOBEC3 antisera. Virus titers were also obtained (Table S1). Both APOBEC3BL6and APOBEC3BALB were packaged in WT and glyco-Gag mutant M-MLV in vivo (Fig. 1A; Fig. S3A). No APOBEC3 protein was detected in supernatants from uninfected spleens, suggesting that it was present in virions and not in exosomes (Fig. S3B). We also assessed the purity of the WT and glyco-Gag mutant M-MLV virus preparations from infected NIH 3T3 fibroblasts or a productively infected T-lymphocyte line by conducting transmission electron microscopy. The majority of the particles in the preparations were virions, with diameters of ∼100–120 nm, and there was little evidence of contaminating exosomes (Fig. S4).
Fig. 1.
APOBEC3 is packaged at similar levels in WT and glyco-Gag mutant viruses in vivo. (A) Western blot of virions isolated from BL/6 and APOBEC3 KO mice infected with WT or glyco-Gag mutant viruses and probed with anti-APOBEC3 (Upper) or anti-MLV (Lower) antisera. (B) Western blot of cores isolated from WT and mutant M-MLV–infected BL/6 and KO mice. This experiment was performed six times with similar results. A3BL6, APOBEC3BL6; NS, nonspecific band (52).
We next tested whether APOBEC3 incorporated into glyco-Gag M-MLV was resistant to detergent, a method used to isolate virus cores. Equal amounts of glyco-Gag-mutant and WT virions isolated from the lymphocyte cultures were sedimented over sucrose step gradients containing 5% (vol/vol) octylphenyl-polyethylene glycol (IGEPAL) and analyzed by Western blots. Both APOBEC3 alleles were packaged at similar levels in mutant and WT viruses shed ex vivo (Fig. 1B; Fig. S3A). Moreover, because the density gradient incorporates detergent, contamination with cellular proteins was reduced. Indeed, a cross-reacting protein detected by the anti-mouse APOBEC3 antisera in virus preparations from M-MLV–infected spleens (Fig. 1A) was not seen in detergent-treated viruses (Fig. 1B). Thus, glyco-Gag does not prevent packaging of APOBEC3 into M-MLV.
Glyco-Gag Mutant Capsids Are Less Stable than WT.
Glyco-Gag mutant M-MLV buds differently from WT virus and contains less membrane cholesterol (13, 14). To determine whether these changes affected stability of viral cores, we treated equal amounts of WT and glyco-Gag mutant virions isolated from NIH 3T3 cells with 0.5% Triton X-100, followed by sucrose density gradient centrifugation; the virion preparations contained the p30CA only found in mature virions. The cores of the mutant viruses were much less stable than those of the WT during centrifugation; cores from the mutant virions were recovered much less efficiently than those from WT virus, with the majority of the p30CA at the top of the gradient (Fig. 2 A and B). Moreover, mutant cores were more sensitive to in vitro trypsin digestion than WT cores (Fig. S5A).
Fig. 2.
Glyco-Gag mutant capsids are less stable than WT. (A) Glyco-Gag mutant and WT virions from infected NIH 3T3 cells were treated with 0.5% Triton-X-100, layered onto sucrose gradients, and subjected to ultracentrifugation. Fractions were subjected to the Western blots with anti-p30 antibodies; the p30 signals were quantified by densitometry. The regions of the gradients corresponding to the cores are indicated by an arrow. Box, WT; diamond, mutant. (B) p30 signals from three independent experiments were quantified by densitometry, and the percentage of p30 in the core fraction vs. the total p30 in the gradient was calculated. The average and SD are shown. Statistical significance was determined by Student t test with equal variances. *P ≤ 0.0002. (C) NIH 3T3 cells were infected with equal amounts of WT and mutant virus (input), and the particulate capsid fraction (pellet) representing intact capsids present in the cytoplasm of infected cells was analyzed. The CA signals from six experiments were quantified by densitometry, and the percentage of protein in the core vs. total viral input was calculated. The average and SD are shown. Statistical significance was determined by paired Student paired t test. *P ≤ 0.002.
To examine core stability after infection, we performed fate of capsid assays in NIH 3T3 cells with in vivo isolated glyco-Gag mutant and WT virions, a method used to study the capsid stability of MLV and HIV-1 in the presence and absence of Trim5α (38, 39). NIH 3T3 cells were incubated on ice with equal amounts of WT and glyco-Gag mutant M-MLV for 30 min to allow virus binding, shifted to 37 °C for 4 h, and after cell lysis, the particulate fraction containing the capsids was analyzed by Western blots. Although the wt p30CA was readily detected, only low levels of p30 were detected in the glyco-Gag mutant particulate fraction (Fig. 2C). Thus, virus capsid stability is enhanced in the presence of glyco-Gag.
APOBEC3 Restricts Endogenous Reverse Transcription of glyco-Gag Mutant but Not WT Virus.
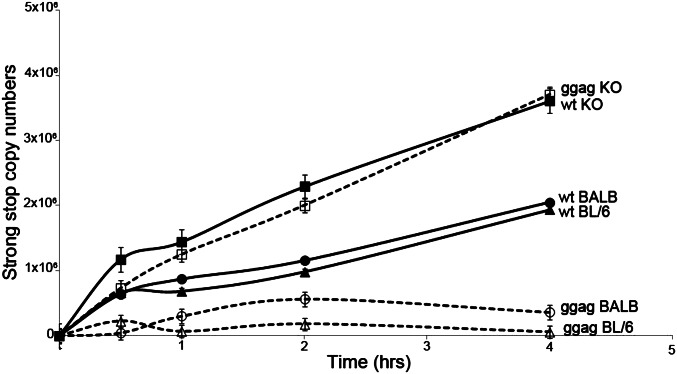
The decreased stability of the glyco-Gag mutant capsids could result in increased access of APOBEC3 to the reverse transcription complex (RTC), which would lead to differential inhibition by APOBEC3 of mutant and WT virus. To examine whether reverse transcription was inhibited by in vivo packaged APOBEC3 and whether this was affected by incorporation of glyco-Gag into virions, we performed endogenous reverse transcription (EnRT) assays with M-MLV WT and glyco-Gag mutant virus isolated from BL/6, BALB, and APOBEC3 KO mice. DNA was isolated at different times after initiation of the reaction, and reverse transcription was assayed by RT-PCR using primers that detect strong stop DNA (ssDNA). There was a significant reduction in ssDNA in glyco-Gag mutant M-MLV compared with WT M-MLV isolated from both BL/6 and BALB mice (Fig. 3). In striking contrast, glyco-Gag mutant virus isolated from APOBEC3 KO mice produced ssDNA at levels similar to WT M-MLV; indeed, either virus isolated from KO mice produced the highest levels of reverse transcripts. This finding suggests that although both WT and glyco-Gag mutant virions package similar amounts of APOBEC3 (Fig. 1), the RTC is more accessible to APOBEC3 in glyco-Gag mutant than WT cores.
Fig. 3.
Virions containing in vivo–packaged APOBEC3 show reduced early reverse transcription in vitro. EnRT assays were performed virus isolated from WT and glyco-Gag (ggag) mutant virus–infected BL/6, APOBEC3 KO, and BALB mice isolated at 16 dpi. Shown is the average of three independent experiments.
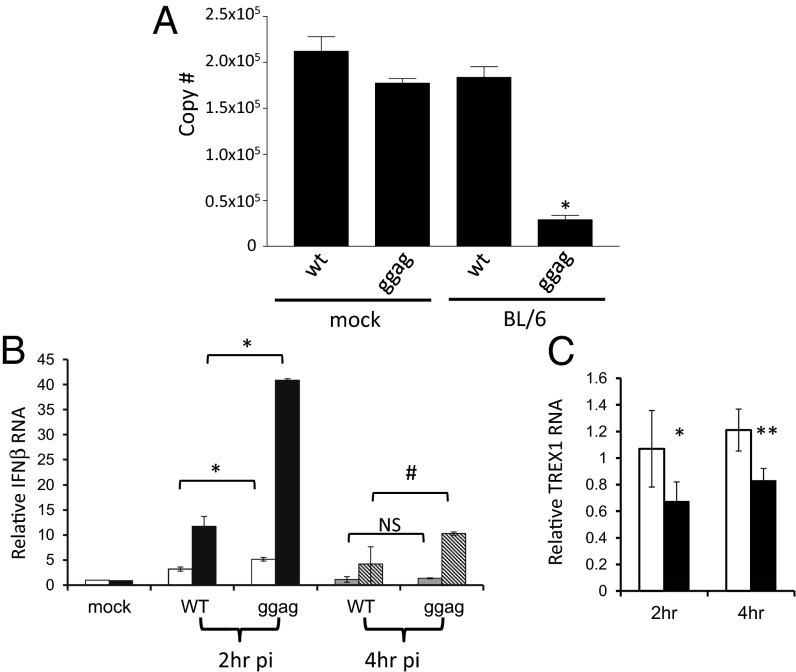
We next tested whether the glyco-Gag mutant viruses were more accessible to exogenous APOBEC3 by adding cytosolic extracts from APOBEC3-transfected cells to EnRT assays of viruses isolated from KO mice. Addition of extracts from APOBEC3-transfected cells but not mock-transfected controls inhibited reverse transcription by the mutant but not WT virions (Fig. 4A). Glyco-Gag mutant viruses were also strongly inhibited by addition of DNase I to the EnRT reaction, presumably because they allowed ready access to the mutant viral RTCs (Fig. S5B).
Fig. 4.
Glyco-Gag–containing virions are resistant to APOBEC3 and cytosolic sensors of virus infection. (A) EnRT assays were performed after supplementing with mock- or APOBEC3BL6-transfected lysates. Samples were harvested at 4 h and analyzed by RT-qPCR. Shown is the average of three independent experiments with the standard mean. Statistical significance was determined by one-way ANOVA. (B) RNA from the control or TREX1 siRNA-treated cells was isolated at the indicated time points after infection with WT or glyco-Gag mutant (ggag) virus and analyzed by RT-qPCR for IFNβ RNA. (C) qPCR analysis of RNA isolated from NR-9456 cells transfected with control (white bars) or TREX1-specific (closed bars) siRNAs and amplified with TREX1 primers. Statistical significance was determined by paired Student t test. *P ≤ 0.05; **P ≤ 0.005; #P ≤ 0.005; NS, no statistical difference.
We showed previously that endogenous APOBEC3 in dendritic cells restricted infection by M-MLV (28) and MMTV (40) that lacked packaged APOBEC3 protein. We thus tested whether APOBEC3 expressed in target cells could differentially restrict reverse transcription of WT vs. glyco-Gag mutant M-MLV. We transiently transfected 293T cells expressing the ecotropic MLV receptor mCAT-1 with APOBEC3BALB or APOBECBL6 expression vectors (34). The cells were then infected with either WT or glyco-Gag mutant virus isolated from APOBEC3 KO mice. At 22 h after infection, DNA was isolated from the infected cells and quantified by qPCR. Both alleles of APOBEC3 inhibited infection by glyco-Gag mutant M-MLV, but had little effect on infection by wt virus (Fig. S6). Thus, APOBEC3 expressed in target cells more effectively inhibited incoming glyco-Gag mutant viruses.
Glyco-Gag–Negative Virus Is Sensitive to Another Cytosolic Sensor of Viral Infection.
Viral capsids are thought to shield incoming retroviral replication complexes from cellular sensors of infection. The t1/2 for MLV attachment through completion of reverse transcription is ∼3–4 h (41), and capsid disassembly is believed to occur either simultaneously with or just before reverse transcription (42, 43). Thus, the less stable glyco-Gag mutant capsid could allow intracellular sensors to more readily sense viral RNA or DNA. TREX1, an exonuclease present in the cytosol of macrophages and T cells, digests reverse-transcribed retroviral and retrotransposon DNA, thereby down-modulating the innate immune response to viral DNA by an as-of-yet unknown cytosolic sensor; in its absence, the innate immune response to retroviral RTCs or viral DNA is greatly increased (44, 45). To determine whether the RTC of glyco-Gag mutant virions was more accessible to this cytosolic sensor, as well as to APOBEC3, NR-9456 macrophages were transfected with TREX1 siRNA and infected with WT and glyco-Gag mutant M-MLV. At 2 and 4 h after infection, RNA was analyzed by reverse-transcribed RT-qPCR for IFNβ (Fig. 4B) and TREX1 RNA to ensure knockdown (Fig. 4C). At both time points, the glyco-Gag mutant virus induced significantly higher levels of IFNβ RNA expression (Fig. 4B). Moreover, induction was dramatically increased in TREX1 knockdown cells infected with either WT or mutant virus, and the mutant virus induced much higher levels of IFNβ RNA. Viral RNA levels were also decreased at 4 h after infection in the TREX1-treated WT virus–infected cells, and this was further diminished in glyco-Gag mutant virus–infected cells (Fig. S7). These data suggest that the less stable capsid structure of glyco-Gag mutant viruses renders the incoming viral RTC and nucleic acid more accessible to both APOBEC3 and cytosolic nucleic acid sensors.
Glyco-Gag Is Critical for Counteracting APOBEC3 In Vivo.
Both FrCasE MLV and M-MLV glyco-Gag-negative viruses revert to WT in mice (10, 12, 13). If the principal function of glyco-Gag was to counteract APOBEC3, then revertant viruses that cause persistent infection should only be selected in WT and not KO mice. We infected neonatal BL/6, KO, and BALB/c with glyco-Gag mutant virus, and at 3 and 6 wk after infection, DNA was isolated from the spleens and thymi. PCR was performed using primers that target the glyco-Gag region, and the PCR products were cloned and sequenced. Glyco-Gag mutant viral sequences isolated at 3 wk after infection did not show reversion, regardless of genetic background (Table 1). However, glyco-Gag mutant viruses isolated at 6 wk after infection reverted to WT in six of eight BL/6 mice and five of five BALB mice (Table 1). In contrast, glyco-Gag mutant virus from KO mice showed no reversion (0/4; Table 1; P = 0.0063 using Fisher’s exact test). Thus, glyco-Gag–containing viruses have a significant replicative advantage in WT mice and a major function of glyco-Gag is counteracting APOBEC3.
Table 1.
Glyco-Gag mutant virus reverts to WT in C57BL/6 and BALB/c but not APOBEC3 KO mice
| Strain | No. mice with revertants |
Codons | Amino acids | |
| 3 wk | 6 wk | |||
| C57BL/6 | 0/10 | 6/8 | TGG/CAG/TAC/TAT | W/Q/Y/Y |
| BALB/c | ND | 5/5 | CAG/TTG/TGG | Q/L/W |
| −/− | 0/6 | 0/4 | TAG | STOP |
WT M-MLV has a TAT(Y) codon.
Discussion
In this study, we provide insights into the function and mechanism of action of the gammaretrovirus glyco-Gag protein and provide a mechanism by which glyco-Gag counteracts the restrictive effects of APOBEC3 on MLV. Comparisons between WT and glyco-Gag mutant M-MLV produced from APOBEC3-positive or -negative cells or animals, and infection of APOBEC3-positive or -negative cells, revealed the following points: (i) glyco-Gag prevents access of APOBEC3 to the RTC both in virions and infected cells; (ii) glyco-Gag reduces the ability of a cytosolic sensor of viral infection to respond to MLV infection; (iii) glyco-Gag enhances the stability of viral cores/capsids; and (iv) the major in vivo target of glyco-Gag is APOBEC3.
Viruses that persist are in a continuous “tug of war” with their host and must be able to counteract the antiviral responses continuously imposed on the virus. Among the multiple factors used by hosts to defend against viruses is the APOBEC3 family of restriction factors (46). Some retroviruses, such as HIV, simian immunodeficiency virus (SIV), and feline immunodeficiency virus (FIV), encode Vif proteins that bind APOBEC3 proteins and target them for degradation in virus-producing cells. Additionally, the Bet protein of feline foamy viruses inhibits APOBEC3 by as yet undetermined means (16) and human T-cell leukemia virus I prevents packaging of APOBEC3G, although not APOBEC3A, 3B, or 3H (47, 48). However, APOBEC3 inhibits infection by a number of viruses that encode no apparent vif- or bet-like genes, and how these viruses overcome restriction is not known. Here, we show that MLV glyco-Gag protein uses a unique mechanism to counteract the antiviral action of APOBEC3. Namely, glyco-Gag limits accessibility of APOBEC to the RTC and thereby prevents it from inhibiting reverse transcription.
Glyco-Gag has long been known to be important for MLV-induced pathogenesis, and its abrogation leads to decreased virus replication and pathogenesis in vivo (4, 9–12). However, previous studies carried out primarily in murine fibroblast lines such as NIH 3T3 that do not express APOBEC3 suggested that glyco-Gag was dispensable for in vitro infection. Here we show that in a macrophage cell line that expresses high levels of APOBEC3, glyco-Gag plays a critical role in virus spread in vitro (Fig. S2). Thus, the requirement for glyco-Gag is not specific for in vivo infection, but instead is cell type-specific. That glyco-Gag mutant virus infection was restricted in a macrophage cell line suggests that in vivo M-MLV infects APOBEC3-expressing cells such as dendritic cells and macrophages in addition to its lymphoid targets, because it has evolved a mechanism to circumvent restriction by this host factor. Although the initial targets of MLV in vivo infection are not well-characterized, we previously found that M-MLV infects primary dendritic cells and that some of the first cells infected when neonatal mice are infected with a M-MLV vector are osteoclasts derived from monocytic cells (28, 49).
Glyco-Gag mutant MLVs revert to WT after in vivo infection, and here we show that this reversion overcomes APOBEC3-mediated restriction in both BL/6 and BALB/c mice, which express different APOBEC3 alleles. Several studies have suggested that APOBECBALB is less effective that APOBECBL6 at restricting infection by F-MLV (27, 30, 50). However, clearly both alleles are important anti–M-MLV restriction factors, because reversion occurred with similar kinetics and frequency in the two mouse strains, and both alleles effectively inhibited reverse transcription in EnRT assays. Interestingly, although APOBEC3 is a cytidine deaminase, we saw no evidence that this activity played a role in reversion (e.g., there were no G to A mutations); it is likely that reversion occurred through reverse transcription errors. Although our in vitro studies also show that glyco-Gag prevents other host cytosolic factors from accessing the viral RTC, the lack of revertants seen in APOBEC3 KO mice suggests that in vivo APOBEC3 is the major intrinsic antiviral factor targeting the early steps of virus infection.
Glyco-Gag is incorporated into MLV virions (4, 14), and we found that glyco-Gag mutant viruses have less stable capsids. Although it is possible that capsid stability and resistance to restriction are unlinked, our data suggest that by increasing the stability of the capsid, glyco-Gag renders these viruses more resistant to target cell APOBEC3 and other cytosolic factors that recognize the retroviral genome, the RTC, or reverse transcribed DNA. We observed that reverse transcription was more inhibited by packaged APOBEC3 in mutant than WT virions in EnRT assays. A recent study suggests that APOBEC3G interacts with the reverse transcriptase of HIV-1 (51). Whether this is also the case for MLV’s reverse transcriptase and APOBEC3 and whether there is differential accessibility of reverse transcriptase to the viral RNA in mutant viruses remain to be tested. Interestingly, a recent report showed that unlike F-MLV and M-MLV, AKV, which arises from an endogenous MLV, showed G to A mutations, a signature of APOBEC3 deaminase activity (29). AKV also expresses a glyco-Gag protein, but the amino acid sequences of AKV and M-MLV glyco-Gags are different; these differences may explain the disparate effects of APOBEC3 on the different MLVs.
The results presented here show that glyco-Gag can potentially use two mechanisms for protection against APOBEC3 and perhaps other cytosolic sensors: (i) by affecting capsid stability and (ii) by sequestering packaged APOBEC3 away from the RTC. By using these two distinct mechanisms, glyco-Gag protects the viral genome from both APOBEC3 of virus producer and target cells. Analogous mechanisms may be used by other retroviruses that replicate in the presence of APOBEC3 but lack Vif-like proteins.
Materials and Methods
Ethics Statement.
All mice were housed according to the policies of the Institutional Animal Care and Use Committee (IACUC) of the University of Pennsylvania. The experiments performed with mice in this study were approved by this committee (IACUC Protocol 801594).
Mice.
BALB/c and C57BL/6 mice were purchased from the National Cancer Institute (NCI). APOBEC3 KO mice and WT controls were bred at the University of Pennsylvania and were previously described (26). Two-day-old mice were infected i.p. with equal amounts of virus and then harvested at the indicated times.
Cell Culture and Transfection.
NIH 3T3 cells were cultured in DMEM supplemented with 10% (vol/vol) FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin. NR-9456 macrophages (National Institute of Allergy and Infectious Diseases/National Institutes of Health Biodefense and Emerging Infections Research Resources Repository) were cultured in the presence of sodium pyruvate and HC11 cells (ATCC) with insulin (10 μg/mL); 293T cells stably transfected with the mCAT-1 receptor were cultured in DMEM with 8% donor calf serum, G418 (10 μg/mL), and penicillin/streptomycin. All transfections were performed using Lipofectamine 2000 (Invitrogen).
Virus Isolation.
Virus was isolated from WT and glyco-Gag mutant M-MLV harvested from NIH 3T3 fibroblasts stably infected with WT and glyco-Gag–negative Ab-X-M-MuLV, respectively, as previously described (9) or from in vivo–infected splenocytes and thymocytes. Infected mice were killed at 16 dpi. Splenocytes and thymocytes were collected and incubated in RPMI 1640, 10% FCS, nonessential amino acids, and penicillin/streptomycin for 48 h. The media was passed through a 0.4-μm filter, treated with 20 U/mL DNase I (Roche) at 37 °C for 30 min, and pelleted through a 30% sucrose cushion. After resuspension, the viruses were titered on NIH 3T3 cells, as well as quantified by reverse-transcribed RT-qPCR and analyzed on Western blots. The primers used for virus quantification were located in the env of M-MLV: F primer, 5′-CCTACTACGAAGGGGTTG-3′; R primer, 5′-CACATGGTACCTGTAGGGGC-3′. RT-PCR was performed using the Power SYBR Green PCR master mix kit (Applied Biosystems). Viruses isolated from glyco-Gag mutant- and WT virus–infected APOBEC3 KO mice and NIH 3T3 had similar infectivity/particle ratios (Table S1), demonstrating that fully infectious virus was isolated from the organ cultures.
Revertant Analysis.
Two-day-old APOBEC3+/+, APOBEC3−/−, and BALB/c pups were infected with equal amounts of M-MLV glyco-Gag mutant viruses and killed 3 and 6 wk after infection. DNA was isolated from spleens and thymuses by the NucleoSpin Tissue kit (Machery-Nagel). The glyco-Gag region in MLV was amplified from 100-ng samples of DNA with the primers 5′-GGTCTCCTCTGAGTGATTGACT and 5′-GGTCAAACTTAAGGGAGTGGTA. The PCR products were purified by the NucleoSpin Extraction kit (Machery-Nagel), treated with ExoSAP-IT (Affymetrix), and subjected to sequencing with the same primers above. The appearance of revertants in samples was assessed by the sequence traces for nucleotides around nt608 (nucleotide accession no. J02255) by Chromas Lite (Technelysium).
Western Blots.
Polyclonal rabbit anti-mouse APOBEC3 antibody has been previously described (52). Because this antisera variably cross-reacts with an unknown nonspecific protein close in size to APOBEC3 (52), the APOBEC3BL6 protein was analyzed on 10% SDS/PAGE and the APOBEC3BALB on 12% SDS/PAGE. Polyclonal goat anti-MLV antibody (NCI Repository), mouse anti-HA (Roche), HRP-conjugated anti-rabbit (Cell Signaling Technology), and anti-goat and -mouse antibody (Sigma Aldrich) were used for detection, using either ECL kits (GE Healthcare Life Sciences) or Supersignal West Femto Chemiluminescent substrate (Thermo Scientific).
EnRT Assay.
Equal amounts of virus isolated from infected BL6, APOBEC3 KO, and BALB mice were incubated in EnRT buffer (1× PBS, 2.5 mM MgCl2, 0.01% Nonidet P-40, 1 mM dNTPs) at 37 °C. Fractions of the reactions were removed at 0, 30 min, 1 h, 2 h, and 4 h and added to 40 μg of sonicated salmon sperm DNA. DNA was isolated from fractions using the Qiagen DNeasy Blood and Tissue Kit (Qiagen). PCR was performed using MLV strong stop primers to measure the level of early reverse transcripts: F primer, 5′-CCTCCGATTGACTGAGTCGCCCC-3′; R primer, 5′-ATGAAAGACCCCCGCTGACGG-3′. For some experiments, 100 μg of cellular lysate from 293T cells transfected with either APOBEC3Δexon5.HA (APOBECBL6) or APOBEC3FL.HA (APOBECBALB) was added to the EnRT reactions. Aliquots of the reaction were taken at 4 h. For the experiments including DNase I, DNase I (Roche) was added in the EnRT reaction buffer at a concentration of 500 U/mL as previously described (53), and samples were harvested at 6 h.
Core Stability Assays.
For analysis of APOBEC3 in cores, virions released from in vivo–infected splenocytes were sedimented through a 10%/30% sucrose step gradient containing 5% IGEPAL (Sigma) in PBS at 108,000 × g for 90 min. The core-containing pellets were resuspended in SDS sample buffer and analyzed by Western blot. For core analysis by sedimentation, medium collected from WT- and glyco-Gag–infected NIH 3T3 cells was clarified by low-speed centrifugation and passed through a 0.45-μm filter. Viral particles were pelleted through a 20% sucrose cushion in a Beckman SW41 rotor at 100,000 × g for 1 h, resuspended in buffer containing 10 mM Tris-HCl (pH 7.4), 100 mM NaCl, 1 mM EDTA, and 0.5% Triton X-100, and incubated at 37 °C for 10 min. The viruses were layered onto a 15–30% sucrose gradient and centrifuged in a Beckman SW41 rotor at 180,000 × g for 90 min. The fractionated samples were analyzed by Western blots with anti-p30 antibodies. For trypsin sensitivity assays, the core fractions from the sucrose gradients were pooled, concentrated by ultracentrifugation, and then digested with trypsin at 37 °C. Samples were subjected to Western blots with anti-p30 antibodies, and the signals were quantified by AlphaEaseFC densitometry software.
Fate of Capsid Assays.
Fate of capsid assays were carried out as previously described (39). Briefly, 1.2 × 106 NIH 3T3 cells were seeded in 10-cm2 dishes. The following day, the cells were incubated with 1 × 109 infectious centers (IC) WT or glyco-Gag mutant APOBEC3− M-MLVs at 4 °C for 30 min to allow viral attachment and then incubated for 2 h at 37 °C. After detachment by pronase treatment (7.0 mg/mL in DMEM; 5 min at 25 °C), the cells were resuspended in 2.5 mL hypotonic lysis buffer (10 mM Tris-HCl, pH 8.0; 10 mM KCl; 1 mM EDTA; 1× Halt Protease & Phosphatase Inhibitor Single-Use Mixture, EDTA-Free; Thermo Inc.) and incubated on ice for 15 min. The cells were lysed in a Dounce homogenizer (B pestle). Cellular debris were cleared by centrifugation for 3 min at 1,600 × g, and an aliquot of the cleared lysate was reserved for Western blots (input in Fig. 2). The remainder of the cleared lysate was layered on a 40% sucrose cushion. Following centrifugation, the pellet was resuspended in SDS sample buffer and subjected to Western blot analysis using anti-MLV antisera.
TREX1 Knockdown and IFNβ Expression Assays.
NR-9456 cells were transfected with siTREX1 Silencer Select siRNA (Ambion S75453) and siControl (Thermacon) using Lipofectamine RNAiMAX reagent (Invitrogen). Cells were incubated at 37 °C for ∼18 h and then infected with WT and glyco-Gag mutant M-MLV (multiplicity of infection = 1,000 IC). Cells were harvested at 2 and 4 h after infection (hpi). RNA was isolated using the RNeasy Mini Kit (Qiagen). cDNA was made using the SuperScript III First Strand Synthesis System for RT-PCR (Invitrogen). RT-PCR was performed using the Power SYBR Green PCR master mix kit (Applied Biosystems). Primers used for the detection of Trex1 were as follows: F′, 5′-CGTCAACGCTTCGATGACA-3′; R′, 5′-AGTCATAGCGGTCACCGTTGT-3′. Primers used for detection of actin: F′, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′; R′, 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′. Primers used for IFNβ detection: F′, 5′-AAGAGTTACACTGCCTTTGCCACT-3′; R′, 5′-CACTGTCTGCTGGTGGAGTTCATC-3.
Statistical Analysis.
Statistical analysis was performed using the GraphPad/PRIZM software.
Supplementary Material
Acknowledgments
We thank Lorraine Albritton, David Derse (deceased), and Masaaki Miyazawa for reagents used in this study; Alice Telesnitzky for advice; and Kristin Blouch and Ferri Soheilian for expert technical assistance. This research was supported by Public Health Service Grants R01-AI-085015 (to S.R.R.) and R01-CA-94188 (to H.F.). S.S. was supported by National Institutes of Health (NIH) Grants T32-CA115299 and F32-AI100512. S.K. was supported by NIH Grant T32-AI07324, and T.N. was supported by a Japan Society for the Promotion of Science Postdoctoral Fellow for Research Abroad and the Barbara Burgess Memorial Fund. This work was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and in part with federal funds from the National Cancer Institute, NIH, under Contract HHSN26120080001E.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1217399110/-/DCSupplemental.
References
- 1.Edwards SA, Fan H. gag-Related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J Virol. 1979;30(2):551–563. doi: 10.1128/jvi.30.2.551-563.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans LH, Dresler S, Kabat D. Synthesis and glycosylation of polyprotein precursors to the internal core proteins of Friend murine leukemia virus. J Virol. 1977;24(3):865–874. doi: 10.1128/jvi.24.3.865-874.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prats AC, De Billy G, Wang P, Darlix JL. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol. 1989;205(2):363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 4.Fujisawa R, McAtee FJ, Zirbel JH, Portis JL. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: Identification of differences in processing in vitro and in vivo. J Virol. 1997;71(7):5355–5360. doi: 10.1128/jvi.71.7.5355-5360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards SA, Fan H. Sequence relationship of glycosylated and unglycosylated gag polyproteins of Moloney murine leukemia virus. J Virol. 1980;35(1):41–51. doi: 10.1128/jvi.35.1.41-51.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pillemer EA, Kooistra DA, Witte ON, Weissman IL. Monoclonal antibody to the amino-terminal L sequence of murine leukemia virus glycosylated gag polyproteins demonstrates their unusual orientation in the cell membrane. J Virol. 1986;57(2):413–421. doi: 10.1128/jvi.57.2.413-421.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujisawa R, McAtee FJ, Favara C, Hayes SF, Portis JL. N-terminal cleavage fragment of glycosylated Gag is incorporated into murine oncornavirus particles. J Virol. 2001;75(22):11239–11243. doi: 10.1128/JVI.75.22.11239-11243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolokithas A, et al. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J Virol. 2010;84(20):10933–10936. doi: 10.1128/JVI.01023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan H, Chute H, Chao E, Feuerman M. Construction and characterization of Moloney murine leukemia virus mutants unable to synthesize glycosylated gag polyprotein. Proc Natl Acad Sci USA. 1983;80(19):5965–5969. doi: 10.1073/pnas.80.19.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbin A, Prats AC, Darlix JL, Sitbon M. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J Virol. 1994;68(6):3857–3867. doi: 10.1128/jvi.68.6.3857-3867.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartzberg P, Colicelli J, Goff SP. Deletion mutants of Moloney murine leukemia virus which lack glycosylated gag protein are replication competent. J Virol. 1983;46(2):538–546. doi: 10.1128/jvi.46.2.538-546.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun R, Fan H. Recovery of Glycosylated gag Virus from Mice Infected with a Glycosylated gag-Negative Mutant of Moloney Murine Leukemia Virus. J Biomed Sci. 1994;1(4):218–223. doi: 10.1007/BF02253305. [DOI] [PubMed] [Google Scholar]
- 13.Low A, et al. Mutation in the glycosylated gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J Virol. 2007;81(8):3685–3692. doi: 10.1128/JVI.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitta T, Kuznetsov Y, McPherson A, Fan H. Murine leukemia virus glycosylated Gag (gPr80gag) facilitates interferon-sensitive virus release through lipid rafts. Proc Natl Acad Sci USA. 2010;107(3):1190–1195. doi: 10.1073/pnas.0908660107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pizzato M. MLV glycosylated-Gag is an infectivity factor that rescues Nef-deficient HIV-1. Proc Natl Acad Sci USA. 2010;107(20):9364–9369. doi: 10.1073/pnas.1001554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullen BR. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J Virol. 2006;80(3):1067–1076. doi: 10.1128/JVI.80.3.1067-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Münk C, Willemsen A, Bravo IG. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol. 2012;12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holmes RK, Koning FA, Bishop KN, Malim MH. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem. 2007;282(4):2587–2595. doi: 10.1074/jbc.M607298200. [DOI] [PubMed] [Google Scholar]
- 19.Bishop KN, Verma M, Kim EY, Wolinsky SM, Malim MH. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 2008;4(12):e1000231. doi: 10.1371/journal.ppat.1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gillick K, et al. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. J Virol. 2013;87(3):1508–1517. doi: 10.1128/JVI.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacMillan AL, Kohli RM, Ross SR. APOBEC3 inhibition of MMTV infection: The role of cytidine deamination versus inhibition of reverse transcription. J Virol. 2013;87:4808–4817. doi: 10.1128/JVI.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goila-Gur R, Strebel K (2008) HIV-1, APOBEC, and intrinsic immunity. Retrovirology 5:51. [DOI] [PMC free article] [PubMed]
- 23.Koning FA, Goujon C, Bauby H, Malim MH. Target cell-mediated editing of HIV-1 cDNA by APOBEC3 proteins in human macrophages. J Virol. 2011;85(24):13448–13452. doi: 10.1128/JVI.00775-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berger G, et al. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog. 2011;7(9):e1002221. doi: 10.1371/journal.ppat.1002221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vetter ML, D’Aquila RT. Cytoplasmic APOBEC3G restricts incoming Vif-positive human immunodeficiency virus type 1 and increases two-long terminal repeat circle formation in activated T-helper-subtype cells. J Virol. 2009;83(17):8646–8654. doi: 10.1128/JVI.00020-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445(7130):927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 27.Santiago ML, et al. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321(5894):1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Low A, et al. Enhanced replication and pathogenesis of Moloney murine leukemia virus in mice defective in the murine APOBEC3 gene. Virology. 2009;385(2):455–463. doi: 10.1016/j.virol.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langlois MA, Kemmerich K, Rada C, Neuberger MS. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J Virol. 2009;83(22):11550–11559. doi: 10.1128/JVI.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeda E, et al. Mouse APOBEC3 restricts friend leukemia virus infection and pathogenesis in vivo. J Virol. 2008;82(22):10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okeoma CM, Petersen J, Ross SR. Expression of murine APOBEC3 alleles in different mouse strains and their effect on mouse mammary tumor virus infection. J Virol. 2009;83(7):3029–3038. doi: 10.1128/JVI.02536-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, et al. Two genetic determinants acquired late in mus evolution regulate the inclusion of exon 5, which alters mouse APOBEC3 translation efficiency. PLoS Pathog. 2012;8(1):e1002478. doi: 10.1371/journal.ppat.1002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Browne EP, Littman DR. Species-specific restriction of apobec3-mediated hypermutation. J Virol. 2008;82(3):1305–1313. doi: 10.1128/JVI.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rulli SJ, Jr, et al. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J Virol. 2008;82(13):6566–6575. doi: 10.1128/JVI.01357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Martinez S, et al. Studies on the restriction of murine leukemia viruses by mouse APOBEC3. PLoS ONE. 2012;7(5):e38190. doi: 10.1371/journal.pone.0038190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogerd HP, Tallmadge RL, Oaks JL, Carpenter S, Cullen BR. Equine infectious anemia virus resists the antiretroviral activity of equine APOBEC3 proteins through a packaging-independent mechanism. J Virol. 2008;82(23):11889–11901. doi: 10.1128/JVI.01537-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zielonka J, et al. Restriction of equine infectious anemia virus by equine APOBEC3 cytidine deaminases. J Virol. 2009;83(15):7547–7559. doi: 10.1128/JVI.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stremlau M, et al. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci USA. 2006;103(14):5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perron MJ, et al. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol. 2007;81(5):2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okeoma CM, et al. Induction of APOBEC3 in vivo causes increased restriction of retrovirus infection. J Virol. 2009;83(8):3486–3495. doi: 10.1128/JVI.02347-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeiffer JK, Topping RS, Shin NH, Telesnitsky A. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J Virol. 1999;73(10):8441–8447. doi: 10.1128/jvi.73.10.8441-8447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roa A, et al. RING domain mutations uncouple TRIM5α restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J Virol. 2012;86(3):1717–1727. doi: 10.1128/JVI.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hulme AE, Perez O, Hope TJ. Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc Natl Acad Sci USA. 2011;108(24):9975–9980. doi: 10.1073/pnas.1014522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nat Immunol. 2010;11(11):1005–1013. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harris RS, Hultquist JF, Evans DT. The restriction factors of human immunodeficiency virus. J Biol Chem. 2012;287(49):40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Derse D, Hill SA, Princler G, Lloyd P, Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc Natl Acad Sci USA. 2007;104(8):2915–2920. doi: 10.1073/pnas.0609444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ooms M, Krikoni A, Kress AK, Simon V, Münk C. APOBEC3A, APOBEC3B, and APOBEC3H haplotype 2 restrict human T-lymphotropic virus type 1. J Virol. 2012;86(11):6097–6108. doi: 10.1128/JVI.06570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okimoto MA, Fan H. Identification of directly infected cells in the bone marrow of neonatal moloney murine leukemia virus-infected mice by use of a moloney murine leukemia virus-based vector. J Virol. 1999;73(2):1617–1623. doi: 10.1128/jvi.73.2.1617-1623.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsuji-Kawahara S, et al. Persistence of viremia and production of neutralizing antibodies differentially regulated by polymorphic APOBEC3 and BAFF-R loci in friend virus-infected mice. J Virol. 2010;84(12):6082–6095. doi: 10.1128/JVI.02516-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X, et al. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J Virol. 2012;86(7):3777–3786. doi: 10.1128/JVI.06594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okeoma CM, Huegel AL, Lingappa J, Feldman MD, Ross SR. APOBEC3 proteins expressed in mammary epithelial cells are packaged into retroviruses and can restrict transmission of milk-borne virions. Cell Host Microbe. 2010;8(6):534–543. doi: 10.1016/j.chom.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Warrilow D, et al. Cell factors stimulate human immunodeficiency virus type 1 reverse transcription in vitro. J Virol. 2008;82(3):1425–1437. doi: 10.1128/JVI.01808-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.