Abstract
Obesity and type 2 diabetes are characterized by altered gut microbiota, inflammation, and gut barrier disruption. Microbial composition and the mechanisms of interaction with the host that affect gut barrier function during obesity and type 2 diabetes have not been elucidated. We recently isolated Akkermansia muciniphila, which is a mucin-degrading bacterium that resides in the mucus layer. The presence of this bacterium inversely correlates with body weight in rodents and humans. However, the precise physiological roles played by this bacterium during obesity and metabolic disorders are unknown. This study demonstrated that the abundance of A. muciniphila decreased in obese and type 2 diabetic mice. We also observed that prebiotic feeding normalized A. muciniphila abundance, which correlated with an improved metabolic profile. In addition, we demonstrated that A. muciniphila treatment reversed high-fat diet-induced metabolic disorders, including fat-mass gain, metabolic endotoxemia, adipose tissue inflammation, and insulin resistance. A. muciniphila administration increased the intestinal levels of endocannabinoids that control inflammation, the gut barrier, and gut peptide secretion. Finally, we demonstrated that all these effects required viable A. muciniphila because treatment with heat-killed cells did not improve the metabolic profile or the mucus layer thickness. In summary, this study provides substantial insight into the intricate mechanisms of bacterial (i.e., A. muciniphila) regulation of the cross-talk between the host and gut microbiota. These results also provide a rationale for the development of a treatment that uses this human mucus colonizer for the prevention or treatment of obesity and its associated metabolic disorders.
Keywords: RegIIIγ, LPS, gut permeability, Lactobacillus plantarum, antimicrobial peptides
Gut microbiota were once characterized as bystanders in the intestinal tract, but their active role in intestinal physiology is now widely investigated. In particular, the mutualism that exists between gut microbiota and the host has received much attention. Obesity and type 2 diabetes are characterized by altered gut microbiota (1), inflammation (2), and gut barrier disruption (3–5). We recently demonstrated an association of obesity and type 2 diabetes with increased gut permeability, which induced metabolic endotoxemia and metabolic inflammation (3–5). Unequivocal evidence demonstrates that gut microbiota influence whole-body metabolism (1, 6) by affecting the energy balance (6), gut permeability (4, 5), serum lipopolysaccharides [i.e., metabolic endotoxemia (7)], and metabolic inflammation (3–5, 7) that are associated with obesity and associated disorders. However, the microbial composition and the exact mechanisms of interaction between these two partners that affect host–gut barrier function and metabolism remain unclear.
The intestinal epithelium is the interface for the interaction between gut microbiota and host tissues (8). This barrier is enhanced by the presence of a mucus layer and immune factors that are produced by the host (9). Antimicrobial peptides for innate immunity are produced by Paneth cells (e.g., α-defensins, lysozyme C, phospholipases, and C-type lectin, primarily regenerating islet-derived 3-gamma, RegIIIγ) or enterocytes (RegIIIγ) (10–12). Adaptive immune system effectors that are secreted into the intestinal lumen, such as IgA, may also restrict bacterial penetration into the host mucus and mucosal tissue (13). These immune factors allow the host to control its interactions with gut microbiota and shape its microbial communities (11).
The endocannabinoid system has also been implicated in the control of the gut barrier and inflammation (5, 14). One lipid in this system, 2-arachidonoylglycerol (2-AG), reduces metabolic endotoxemia and systemic inflammation (15). Another acylglycerol, 2-palmitoylglycerol (2-PG), potentiates the antiinflammatory effects of 2-AG (16). Importantly, 2-oleoylglycerol (2-OG) stimulates the release of gut peptides, such as glucagon-like peptide-1 (GLP-1) and glucagon-like peptide-2 (GLP-2), from intestinal L-cells (17). These peptides are implicated in the control of glucose homeostasis and gut barrier function, respectively (4).
Recently, Akkermansia muciniphila has been identified as a mucin-degrading bacteria that resides in the mucus layer (18), and it is the dominant human bacterium that abundantly colonizes this nutrient-rich environment (18). A. muciniphila may represent 3–5% of the microbial community (18, 19) in healthy subjects, and its abundance inversely correlates with body weight (20–23) and type 1 diabetes (24) in mice and humans, although a recent metagenomic study found that some of the genes belonging to A. muciniphila were enriched in type 2 diabetic subjects (25).
We recently discovered that the administration of prebiotics (oligofructose) to genetically obese mice increased the abundance of A. muciniphila by ∼100-fold (23). However, the direct implications of A. muciniphila for obesity and type 2 diabetes have not been determined, and the precise physiological roles it plays during these processes are not known.
Our previous results and the close proximity of this bacterium to the human intestinal epithelium support the hypothesis that A. muciniphila plays a crucial role in the mutualism between the gut microbiota and host that controls gut barrier function and other physiological and homeostatic functions during obesity and type 2 diabetes. We administered alive or heat-killed A. muciniphila to mice that were fed a high-fat diet and investigated the gut barrier, glucose homeostasis, and adipose tissue metabolism to test this hypothesis.
Results
A. muciniphila Abundance Decreased in Obese and Type 2 Diabetic Mice.
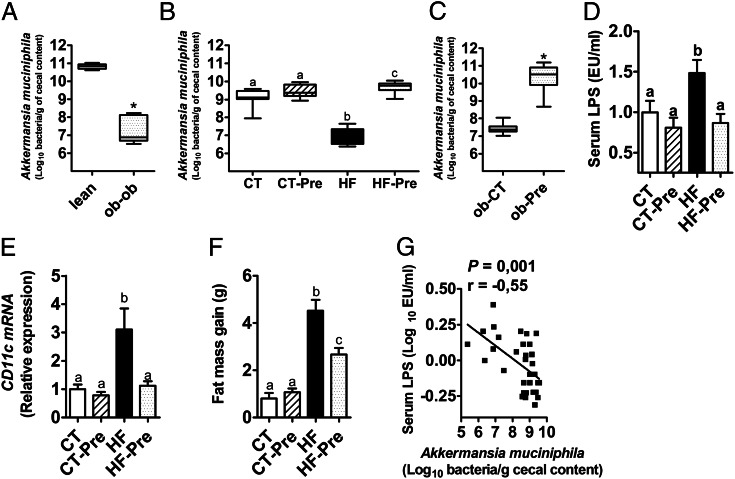
We observed that the abundance of A. muciniphila was 3,300-fold lower in leptin-deficient obese mice than in their lean littermates (Fig. 1A). We also observed a 100-fold decrease of this bacterium in high-fat-(HF)-fed mice (Fig. 1B).
Fig. 1.
A. muciniphila abundance is decreased in obese and diabetic mice, and prebiotic treatment restored A. muciniphila to basal levels and reversed metabolic endotoxemia and related disorders. (A) A. muciniphila abundance (log10 of bacteria per g of cecal content) measured in the cecal content of leptin-deficient (ob-ob) obese mice and their lean littermates (lean) (n = 5). (B) A. muciniphila abundance (log10 of bacteria per g of cecal content) measured in the cecal content of control diet-fed mice (CT) or CT diet-fed mice treated with prebiotics (CT-Pre) added to their drinking water and HF diet-fed mice (HF) or HF diet-fed mice treated with prebiotics (HF-Pre) added to their drinking water for 8 wk (n = 10). (C) A. muciniphila abundance (log10 of bacteria per g of cecal content) measured in the cecal content of obese mice fed a control diet (ob-CT) or treated with prebiotics (ob-Pre) for 5 wk (n = 10). (D) Portal vein serum LPS levels (n = 7–9). (E) mRNA expression of the adipose tissue macrophage infiltration marker CD11c (n = 10). (F) Total fat mass gain measured by time-domain NMR (n = 10). (G) Pearson’s correlation between log values of portal vein LPS levels and A. muciniphila abundance (log10 of bacteria per g of cecal content); (Inset) Pearson’s correlation coefficient (r) and the corresponding P value. Data are shown as means ± SEM; *P < 0.05 by two-tailed Student t test, data with different superscript letters are significantly different (P < 0.05) according to post hoc ANOVA one-way statistical analysis.
Prebiotic Treatment Restored Basal Levels of A. muciniphila and Improved Metabolic Endotoxemia and Related Disorders That Are Associated with HF-Diet-Induced Obesity.
Prebiotics (oligofructose) completely restored A. muciniphila counts in both models (Fig. 1 B and C), therefore supporting the data obtained in our previous study performed in ob/ob mice (23). Administration of prebiotics in HF-fed mice abolished metabolic endotoxemia (Fig. 1D) and normalized the CD11c subpopulation of macrophages in adipose tissue, which is the primary population of increased adipose tissue macrophages in obesity (2) (Fig. 1E). Administration of prebiotics also reduced the total fat mass, the mass of the different fat pads (i.e., s.c., mesenteric, and epididymal), and the body weight (Fig. 1F and Fig. S1 A–C). These results were significantly and inversely correlated with A. muciniphila abundance (Fig. 1G and Fig. S1 D and E). However, the role of the lack of A. muciniphila in the molecular mechanisms that underlie the onset of these disorders has not been demonstrated, and whether an increased abundance of A. muciniphila reverses these disorders must be investigated. A. muciniphila was orally administered to control or HF-fed mice for 4 wk to address these questions.
HF Diet Altered the Gut Microbiota Composition, Whereas A. muciniphila Did Not Significantly Induce Changes.
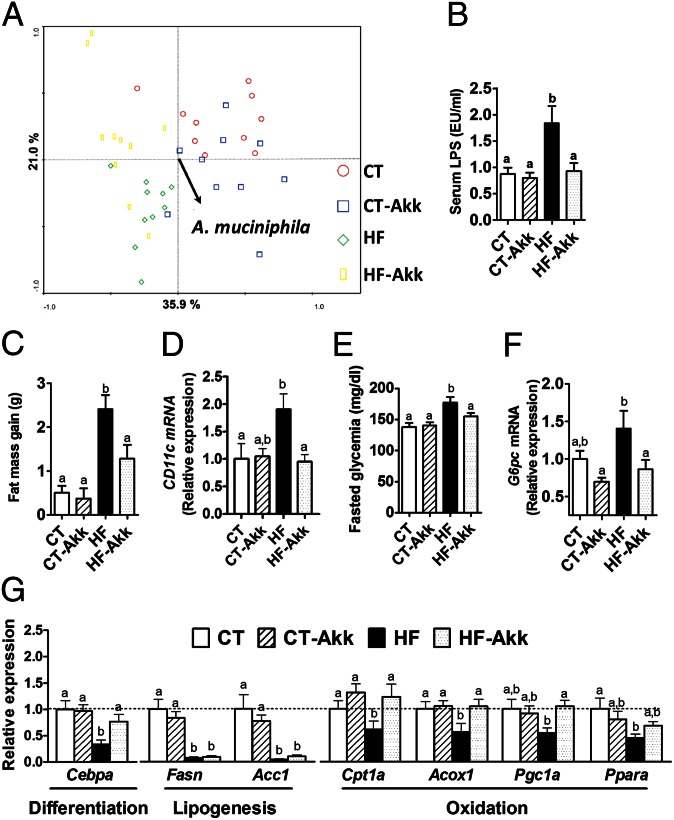
A. muciniphila treatment was associated with an increase in A. muciniphila abundance in the cecal content of mice (Fig. S2A). We also demonstrated that an HF diet significantly changes the gut microbiota using a phylogenetic microarray (Mouse Intestinal Tract Chip, MITChip) (10, 23), as shown by principal component analyses (Fig. 2A), dendrogram clustering, and representational difference analysis (Fig. S2 B and C), whereas A. muciniphila treatment did not modify this profile (Fig. 2A and Fig. S2 B and C).
Fig. 2.
A. muciniphila counteracted metabolic endotoxemia, diet-induced obesity, adipose tissue macrophage infiltration, improved glucose homeostasis, and adipose tissue metabolism in diet-induced obese mice without modifying gut microbiota composition. (A) Principal component analysis using the MITChip phylogenetic fingerprints of the gut microbiota from the cecal contents of control mice treated with a daily oral gavage containing sterile anaerobic PBS for 4 wk and fed a control (CT) or HF diet (HF) (CT in red and HF in green) and in mice treated with a daily oral gavage containing A. muciniphila (2.108 bacterial cells suspended in 200 µL of sterile anaerobic PBS) and fed a control (CT-Akk) or HF diet (HF-Akk) (CT-Akk in blue and HF-Akk in yellow) (n = 10). (B) Portal vein serum LPS levels (n = 6–10). (C) Total fat mass gain measured by time-domain NMR (n = 10). (D) mRNA expression of the adipose tissue macrophage infiltration marker CD11c (n = 10). (E) Fasting glycemia (n = 10). (F) Liver G6pc mRNA (n = 10). (G) mRNA expression of markers of adipocyte differentiation (Cebpa), lipogenesis (Acc1; Fasn), and lipid oxidation (Cpt1; Acox1; Pgc1a; and Ppara) was measured in visceral fat depots (mesenteric fat) (n = 10). Data are shown as means ± SEM. Data with different superscript letters are significantly different (P < 0.05) according to post hoc ANOVA one-way statistical analysis.
A. muciniphila Improved Metabolic Disorders in Diet-Induced Obese Mice.
A. muciniphila treatment normalized diet-induced metabolic endotoxemia, adiposity, and the adipose tissue marker CD11c (Fig. 2 B–D and Fig. S3A). Similarly, A. muciniphila treatment reduced body weight and improved body composition (i.e., fat mass/lean mass ratio) (Fig. S3 B and C) without changes in food intake (Fig. S3D). We demonstrated that A. muciniphila treatment completely reversed diet-induced fasting hyperglycemia (Fig. 2E) via a mechanism that was associated with a 40% reduction in hepatic glucose-6-phosphatase expression (Fig. 2F), thereby suggesting a reduction in gluconeogenesis. Notably, the insulin resistance index was similarly reduced after A. muciniphila treatment (Fig. S3E). These results suggest a key role for A. muciniphila in gut barrier function, metabolic inflammation, and fat storage. Therefore, we hypothesized that A. muciniphila would impact adipose tissue metabolism. We demonstrated that A. muciniphila treatment under an HF diet increased the mRNA expression of markers of adipocyte differentiation (e.g., CCAAT/enhancer–binding protein-α, encoded by Cebpa) and lipid oxidation (e.g., carnitine palmitoyltransferase-1, encoded by Cpt1; acyl-CoA-oxidase, encoded by Acox1; peroxisome proliferator-activated receptor γ coactivator, encoded by Pgc1a; and peroxisome proliferator-activated receptor alpha, encoded by Ppara) without affecting lipogenesis markers (e.g., acetyl-CoA carboxylase, encoded by Acc1 and fatty acid synthase, encoded by Fasn) (Fig. 2G). These data further confirm our hypothesis that A. muciniphila controls fat storage, adipose tissue metabolism, and glucose homeostasis.
A. muciniphila Treatment Exerted Minor Effects on Antibacterial Peptide Content in the Ileum and IgA Levels in the Feces.
Recent data suggest that the intestinal mucosa contributes to the maintenance of the gut barrier by secreting antimicrobial peptides for innate immunity that are produced by Paneth cells (e.g., α-defensins, lysozyme C, phospholipases, and C-type lectin, primarily the RegIIIγ) or enterocytes (RegIIIγ) (10, 12). We measured the expression of Paneth and epithelial cell antibacterial markers in the ileum to elucidate the impact of the HF diet and A. muciniphila treatment on gut barrier function. A. muciniphila increased the expression of Reg3g (RegIIIγ) under the control diet, but this effect was not observed in HF-fed mice (Fig. S4A). Pla2g2a and Defa expression were similar between groups, but Lyz1 expression tended to be lower after bacterial administration (Fig. S4 B–D). We also measured IgA in fecal samples as an adaptive immune system factor (13). Fecal IgA levels were not affected by the treatments (Fig. S4E), which suggests that A. muciniphila controls gut barrier function by other mechanisms of epithelial signaling (26).
A. muciniphila Increased Endocannabinoid (Acylglycerols) Content in the Ileum.
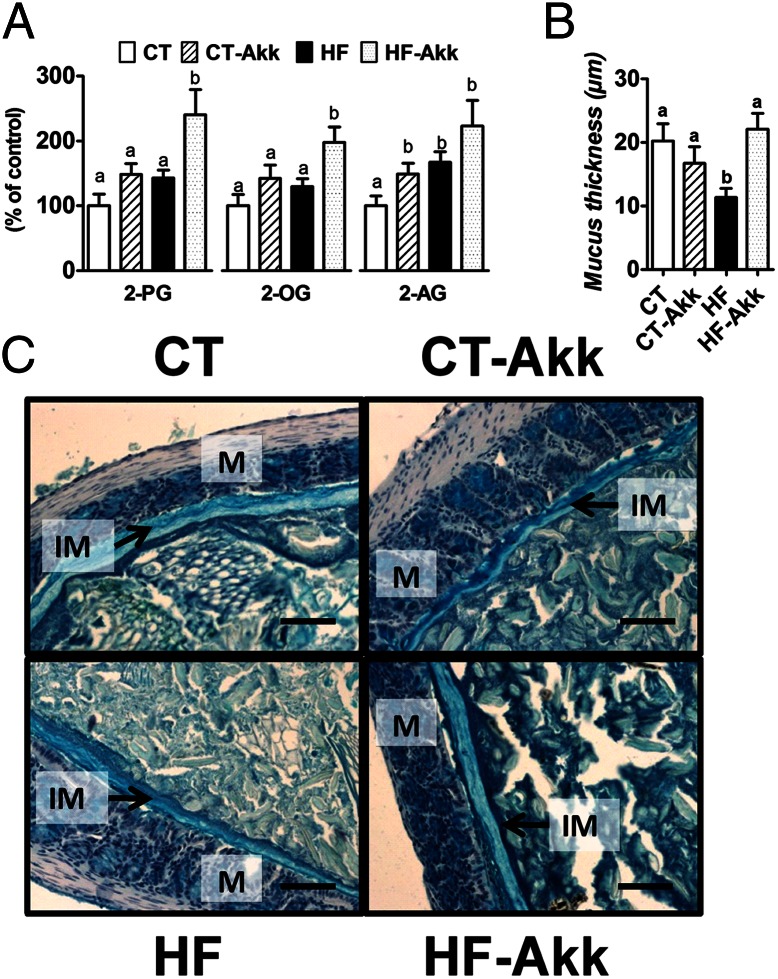
We previously observed a link between gut microbiota and intestinal endocannabinoid system tone (5). We demonstrated an association of decreased monoacylglycerol lipase expression with improved gut barrier function and decreased metabolic inflammation (5). We also demonstrated previously that the pharmacological inhibition of monoacylglycerol lipase reduced metabolic endotoxemia and systemic inflammation (15), which suggests a direct link between acylglycerols and gut barrier function. Therefore, we measured intestinal acylglycerol levels and demonstrated that A. muciniphila treatment increased the levels of 2-OG, 2-AG, and 2-PG (Fig. 3A). These results support a direct link between A. muciniphila administration and intestinal levels of acylglycerols that are involved in glucose and intestinal homeostasis.
Fig. 3.
A. muciniphila colonization restored gut barrier function and increased intestinal endocannabinoids in diet-induced obese mice. (A) Ileum 2-PG, 2-OG, and 2-AG (expressed as percentage of the control) (n = 10). (B) Thickness of the mucus layer measured by histological analyses After alcian blue staining (n = 7–8). (C) Representative alcian blue images that were used for mucus layer thickness measurements. M, mucosa; IM, inner mucus layer. (Scale bars, 40 µm.) Data are shown as means ± SEM. Data with different superscript letters are significantly different (P < 0.05) according to post hoc ANOVA one-way statistical analysis.
A. muciniphila Counteracted Diet-Induced Colon Mucosal Barrier Dysfunction During Obesity.
Recent evidence supports that interactions between the gut microbiota and mucus layer are dynamic systems that affect mucus barrier biology (9, 27). Therefore, we investigated the impact of A. muciniphila treatment on the thickness of the inner mucus layer. We demonstrated a 46% thinner mucus layer in HF-fed mice, and A. muciniphila treatment counteracted this decrease (Fig. 3 B and C).
Viable but Not Heat-Killed A. muciniphila Counteracted Diet-Induced Metabolic and Mucosal Barrier Dysfunction During Obesity.
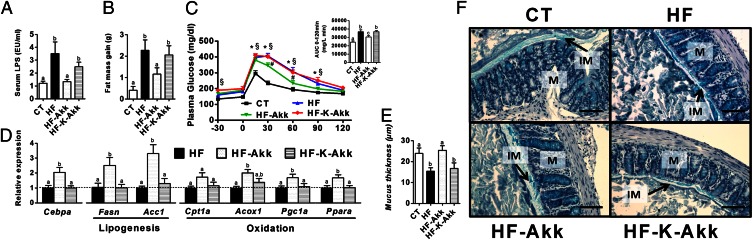
To further demonstrate whether A. muciniphila has to be alive to exert its metabolic effects, we have compared the impact of viable A. muciniphila administration with that of heat-killed A. muciniphila. We found that viable A. muciniphila counteracted diet-induced metabolic endotoxemia, fat mass development, and altered adipose tissue metabolism (Fig. 4 A, B, and D and Fig. S5A) to a similar extent as observed in the first set of experiments (Fig. 2 B, C, and G and Fig. S3A). Importantly, these effects were not observed after administration of heat-killed A. muciniphila (Fig. 4 A, B, and D and Fig. S5A). In addition, we found that viable A. muciniphila significantly reduced plasma glucose levels after an oral glucose tolerance test (Fig. 4C), whereas heat-killed A. muciniphila exhibited glucose intolerance similar to that of HF-fed mice (Fig. 4C). Finally, we confirmed that viable A. muciniphila restored mucus layer thickness upon HF-diet, whereas we found that heat-killed A. muciniphila did not improve mucus layer thickness compared with HF (Fig. 4 E and F). It is worth noting that we found 100-fold more viable A. muciniphila recovered from the cecal and colonic content of A. muciniphila-treated mice compared with the HF and heat-killed bacteria groups (HF-Akk: 9.5 ± 1.02 log10 cells/mg of content; HF and HF-K-Akk: 6.8 ± 0.51 log10 cells/mg of content; P = 0.0059), thereby evidencing the viability of A. muciniphila after oral administration.
Fig. 4.
Heat-killed A. muciniphila did not counteract metabolic endotoxemia, diet-induced obesity, oral glucose intolerance, and did not improve adipose tissue metabolism and gut barrier function in diet-induced obese mice. Control mice were fed a control (CT) or HF diet (HF) and treated with a daily oral gavage containing sterile anaerobic PBS and glycerol for 4 wk daily. Treated mice received an oral gavage of alive A. muciniphila (HF-Akk) or killed A. muciniphila (HF-K-Akk) (2.108 bacterial cells suspended in 200 µL of sterile anaerobic PBS) and fed an HF diet (n = 8). (A) Portal vein serum LPS levels (n = 6–7). (B) Total fat mass gain measured by time-domain NMR (n = 7–8). (C) Plasma glucose profile after 2 g/kg glucose oral challenge in freely moving mice. (Inset) Mean area under the curve (AUC) measured between 0 and 120 min after glucose load (n = 7–8). (D) mRNA expression of markers of adipocyte differentiation (Cebpa), lipogenesis (Acc1; Fasn), and lipid oxidation (Cpt1; Acox1; Pgc1a; and Ppara) was measured in visceral fat depots (mesenteric fat) (n = 8). (E) Thickness of the mucus layer measured by histological analyses after alcian blue staining (CT n = 4, HF n = 6, HF-Akk and HF-K-Akk n = 5). (F) Representative alcian blue images that were used for mucus layer thickness measurements. M, mucosa; IM, inner mucus layer. (Scale bars, 40 µm.) Data are shown as means ± SEM. Data with different superscript letters are significantly different (P < 0.05) according to post hoc ANOVA one-way statistical analysis.
This study confirms that that HF diet-induced obesity is associated with changes in gut microbiota composition (7) (28) (Fig. 2A and Fig. S2 B and C). However, antimicrobial peptides in the ileum were not affected by the treatments. In contrast, Reg3g expression in colon epithelial cells was significantly reduced, by ∼50%, in HF and heat-killed A. muciniphila treated mice, whereas viable A. muciniphila treatment completely blunted this effect and increased Reg3g expression upon HF diet (Fig. S5B).
Discussion
This study demonstrated a dramatic decrease in A. muciniphila in genetically and diet-induced obese mice. We demonstrated that prebiotic (oligofructose) treatment restored A. muciniphila abundance and improved gut barrier and metabolic parameters. However, the mechanisms that were responsible for the bloom in A. muciniphila caused by prebiotic administration are not clear. A. muciniphila does not grow on oligofructose-enriched media (in vitro), which suggests that complex cross-feeding interactions contributed to this effect. However, it has been previously shown in rats that oligofructose feeding increases the number of goblet cells and mucus layer thickness (29). Thus, whether oligofructose feeding increases A. muciniphila by providing the main source of energy for this bacterium and thereby favoring its growth or whether the increase of A. muciniphila increases mucus production and degradation (i.e., turnover) remain to be demonstrated. Oligofructose changes more than 100 different taxa in mice (23). Therefore, we cannot exclude that oligofructose induces specific changes in the gut bacteria and cross-feeding promoting the growth of A. muciniphila. In the present study, we investigated the direct impact of A. muciniphila. We reversed the pathological phenotype by restoring the physiological abundance of this strain in obese and diabetic mice. These results demonstrated the key role of A. muciniphila in the physiopathology of obesity, type 2 diabetes, and metabolic inflammation. These experiments clearly demonstrate that viable A. muciniphila controls gut barrier function, fat mass storage, and glucose homeostasis in obese and type 2 diabetic mice via several mechanisms. These results provide proof of this concept in this context. The major weaknesses in investigations of the role of gut microbiota in the etiology of obesity and type 2 diabetes is the reliance on conclusions that are based on correlative data between bacteria (or one genus) and physiological parameters, because most of the gut bacteria have been identified at the phylogenetic level (i.e., through metagenomic approaches) but have never been cultured.
Several reports have demonstrated the importance of selected bacteria [i.e., Lactobacillus spp (30, 31), Bifidobacterium spp (32, 33), and Bacteroides uniformis CECTT 7771 (34)] on fat mass development during diet-induced obesity, but the aims of these studies were different from that of the present study. These studies investigated the impact of supplementation with one specific probiotic strain or strains that were isolated from healthy infants on physiological parameters. Here we investigated the strain that is affected during obesity and type 2 diabetes in humans and rodents (18, 23). Probiotics have far fewer opportunities for direct contact with the mucosa, but A. muciniphila may induces differential host responses because of more intensive contact with the host mucosa (26). To further confirm this hypothesis, we have treated HF-fed mice with a probiotic (i.e., Lactobacillus plantarum WCFS1). We found that L. plantarum administration did not change fat mass development, adipose tissue metabolism, mucus layer thickness, colon Reg3g mRNA, and metabolic endotoxemia (Fig. S6 A–E). Therefore, these data suggest that A. muciniphila induces specific host responses compared with other putative beneficial microbes.
A. muciniphila is a Gram-negative bacteria (i.e., it contains LPS) that constitutes 3–5% of the gut microbial community. However, our study clearly demonstrated the lack of a direct relationship between the abundance of Gram-negative bacteria within the gut and metabolic endotoxemia (i.e., that is caused by serum LPS) because gut colonization by A. muciniphila decreased metabolic endotoxemia arising on an HF diet. One explanation for this counterintuitive result may be that A. muciniphila regulates gut barrier function at different levels. Previous data suggest that gut microbiota contribute to gut barrier alterations during obesity and metabolic endotoxemia (4). However, the different mechanisms of interaction between bacteria and the host that affect gut barrier function during obesity and type 2 diabetes have not been elucidated. This study identified an association of obesity with a decrease in mucus thickness, which supports an additional mechanism of increased gut permeability (i.e., metabolic endotoxemia) that is characteristic of obesity and associated disorders. Furthermore, we demonstrated that A. muciniphila restored this mucus layer, which suggests that this mechanism contributes to the reduction in metabolic endotoxemia that was observed during A. muciniphila treatment. Moreover, we found that viable A. muciniphila induces these effects, whereas heat-killed A. muciniphila did not protect the mice from diet-induced obesity and associated disorders.
These results suggest that the presence of viable A. muciniphila within the mucus layer is a crucial mechanism in the control of host mucus turnover (19), which improves gut barrier function. However, we cannot exclude additional mechanisms that have been implicated in the regulation of gut barrier. For example, we previously demonstrated that gut microbiota control gut peptides (e.g., GLP-2) that regulate epithelial cell proliferation and gut barrier function (4). Prebiotics stimulate GLP-1 and GLP-2 secretion by acting on the enteroendocrine L-cells that are primarily in the ileum and colon (6). The abundance of A. muciniphila is associated with higher L-cell activity (i.e., GLP-1 and GLP-2 secretion) (4, 23), but the mechanisms underlying this relationship are not known. Here, we demonstrated that A. muciniphila administration significantly increased intestinal levels of 2-OG, which stimulates glucagon-like peptide secretion from intestinal L-cells (17). Altogether our data suggest that this could be a key mechanism by which A. muciniphila controls gut barrier function, metabolic endotoxemia, and metabolism. We also demonstrated that A. muciniphila administration increased 2-AG intestinal levels. We recently demonstrated that an increase in 2-AG endogenous levels induced by selective monoacylglycerol lipase inhibitor protects against trinitrobenzene sulfonic acid-induced colitis in mice (15) and reduces metabolic endotoxemia as well as the level of circulating inflammatory cytokines and peripheral and brain inflammation. Therefore, the increased 2-AG levels that were observed after A. muciniphila treatment may have also contributed to the reduced inflammation. However, whether the induction of these endocannabinoids after A. muciniphila treatment constitutes the molecular event that links these metabolic features warrants further investigation.
Specifically, we demonstrated that the restoration of the physiological abundance of A. muciniphila reduced diet-induced body weight gain, fat mass development, and fasting hyperglycemia without affecting food intake. This variation in energy storage is explained by the normalization of adipose tissue adipogenesis (i.e., differentiation and lipogenesis) and fatty acid oxidation. We have previously demonstrated that higher circulating LPS levels inhibit adipose tissue differentiation and lipogenesis, thereby contributing to altered adipose tissue metabolism characterizing obesity (5). Thus, we postulate that A. muciniphila restores gut barrier function and thereby contributes to normalize metabolic endotoxemia and adipose tissue metabolism. We found that A. muciniphila improved glucose tolerance and decreased endogenous hepatic glucose production. These findings are not in agreement with the apparent but low association of A.muciniphila genes with type 2 diabetes-associated metagenome-wide associated studies (25). Nevertheless, the data by Qin et al. remain to be confirmed because this related to only 337 of the 2,176 A.muciniphila genes (35) and may be confounded by dietary or pharmaceutical treatments specifically favoring its growth in the human intestine.
Dynamic insulin resistance assessments and the present results suggest improved insulin sensitivity. However, we cannot exclude the possibility that the improvements in glucose and lipid metabolism occurred via an LPS-dependent mechanism, as demonstrated previously (5, 7). We confirmed (7, 36) that an HF diet profoundly affected the gut microbiota composition, whereas A. muciniphila administration did not significantly affect this profile. Therefore, it is tempting to extrapolate our findings as a single-species-dependent modulation of the gut microbiota. Moreover, because heat-killing of A. muciniphila completely abolished the metabolic effects it is unlikely that specific A. muciniphila-derived cell-envelope components may directly contribute to the phenotype observed with viable A. muciniphila. It is worth noting that this observation also minimizes the possibility that the host response was caused by a substance in the culture media. However, although not directly fitting with the aim of the present study, follow-up studies of the gut microbiome after viable A. muciniphila administration may identify the components that contribute to disease or the host physiological response (37).
Finally, we demonstrated that A. muciniphila regulates intestinal antimicrobial peptides in the colon (e.g., RegIIIγ). A. muciniphila exerted minor effects on antimicrobial peptide production in the ileum. RegIIIγ exerts direct bactericidal activity against Gram-positive bacteria in the intestine. Therefore, A. muciniphila may manipulate host immunity to favor its own survival through an increase in RegIIIγ expression, which reduces the competition for resources and induces long-term tolerance for the development in the mucus layer. Here, we clearly found that viable A. muciniphila significantly increased RegIIIγ, whereas heat-killed A. muciniphila did not affect this parameter. Whether the effect on RegIIIγ should be considered as beneficial or harmful for the host remain to be determined. These results link the colonization of the colon, but not the ileum, by A. muciniphila with the fundamental immune mechanisms through which RegIIIγ promotes host–bacterial mutualism and regulates the spatial relationships between the microbiota and host (38). Finally, A. muciniphila is known to degrade human mucus (18). However, whether the beneficial effects observed here may be extended to other pathological situations in which the mucus layer is altered (e.g., intestinal inflammatory diseases) (39) remain to be elucidated.
We recently demonstrated that germ-free mice that were monoassociated with A. muciniphila exhibit important modulations of gene expression; the most marked changes were observed in the colon (442 genes), followed by the ileum (253 genes) and the cecum (211 genes) (26). In the colon, 60 genes, including 16 genes encoding CD antigen markers and 10 genes encoding immune cell membrane receptors, were up-regulated after A. muciniphila colonization (26). Several pathways that regulate lipid metabolism, cell signaling, and molecular transport are mostly affected in the ileum (26). These data have uncovered mechanisms of bacterial interaction with the host to control gut permeability and metabolism. Further studies should explore the cellular processes and identify the bacterial products that regulate the host cell responses and metabolic effects of A. muciniphila.
In summary, this study provided unique and substantial insights into the intricate regulation of the cross-talk between the host and A. muciniphila bacteria. These results provide a rationale for the development of a treatment that uses this human mucus colonizer for the prevention or treatment of obesity and its associated metabolic disorders.
Materials and Methods
Mice.
Male C57BL/6 mice were used in the four series of experiments. Cecal contents from genetic (ob/ob) and HF-fed obese and type 2 diabetic mice treated or not with prebiotics (oligofructose, 0.3 g per mouse per day) were harvested, immersed in liquid nitrogen, and stored at −80 °C for further A. muciniphila analysis. A subset of 10-wk-old C57BL/6J was fed a control diet (CT) or an HF diet (60% fat). The mice were treated with A. muciniphila by oral gavage at a dose 2.108 cfu/0.2 mL suspended in sterile anaerobic PBS (CT-Akk and HF-Akk), or heat-killed A. muciniphila (HF-K-Akk). Control groups were orally administered an equivalent volume of sterile anaerobic PBS containing a similar end concentration of glycerol (2.5% vol/vol) (CT and HF). Treatments were continued for 4 wk. A. muciniphila MucT (ATTC BAA-835) was grown anaerobically in a mucin-based basal medium as described previously (18). The cultures were washed and concentrated in anaerobic PBS that included 25% (vol/vol) glycerol to an end concentration of 1.1010 cfu/mL under strict anaerobic conditions. Body composition was assessed using a 7.5-MHz time-domain NMR. Blood, adipose depots, liver, cecal content, and intestinal segments (ileum, cecum, and colon) were collected at death and analyzed. A complete description of the mouse experiments and bacteria preparation is provided in SI Material and Methods.
Gut Microbiota Analysis.
Gut microbiota analyses were performed using real-time quantitative PCR (qPCR) analysis and the MITChip, which is a phylogenetic microarray consisting of 3,580 different oligonucleotide probes that target two hypervariable regions of the 16S rRNA gene (the V1 and V6 regions). Analyses of the MITChip were performed as described previously (23, 40) and in SI Material and Methods.
Gene Expression Analysis.
The expression of metabolic genes of interest and RNA expression profiles were analyzed using real-time qPCR analysis as described in SI Material and Methods.
Measurement of Endocannabinoid Intestinal Levels.
Intestinal endocannabinoids were measured using an LTQ Orbitrap mass spectrometer as described in SI Material and Methods.
Biochemical Analysis.
Plasma insulin and fecal IgA were analyzed using ELISA as described in SI Material and Methods. The thickness of the mucus layer was measured in proximal colon segments that were fixed in Carnoy’s solution and in 5-µm paraffin sections stained with alcian blue as described in SI Material and Methods. LPS concentrations in portal vein blood were measured using Endosafe-Multi-Cartridge System based on the limulus amebocyte lysate kinetic chromogenic methodology as described the in SI Material and Methods.
Statistical Analysis.
Data are expressed as means ± SEM. Differences between two groups were assessed using the unpaired two-tailed Student t test. Data sets that involved more than two groups were assessed using ANOVA followed by Newman-Keuls post hoc tests. Correlations were analyzed using Pearson’s correlation. In the figures, data with different superscript letters are significantly different at P < 0.05, according to post hoc ANOVA statistical analyses. Data were analyzed using GraphPad Prism version 5.00 for Windows (GraphPad Software). The results were considered statistically significant when P < 0.05.
Supplementary Material
Acknowledgments
We thank R. M. Goebbels for histological assistance, and B. Es Saadi and R. Selleslagh for technical assistance. P.D.C. is a research associate from the Fonds de la Recherche Scientifique (FRS-FNRS, Belgium) and is the recipient of FSR and FRSM (Fonds Spéciaux de Recherches, Université catholique de Louvain, Belgium; Fonds de la Recherche Scientifique Médicale, Belgium) and Société Francophone du Diabète (France) subsidies. A.E. is a doctoral fellow from the FRS-FNRS. G.G.M. is the recipient of subsidies from the FSR and FRSM and from FRS-FNRS Grant FRFC 2.4555.08. J.P.O. and C.B. were funded by European Research Council Advance Grant 250172-MicrobesInside, awarded to W.M.d.V., whose work was further supported by an unrestricted Spinoza Award of the Netherlands Organization for Scientific Research.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219451110/-/DCSupplemental.
References
- 1.Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 2.Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18(3):363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- 3.Cani PD, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 4.Cani PD, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muccioli GG, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. 2010;6:392. doi: 10.1038/msb.2010.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3(4):279–288. doi: 10.4161/gmic.19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cani PD, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 8.Wells JM, Rossi O, Meijerink M, van Baarlen P. Epithelial crosstalk at the microbiota-mucosal interface. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4607–4614. doi: 10.1073/pnas.1000092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9(5):356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 11.Pott J, Hornef M. Innate immune signalling at the intestinal epithelium in homeostasis and disease. EMBO Rep. 2012;13(8):684–698. doi: 10.1038/embor.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10(3):159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 13.Macpherson AJ, Geuking MB, Slack E, Hapfelmeier S, McCoy KD. The habitat, double life, citizenship, and forgetfulness of IgA. Immunol Rev. 2012;245(1):132–146. doi: 10.1111/j.1600-065X.2011.01072.x. [DOI] [PubMed] [Google Scholar]
- 14.Cani PD. Crosstalk between the gut microbiota and the endocannabinoid system: impact on the gut barrier function and the adipose tissue. Clin Microbiol Infect. 2012;18(Suppl 4):50–53. doi: 10.1111/j.1469-0691.2012.03866.x. [DOI] [PubMed] [Google Scholar]
- 15.Alhouayek M, Lambert DM, Delzenne NM, Cani PD, Muccioli GG. Increasing endogenous 2-arachidonoylglycerol levels counteracts colitis and related systemic inflammation. FASEB J. 2011;25(8):2711–2721. doi: 10.1096/fj.10-176602. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Shabat S, et al. An entourage effect: Inactive endogenous fatty acid glycerol esters enhance 2-arachidonoyl-glycerol cannabinoid activity. Eur J Pharmacol. 1998;353(1):23–31. doi: 10.1016/s0014-2999(98)00392-6. [DOI] [PubMed] [Google Scholar]
- 17.Hansen KB, et al. 2-Oleoyl glycerol is a GPR119 agonist and signals GLP-1 release in humans. J Clin Endocrinol Metab. 2011;96(9):E1409–E1417. doi: 10.1210/jc.2011-0647. [DOI] [PubMed] [Google Scholar]
- 18.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(Pt 5):1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 19.Belzer C, de Vos WM. Microbes inside—from diversity to function: The case of Akkermansia. ISME J. 2012;6(8):1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Santacruz A, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104(1):83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson CL, et al. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity (Silver Spring) 2012;20(11):2257–2261. doi: 10.1038/oby.2012.110. [DOI] [PubMed] [Google Scholar]
- 22.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88(4):894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 23.Everard A, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60(11):2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen CH, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia. 2012;55(8):2285–2294. doi: 10.1007/s00125-012-2564-7. [DOI] [PubMed] [Google Scholar]
- 25.Qin J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 26.Derrien M, et al. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ottman N, Smidt H, de Vos WM, Belzer C. The function of our microbiota: Who is out there and what do they do? Front Cell Infect Microbiol. 2012;2:104. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleessen B, Hartmann L, Blaut M. Fructans in the diet cause alterations of intestinal mucosal architecture, released mucins and mucosa-associated bifidobacteria in gnotobiotic rats. Br J Nutr. 2003;89(5):597–606. doi: 10.1079/BJN2002827. [DOI] [PubMed] [Google Scholar]
- 30.Mohamadzadeh M, et al. Regulation of induced colonic inflammation by Lactobacillus acidophilus deficient in lipoteichoic acid. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4623–4630. doi: 10.1073/pnas.1005066107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fåk F, Bäckhed F. Lactobacillus reuteri prevents diet-induced obesity, but not atherosclerosis, in a strain dependent fashion in Apoe-/- Mice. PLoS ONE. 2012;7(10):e46837. doi: 10.1371/journal.pone.0046837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JJ, Wang R, Li XF, Wang RL. Bifidobacterium longum supplementation improved high-fat-fed-induced metabolic syndrome and promoted intestinal Reg I gene expression. Exp Biol Med (Maywood) 2011;236(7):823–831. doi: 10.1258/ebm.2011.010399. [DOI] [PubMed] [Google Scholar]
- 33.An HM, et al. Antiobesity and lipid-lowering effects of Bifidobacterium spp. in high fat diet-induced obese rats. Lipids Health Dis. 2011;10:116. doi: 10.1186/1476-511X-10-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gauffin Cano P, Santacruz A, Moya A, Sanz Y. Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS ONE. 2012;7(7):e41079. doi: 10.1371/journal.pone.0041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Passel MW, et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE. 2011;6(3):e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cani PD, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–2383. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 37.Greenblum S, Turnbaugh PJ, Borenstein E. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci USA. 2012;109(2):594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334(6053):255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. 2013;19(3):481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 40.Geurts L, et al. Altered gut microbiota and endocannabinoid system tone in obese and diabetic leptin-resistant mice: impact on apelin regulation in adipose tissue. Front Microbiol. 2011;2:149. doi: 10.3389/fmicb.2011.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.