Significance
Peptides and their specific receptors form molecular peptidergic systems (PSs) that are essential components of neuronal communication in the animal brain. Many PSs have been characterized in insects and mammals but their precise evolutionary relationship is not fully understood. We interrogated genomic sequence databases and used phylogenetic reconstruction tools to show that a large fraction of human PSs were already present in the last common ancestor of flies, mollusks, urchins, and mammals. Our analysis provides a comprehensive view of animal PSs that will pave the way for comparative studies leading to a better understanding of animal physiology and behavior.
Keywords: neuropeptide evolution, GPCR evolution, bilaterian CNS cell types, bilaterian brain evolution
Abstract
Peptide hormones and their receptors are widespread in metazoans, but the knowledge we have of their evolutionary relationships remains unclear. Recently, accumulating genome sequences from many different species have offered the opportunity to reassess the relationships between protostomian and deuterostomian peptidergic systems (PSs). Here we used sequences of all human rhodopsin and secretin-type G protein-coupled receptors as bait to retrieve potential homologs in the genomes of 15 bilaterian species, including nonchordate deuterostomian and lophotrochozoan species. Our phylogenetic analysis of these receptors revealed 29 well-supported subtrees containing mixed sets of protostomian and deuterostomian sequences. This indicated that many vertebrate and arthropod PSs that were previously thought to be phyla specific are in fact of bilaterian origin. By screening sequence databases for potential peptides, we then reconstructed entire bilaterian peptide families and showed that protostomian and deuterostomian peptides that are ligands of orthologous receptors displayed some similarity at the level of their primary sequence, suggesting an ancient coevolution between peptide and receptor genes. In addition to shedding light on the function of human G protein-coupled receptor PSs, this work presents orthology markers to study ancestral neuron types that were probably present in the last common bilaterian ancestor.
In animals the regulation of complex homeostatic processes and their behavioral output relies on the modulation of neuronal activity in well-defined circuits of the brain. Groups of neurons can influence the activity of other groups of neurons by releasing in the extracellular milieu short peptide hormones, called neuropeptides, which, with few exceptions, notably insulin-like peptides, bind G protein-coupled receptors (GPCRs) that are expressed at the surface of target cells. GPCRs are seven-pass membrane receptors that can bind to a wide variety of ligands (1) and form the largest family of integral membrane proteins in the human genome (2). The majority of GPCRs that are activated by peptide ligands are thought to be evolutionarily related and belong to the rhodopsin β and γ classes of rhodopsin (r) GPCRs, or to the secretin (s) family of GPCRs (3). Peptides are short (<40 amino acids) secreted polypeptides derived from larger precursor proteins that are encoded by the genome and processed by specialized enzymes (4). They share defining features at the level of their primary sequence, including the presence of signal peptides at their N terminus, of canonical dibasic processing sites that are recognized by prohormone convertase cleaving enzymes (5), and of a C-terminal glycine that is the target of amidation enzymes (6).
Research on peptide hormones has a long history (7, 8), and during the last decades a substantial number of peptide–receptors systems, or peptidergic systems (PSs), have been characterized by reverse pharmacology methods in both insects (9) and mammals (10). In parallel, the first genome sequencing projects have enhanced our knowledge of PS diversity in protostomian species, notably in the fly Drosophila melanogaster (11), the nematode Caenorhabditis elegans (12), and the mosquito Anopheles gambiae (13). In these species, original genome-wide searches have revealed the existence of a large number of GPCRs that resembled vertebrate GPCRs (11), but comparatively few vertebrate-type peptides (11, 12, 14).
Before the genomic era, some researchers had postulated a deep orthology between PSs from distant animals on the basis of peptide primary sequence similarity (15), functional analogies (16), and immunoreactivity of invertebrate tissues to mammalian hormone antibodies (17), but the idea that it could be a general feature of PSs remained controversial. Now, with the accumulation of molecular sequence data and the characterization of a growing number of PSs from insects and mammals, the concept of a bona fide orthology between protostome and deuterostome PSs has garnered new support (18, 19). Recently, Schoofs and coworkers have added new weight to this theory by showing that some arthropod-type PSs [adipokinetic hormone (AKH), pyrokinin (PK), and sulfakinin (SK)] occurring in C. elegans were orthologous to vertebrate PSs [gonadoliberin (GnRH), neuromedin U (NMU), and cholecystokinin (CCK)] (20–22).
In an effort to clarify the relationships between protostomian and deuterostomian PSs, we set out to reconstruct the full evolutionary history of bilaterian peptide and receptor genes. We used data from publicly available genomes from Ensembl (23), the Joint Genome Institute (JGI) (24), the Ghost database (25), and the Baylor College of Medicine (BCM), including data from key lophotrochozoan and ambulacrarian phyla that are thought to have retained ancestral features of the bilaterian brain (26–28). We performed phylogenetic reconstructions (29, 30) and used a hidden Markov model (HMM)-based program, which predicts precursor hormone sequences (31).
Our analysis suggests that 29 PSs were present in the last common ancestor of bilaterians (the urbilaterian) and that, in the general case, peptide and receptor genes coevolved in the different lineages leading to present-day bilaterians. We present a comprehensive list of PSs that are common to bilaterian species, so that these orthology markers can be used to reveal the origin and function of ancient peptidergic cell types and circuits. All sequences, phylogenetic trees, and annotations derived from these analyses can be found at http://neuroevo.org.
Results
Phylogenetic Analysis of Bilaterian Receptors Reveals Ancestral Receptors.
By following the strategy described in Fig. 1A (Methods) we were able to retrieve a set of 592 bilaterian β rGPCRs, 166 bilaterian γ rGPCRs, and 115 sGPCRs. Maximum likelihood phylogenetic analysis with SH-like likelihood ratio tests (LRTs) as branch support values (Methods) applied to β rGPCRs (respectively γ rGPCRs and sGPCRs) revealed, to our surprise, 22 (respectively 2 and 5) well-supported subtrees, denoted AncBILAT, that contained a diversified set of deuterostome and protostome GPCRs, suggesting that the receptors forming these subtrees evolved from distinct ancestral bilaterian receptors (Fig. 2).
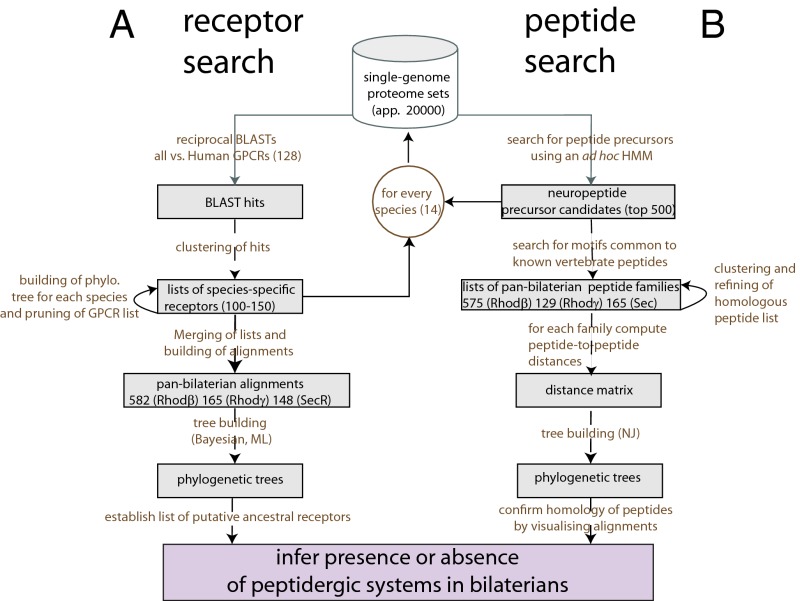
Fig. 1.
Phylogenomics pipeline for the study of PS evolution. A standard phylogenomics strategy was used to derive sets of potential peptide GPCRs (receptor search, A) for all of the species considered (Methods). The final three phylogenetic trees of bilaterian rhodopsin GPCRs (β and γ rGPCRs and sGPCRs) were used to derive potential ancestral PSs. Then, to isolate potential peptide precursor sequences (peptide search, B) a noncanonical strategy was used, which involved the use of an HMM designed to find candidate peptide precursors in each of the bilaterian species and the construction of peptide phylogenetic trees using the neighbor-joining method and a normalized kernel-based distance (Methods).
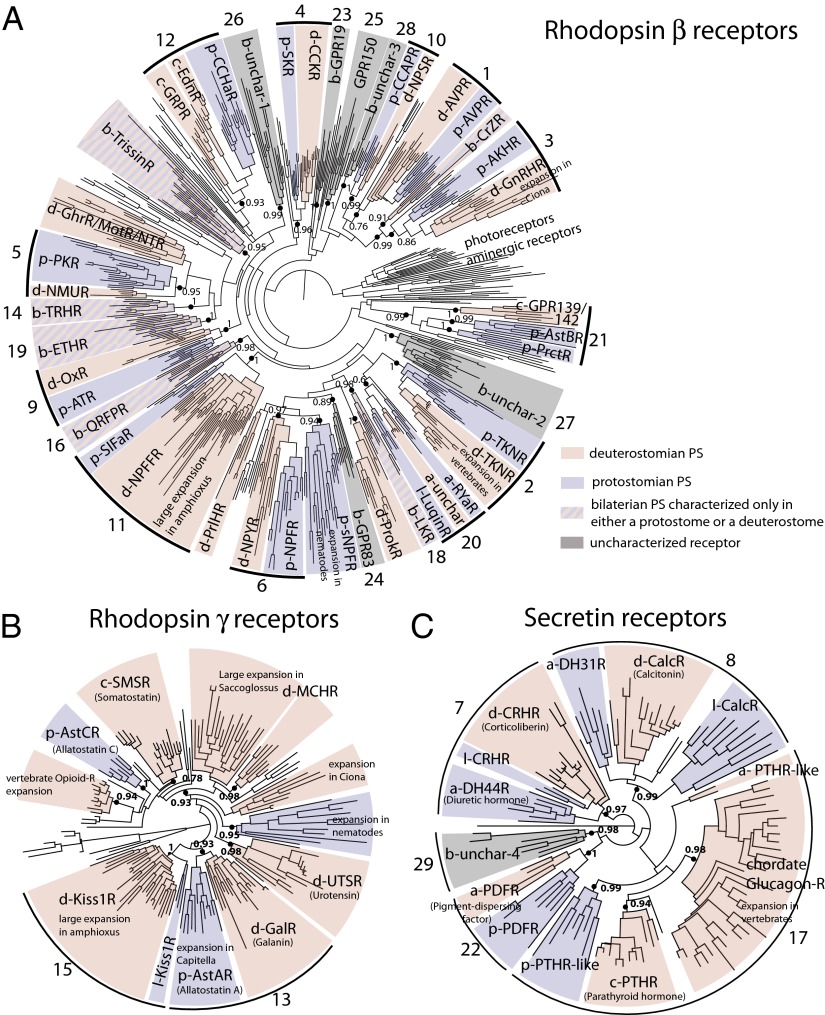
Fig. 2.
Phylogenetic analysis of bilaterian rhodopsin and secretin receptors. Maximum likelihood tree of bilaterian rhodopsin β (A), γ-type (B), and secretin (C) receptors, according to the GRAFS classification established in ref. 3. The tree is structured in well-supported subtrees containing both clusters of protostome (blue) and deuterostome (pink) groups of sequences. At the root of blue-pink subtrees (shown as black or green solid circles), a prototypic receptor of each subtype was already present in the urbilaterian. Black solid circles indicate well-supported bilaterian GPCR families, and green solid circles show hypothetical evolutionary relationships among bilaterian families. The bilaterian (b-), protostomian (p-), deuterostomian (d-), chordate (c-), lophotrochozoan (-l), or arthropod (a-) origin is indicated by an initial letter before each peptide GPCR acronym. Ancestral bilaterian clusters containing receptors characterized only in either protostomes or deuterostomes (e.g., b-TRHR and b-ETHR) were colored with alternating blue and pink bands, and bilaterian clusters containing no characterized receptors were shaded in gray. Photoreceptors and aminergic receptors were used as an outgroup for rhodopsin β receptors (A), and human adhesion GPCRs were used as an outgroup for the secretin receptors (C).
To test whether these associations of protostomian and deuterostomian peptidergic GPCRs were statistically robust we performed complementary computations including nonparametric bootstrapping and Bayesian phylogenetic analyses (Methods). We saw general agreement between the topology of maximum likelihood and Bayesian trees, and between SH-like LRT P values (PvalSH) and Bayesian posterior probabilities (PPBayes) supporting AncBILAT. However, bootstrap values (btspML) generally gave weaker support, as did the other two branch support values (BSVs), and in some cases they did not give firm support for AncBILAT.
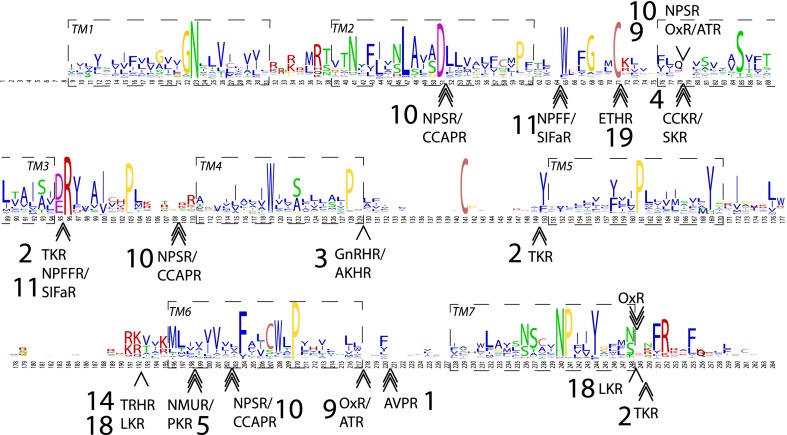
Conserved introns have been shown to be reliable markers of evolutionary homology (32) in eukaryotes. To consolidate our hypotheses, we asked whether receptors forming each AncBILAT shared introns at identical position and phase relative to our protein alignments. Our analysis of the intronic structure of human, lophotrochozoan, ambulacrarian, and arthropod genes forming members of AncBILAT suggest that gene members of several bilaterian subtrees including neuropeptide S (NPS)-R/crustacean cardioactive peptide (CCAP)-R, neuropeptide FF (NPFF)-R/ SIF-amide (SIFa)-R, ecdysis-triggering hormone (ETH)-R, CCK-R/SK-R, GnRH-R/AKH-R, tachykinin-R (TKR), orexin (Ox)-R/allatotropin (AT)-R, vasopressin (AVP)-R, and leucokinin (LK)-R share orthologous introns (Fig. 3) and are likely to have evolved from a common ancestral bilaterian receptor gene. In all abbreviations of protein names the suffix R stands for receptor. Note that because all of these receptors are members of a family, we chose to use the name of one of the members to designate the group of closely related receptors [e.g., “arginine-vasopressin receptor” (AVPR) was used to denominate both vasopressin and oxytocin receptors].
Fig. 3.
Conserved introns in β-rhodopsin receptor genes. Motif logo of rhodopsin β receptor alignment showing the introns that have a conserved position across bilaterians. Names of deuterostome, protostome, or bilaterian PSs were used, as defined in Fig. 2. The seven transmembrane domains (TM1–7) are indicated by dashed boxes. Single, double, and triple arrows indicate that the intron phase is 0, 1, or 2, respectively.
Ligands from Orthologous Receptors Are Orthologous Peptides.
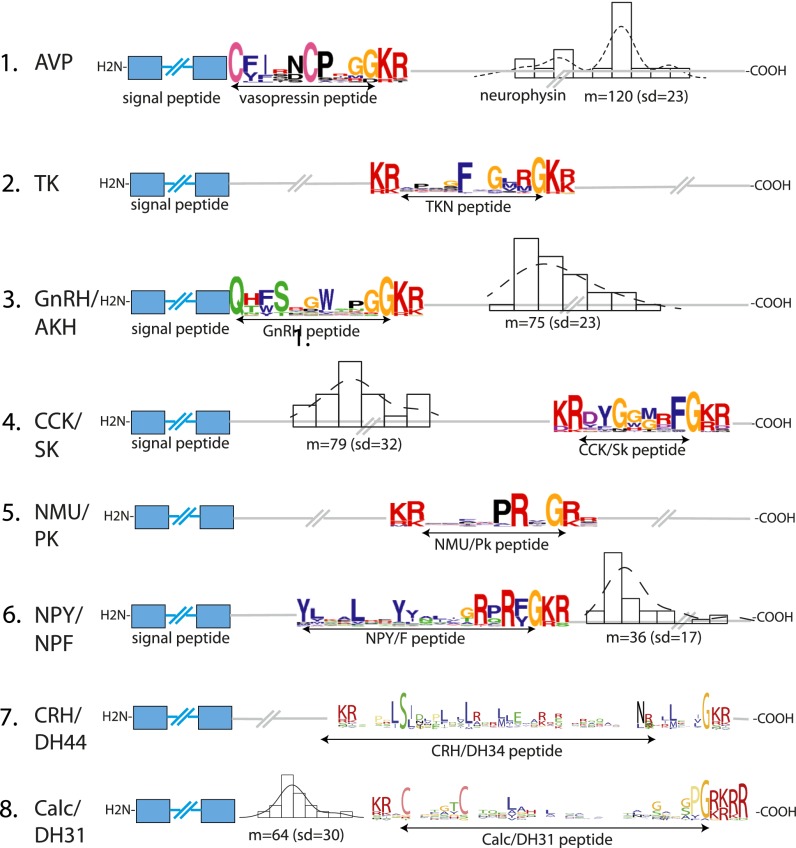
We next used our receptor orthology hypotheses as a guide to derive orthology hypotheses for peptides and reconstruct bilaterian, protostomian, deuterostomian, and chordate alignments of homologous peptides (Dataset S1). For that we first screened public databases for the presence of putative peptide precursor sequences using an HMM-based program (Methods and Fig. 1B). For each species we screened the top 500 candidate sequences for degenerate motifs specifically found in peptide families that were known ligands for receptors in AncBILAT. For instance, we looked for the motif QxG[KR]R just C-terminal to the signal peptide of candidate precursor sequences to find GnRH and adipokinetic hormone (AKH)-like sequences (Fig. 4). For eight families of bilaterian peptides, namely AVP, neuropeptide Y (NPY)/neuropeptide F (NPF), tachykinin (TK), GnRH/AKH, CCK/SK, neuromedin U (NMU)/PK, corticoliberin (CRH), and calcitonin (Calc), we could detect similarity at the level of the primary sequence between protostomian and deuterostomian peptides and reconstruct the entire bilaterian families (Fig. 4 and Dataset S1), including most members of these peptide groups in lophotrochozoan and ambulacrarian species for which comparatively little biochemical characterization of PSs has been made. For the other peptide families, obvious primary sequence similarity was restricted to protostomes [CCAP, CCH-amide peptide (CCHa), allatostatin A, luqin, ETH, allatostatin B, proctolin, and pigment-dispersing factor (PDF)], deuterostomes [NPS and thyrotropin-releasing hormone (TRH)], or chordates [NPFF, endothelin, gastrin-releasing peptide (GRP), galanin, kisspeptin (Kiss1), pyroglutamylated RF-amide peptide (QRFP), parathyroid hormone (PTH), and glucagon + pituitary adenylate cyclase-activating polypeptide (PACAP)] (see Fig. 6D and Dataset S1). In most cases peptides that were ligands to orthologous receptors showed similarity in their primary sequence, suggesting systematic coevolution between peptides and receptors.
Fig. 4.
Conservation of eight ancestral bilaterian peptide precursor families. Conserved features of these eight peptide precursor families include key residues shown in the peptide logo. For example, the N-terminal glutamine in GnRH/AKH sequences, pairs of cysteines in AVP and Calc/DH31 sequences, the position of peptide(s) and other domains inside the precursor sequence, and constrained length distribution of spacer sequences in the precursor (shown by histograms).
Fig. 6.
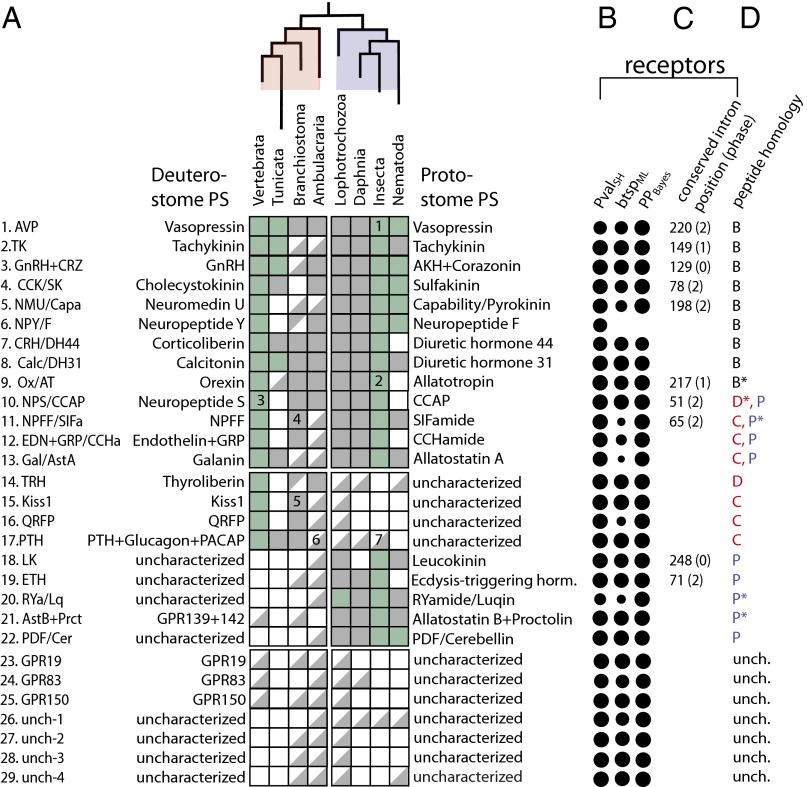
Ancestral bilaterian peptidergic systems. Inferred evolutionary relationships between the different ancestral bilaterian PSs. (A) Names of characterized deuterostomian systems (Left) with their orthologous protostomian systems (Right). A top-left half-square indicates the presence of the peptide in a given phylogenetic group, or single species, in the case of Branchiostoma (floridae) and Daphnia (pulex). Subphylum Vertebrata is composed of H. sapiens and Takifugu rubripes, phylum Tunicata of Ciona intestinalis and Ciona savignyi, superphylum Ambulacraria of S. purpuratus and S. kowalevskii, Lophotrochozoa of C. teleta and L. gigantea, class Insecta of D. melanogaster, Tribolium castaneum and Acyrthosiphon pisum and phylum Nematoda of C. elegans and Pristionchus pacificus. A bottom-right half-square indicates the presence of the receptor in a group. A receptor was considered to be present in a given group of animals when it was positioned inside a well-supported subtree (branch support value >0.95) including at least one characterized receptor. A full square denotes the presence of both peptides and receptors from a given PS. A green full square indicates that, for at least one member of that group, binding between a peptide and its receptor was biochemically demonstrated. For both peptides and receptors, the presence in a phylogenetic group implied most of the time the presence of at least two members of that group clustered together in the peptidergic system-specific subtree. The plus symbol refers to the last common ancestor of the given systems and a slash indicates the different names that were given to orthologous systems in distinct species. (B) Branch support values for each of the subclasses of peptide receptors are represented as solid circles of area scaled according to the three statistical test values: likelihood ratio test (PValSH) and bootstrap (btspML) values for maximum likelihood trees and posterior probability values (PPBayes) for the Bayesian reconstruction trees. (C) For each bilaterian PS, we wrote down the positions (relative to the global alignment of rhodopsin β receptors) and phases of introns that were conserved in, and specific to, that PS (Fig. 3). (D) For each peptide family we reported the largest phylogenetic group level (A, arthropods; B, bilaterians; C, chordates; D, deuterostomes; P, protostomes) for which peptide or precursor similarity could be detected in alignments (see also Dataset S1). An asterisk after the letter indicates the presence of a conserved domain outside of the peptide region that was used to establish our orthology hypotheses. Notable phyla-specific losses, expansions, and appearances of known PS: (1) AVP was lost in Drosophila. (2) AT was lost in Drosophila. (3) NPS was lost in teleosts. (4) A large expansion of both NPFF peptide and receptor genes is observed in amphioxus. (5) Large expansion of both Kiss1 peptide and receptor genes in amphioxus. (6) PTH-like peptides and glucagon-like peptides are found in Ciona and Branchiostoma.( 7) Receptors from the PTH + glucagon + PACAP family are absent from the genome of Drosophila.
To verify that the similarity we observed in single-family alignments was likely to reflect real homology we constructed a neighbor-joining tree using all characterized and predicted bilaterian peptides from each of the three groups (β and γ rhodopsin and secretin-like peptides) using a nonstandard distance adapted for measuring short-peptide similarity (Methods). This unbiased procedure was able to group together entire families of peptides (Fig. S1) that we previously had recognized as homologous (Fig. 4), either through our literature search or by visual inspection of alignments, indicating that this method can accurately recover distant peptide homology and suggesting a true common evolutionary origin for these bilaterian peptide genes.
Conserved Domains in Peptide Precursor Genes Inform Us of Their Origin.
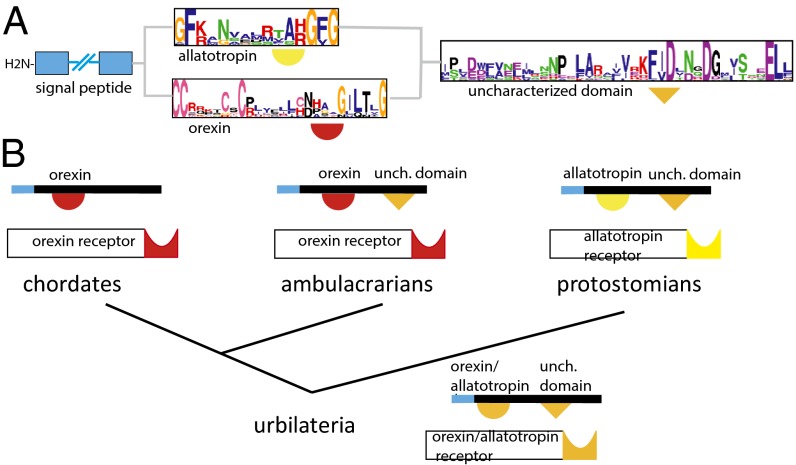
In a few interesting cases where it was not possible to deduce homology on the basis of the peptide sequence, we noted the existence of conserved domains outside of the peptide region. Such a conserved cryptic domain was found in all protostomian allatotropin (p-AT) and a Saccoglossus Ox-like precursor sequences, suggesting that Ox and AT precursor genes are orthologous, just as their receptors are. This observation supports an evolutionary model whereby Ox and AT precursors are orthologous and this cryptic domain was lost in chordates and retained in other extant phyla (Fig. 5).
Fig. 5.
Evolutionary scenario for the Ox and AT precursors. Structure of the hypothetical bilaterian ancestral Ox/AT precursor. (A) The hypothetical ancestral Ox/AT bilaterian precursor is composed of an N-terminal signal peptide (blue box), an Ox or AT peptide, represented by the two logo motifs just C-terminal to the signal peptide, and a C-terminal domain of unknown function that is found in most protostomes and in a deuterotostome, the acorn worm. However, we cannot conclude whether this precursor was more closely related to the extant deuterostome Ox neuropeptides bearing prototypic cysteine patterns or to the extant protostomian AT neuropeptides. (B) Probable scenario describing Ox/AT precursor evolution. Even though Oxs (red half-circle) and ATs (yellow half-circle) display no obvious similarity, their receptors are orthologous to each other and the last common ancestor of bilaterians possessed a C-terminal domain (orange triangle) that was retained in present-day ambulacrarians and protostomes and was lost in the lineage leading to chordates.
In another case, we saw sequence similarity within deuterostomian neuropeptide S (d-NPS) peptide candidates and within protostomian CCAP (p-CCAP) (Dataset S1), but only little (FxN motif) between NPS and CCAP peptides, although their receptors were clearly found to be orthologous to one another. However, as already noted in ref. 33, neurophysin, a vasopressin-associated peptide, is present at the C terminus of amphioxus, acorn worm, and urchin NPS-like precursor sequences, suggesting that the d-NPS gene family is evolutionarily related to the AVP gene family and that the ancestral NPS/CCAP precursor contained a neurophysin carrier domain (Fig. S2A). This is in line with both our receptor analysis, which shows an association between bilaterian AVPR and NPSR (PvalSH = 0.88), and the neighboring tandem position of AVP and NPS in the amphioxus genome (Fig. S2B).
In several other instances including for luqin/Arginine-Tyrosine-amides (RYa), SIFa, AKH, and proctolin, alignments of precursor sequences revealed domains of unknown function C-terminal to the peptide domain that were conserved across protostomes (Dataset S1). In vertebrates, a domain common to gastrointestinal peptides ghrelin and motilin, corresponding to obestatin and motilin-associated peptides, mirrors the tight evolutionary relationship between these two vertebrate receptors (Fig. 2A and Fig. S1).
Establishing the List of Putative Ancestral PSs.
Owing to their greater statistical robustness, we chose to take the conclusions from our receptor phylogenetic analysis to derive the set of probable ancestral bilaterian PSs (PSbilat). Our initial criteria for including a PS in our final list (Fig. 6) was that that both maximum likelihood and Bayesian analyses supported this ancestrality and that at least one of the BSVs defining the receptor subtree—PvalSH, PPBayes, or btspML—was over 0.95. In 12 cases [TK, GnRH/AKH, NPS/CCAP, calcitonin/diuretic hormone 31 (DH31), TRH, Kiss1, PTH + glucagon + PACAP, leucokinin, ETH, human orphan GPCR 19 (GPR19), and unch-3 and -4] the receptor subtrees received almost maximal statistical support by all three BSVs. In other cases where statistical support of subtrees was weaker, a specific conserved intron (AVP, CCK/SK, NMU/PK/Capability Ox/AT) (Fig. 3) and/or clear similarity of bilaterian peptides (AVP, CCK/SK, NPY/NPF, and CRH) (Fig. 4 and Fig. S1) convinced us that they had evolved from ancestral bilaterian PSs. We also found six well-supported ancestral bilaterian subtrees that lacked ecdysozoan or vertebrate members and contained no characterized receptors (Fig. 6). For each PSbilat we reported on the left (right) side of the table its deuterostome (protostome) denomination (Fig. 6A), whenever it was known. When it had not been characterized in any of the species from a given group, we indicated it as such.
For each of the PSs we reported the presence and absence of receptor and peptide genes in the different phyla and noted in panels (Fig. 6 B–D) the different lines of evidence that were used to establish the ancestrality of the PS. For PSs for which both deuterostomian and protostomian characterization was available (first 13 PSs from Fig. 6) the presence/absence of receptors in a given taxonomic group correlated well with that of peptides (ρ = 92/104 = 0.88). For every characterized insect PS we could successfully find all other expected orthologous protostomian peptides (PSs 1–13 and 18–22, all squares are full in the protostomian half, Fig. 6).
Description of Ancestral Bilaterian PSs.
Eight conserved ancestral bilaterian PS (families 1–8).
We found that eight PSs, vasopressin, NPY/NPF, tachykinin, GnRH/AKH, cholecystokinin/sulfakinin, neuromedin U/pyrokinin, CRH/diuretic hormone 44 (DH44), and Calc/DH31, for which we have the strongest conservation of peptides across bilaterians (Dataset S1) and a clear coevolution of peptides and receptors (Fig. S3), are present in all of the major phylogenetic groups we have looked at: chordates, ambulacrarians, lophotrochozoans, and ecdysozoans (systems 1–8, Fig. 6A). This likely reflects their importance in the biology of all bilaterian animals, as demonstrated by the number of studies devoted to their function in insects and mammals.
Five associations between protostomian and deuterostomian PSs (families 9–13).
We inferred five unique associations between deuterostomian and protostomian PSs that had been discovered and studied independently in mammals and insects. Two of them, d-Ox/p-AT and d-NPS/p-CCAP, are supported by high PvalSH (1.0, 0.99), PPBayes (1.0, 1.0), and btspML (0.87, 0.96) values in receptor trees (Fig. 6B) and conserved and specific position and phase of at least one intron (Fig. 6C). In both cases we have no clear similarity at the level of the primary peptide sequence; however, we noted the presence of a conserved domain of unknown function in p-ATs and Ox-like–containing precursor sequence of the acorn worm (Fig. 6 and Fig. S1) that provided the link between the two gene families. The third association we put forward is between deuterostomian NPFF (d-NPFF) and protostomian SIF-amide (p-SIFa) systems. In this case we have solid support from PvalSH (0.99) and PPBayes (1.0), but poor btspML (0.29). However, we found that both d-NPFFR and p-SIFaR genes share a phase-2 intron at position 65 in the protein alignment that is only present in these genes, strongly suggesting a common evolutionary origin for these two systems. On the peptide side, chordate NPFF and p-SIFa only share a common phenylalanine at their C terminus, and that similarity was not sufficient to group them together in our phylogenetic study. The fourth association is between vertebrate gastrin-releasing peptide (GRP) and endothelin and protostomian CCHa systems. This association is supported by high PvalSH (0.93) and PPBayes (1.0) values but low btspML (0.5) in the receptor phylogenetic analysis. GRP, endothelin and CCHa peptides are all encoded at the N-terminal part of their precursor but exhibit no obvious similarity among them. However, we could reconstruct the full protostomian CCHa peptide precursor family, and we could find a GRP-like peptide in Branchiostoma (Dataset S1). Finally, the association between d-galanin (Gal)R and p-allatostatin A (AstA)R is supported by high PvalSH (0.93) and PPBayes (0.93), but not by bootstrap values. A Gal-like peptide was found in the genome of Ciona, where it is encoded, like vertebrate Gals, right after the signal peptide (Dataset S1), but no good Gal candidate was found in ambulacrarian genomes. We could see a clear similarity between protostomian AstA peptides (Dataset S1) but could not bring out a clear motif in alignments of p-AstA and d-Gal.
Nine partially characterized bilaterian systems (families 14–22).
On the basis of our receptor analysis (Figs. 2A and 3) we posit the existence of nine PSbilat that have only been characterized in either a deuterostome (TRH, Kiss1, QRFP, and PTH + glucagon + PACAP) or a protostome [ETH, LK, allatostatin B (AstB) + proctolin, RYa/luqin, and PDF]. The receptor subtree is well supported by all three BSVs (Fig. 6B) and TRHRs are present in ambulacrarians, lophotrochozoans, and nematodes. We were able to find TRH-like peptides (Dataset S1) in the amphioxus and urchin gene sets, suggesting that they were present in ancestral deuterostomes. However, no TRH-like ligand could be found in nematodes and lophotrochozoans, where TRHRs are present.
Kiss1 receptors are present in vertebrates, Branchiostoma, ambulacrarians, and lophotrochozoans and were lost in lineages leading to tunicates and ecdysozoans. We found four Kiss1 peptide genes in Branchiostoma (Dataset S1), mirroring the large expansion of Branchiostoma Kiss1 receptors observed in the rhodopsin γ tree (Fig. 2B), indicative of a codiversification of Kiss1 peptide and receptor genes in the Branchiostoma genome. However, we could not find any Kiss1-like genes in ambulacrarian and lophotrochozoan genomes (Fig. 6A), where Kiss1 peptides are expected to occur, based on our analysis on receptors. The bilaterian ancestrality of QRFP receptors is supported by good LRT (PvalSH = 0.98) and Bayesian (btspML = 1) values but not by bootstrap values (btspML < 0.5). The QRFPR subtree contains deuterostomian and lophotrochozoan receptors; three QRFP peptide genes were found in the genome of Branchiostoma (Dataset S1), but no clear orthologs could be detected in ambulacrarians or lophotrochozoans. The fourth deuterostomian system with no known protostomian homolog is the PTH + glucagon + PACAP system. This notation designate the ancestral bilaterian system that in vertebrates diversified into several systems, including the parathyroid hormone, glucagon-like, and pituitary adenylate cyclase-activating peptide systems. The PTH + glucagon + PACAP bilaterian receptor subtree is highly supported by all three BSVs (1, 1, 1). No known peptide homolog is known outside vertebrates, yet we found glucagon-like and PTH-like peptide genes in Branchiostoma and Ciona (Dataset S1); however, we did not find obvious peptide candidates from that family in ambulacrarians or lophotrochozoans.
Likewise, the existence of ancestral bilaterian LK and ETH receptors is well supported by all three BSVs and by our intron conservation analysis (Fig. 6 B and C). ETHR and LKR occur in ambulacrarians and ETHR is found in ambulacrarians and Branchiostoma; however, both of these PSs have been lost in the lineage leading to vertebrates, which likely explains their lack of characterization in deuterostomes. The bilaterian RYa/Lq receptor subtree is strongly supported by Bayesian BSV (0.98) but only weakly by LRT (0.59) and bootstrap values (<0.5). When we aligned the arthropod RYa and lophotrochozoan Lq peptide precursors we saw that peptides were all encoded right after the signal peptide and that an uncharacterized domain containing two cysteines was present in all of the peptide precursors, further confirming the orthology between these two protostomian PSs. In the β-RhodR tree we noted one well-supported subtree (PvalSH = 1, PPBayes = 1, btspML = 0.83) containing two human orphan receptors (GPR139/142) and two groups of characterized protostomian receptors, proctolin-R and AstBR. We could reconstruct the entire family of peptide precursors of proctolin and AstB peptides but could not find orthologs of these peptides in human or in the amphioxus that could be potential novel ligands for GPR139 or GPR142. We found PDF receptors in protostomes and ambulacrarians (with all three BSVs >0.95) and PDF peptides in all protostomian genomes that we have screened; a PDF-like candidate peptide was even found in the acorn worm, further strengthening the case for the bilaterian ancestrality of this PS.
Uncharacterized receptors (families 23–29).
We have also included in our analysis human orphan receptors that are expected to be peptide receptors. We found that three human orphan receptors, GPR83, GPR19, and GPR150, showed up in distinct well-supported bilaterian subtrees (Fig. 1A) and that all three were probably lost in a linage leading to insects and nematodes (Fig. 6A). In addition, we found a group of four well-supported subtrees containing bilaterian receptors that did not belong to genomes from common animal models such as the fly, worm, and human and that lacked a characterization in other species (Fig. 2A, in gray). Two of them seem to be members of known families; unchar-3 is related to AVP + NPS + GnRH (Fig. 2A) and unchar-4 to PDF receptors (Fig. 2C).
Large-scale coevolution between peptides and receptors.
To test the hypothesis of coevolution between known peptides and their receptors, we made a plot in which x was the phylogenetic distance between any two characterized receptors and y the phylogenetic distance between their corresponding peptides (Fig. S3). We found a statistically significant correlation (P = 3.1e-11) between these two distances, indicating that there is coevolution of peptides with their receptors, in the general case. Furthermore, we noted that each time we saw large species or phylum-specific expansions in receptors, including for vertebrate opioid, tunicate GnRH, Lottia AT, nematode sNPF, and allatostatin C and Branchiostoma NPFF, QRFP, and Kiss1 receptors (Fig. 2), we also had multiple related peptide precursors (Dataset S1), suggesting that expansions of receptor and corresponding peptide genes often happen in conjunction with each other. Finally, peptide position within the precursor is often conserved across bilaterians for a given PS family; it is often just after the signal peptide (AVP, GnRH/AKH, or NPY/F), or near the C terminus of the precursor (CCK/SK and Calc/DH44). This observation excludes models whereby a ligand encoded from an unrelated peptide precursor gene would have outcompeted, for a given receptor, the existing ligand.
Losses of PSs in the different taxons.
A benefit of our comprehensive approach is that we could make deductions about losses of the ancestral bilaterian PSs in the different groups of species. When we lacked both receptors and peptides (empty squares in Fig. 6A) in a protostomian lineage where we normally would have expected to find them, we could reasonably claim that it was lost in this lineage. This was notably the case for NPS in teleosts, NMU, NPY, NPS, NPFF, (endothelin + GRP), TRH, Kiss1, and QRFP in Ciona, CCK in Branchiostoma, Kiss1 and QRFP in arthropods, LK in Daphnia, AVP and AT in Drosophila, and DH31, AT, CCAP, CCHa, and ETH in nematodes.
Taxon-specific systems.
We went one step further in the analysis and tried to define which systems were likely to be taxon-specific. Prolactin-releasing hormone (PrlH)R (Fig. 2A), urotensin, and melanin-concentrating hormone (MCH) (Fig. 2B) are likely to be deuterostomian-specific, and sNPF and allatostatins C (AstC) are likely to be protostomian (Fig. 2A). Furthermore, our receptor analysis indicates a chordate origin for somatostatin (SMS), endothelin, GRP, and ghrelin/motilin (Fig. 2A), and we could find SMS and GRP-like peptides in the amphioxus (Dataset S1). Our peptide analysis revealed significant similarity between SMS and AstC (Dataset S1), suggesting that these two systems are orthologous. Of the remaining well-studied systems, according to our receptor analysis, the opioid and neuropeptide B/W systems, as well as neurotensin, seem to have emerged and diversified in vertebrates (Fig. 2A). However, we found opioid-like precursor genes and neurotensin-like genes in Ciona (Dataset S1), suggesting that these systems could have emerged in an ancient chordate lineage.
Discussion
We have annotated the rhodopsin-type GPCRs and their associated peptides in several bilaterians. In the process we discovered that a greater number of vertebrate PSs than expected was conserved in bilaterian species including Capitella, an annelid, and Saccoglossus, a hemichordate. With our phylogenetic analyses we provide a complete annotation of both vertebrate-type and arthropod-type GPCRs and peptides in these animals. Out of the 13 ancestral bilaterian PSs that have been characterized to date in both deuterostomes and protostomes, 8 exhibited a clear resemblance in their peptide sequences (families 1–8). Among these eight peptide families, AVP, TK, CCK/ SK, NPY/NPF, CRH, and Calc have been hypothesized before to be of bilaterian origin (11, 18, 34, 35), and data supporting a deep orthology between deuterostomian GnRH (NMU) and protostomian AKH (PK) PSs have been recently presented (20, 21). For the other five protostomian–deuterostomian associations, Ox/ AT, NPS/CCAP, NPFF/SIFa, (endothelin + GRP)/CCHa and Gal/ AstA, we found no obvious similarity between the peptides, and orthology hypotheses had previously been restricted to receptors (11, 36, 37). However, we found one domain that was common to protostomian AT and Saccoglossus Ox-like precursors; this observation provided the missing evolutionary link between the two gene families and illustrates the importance of including underrepresented phyla from ambulacraria and lophotrochozoa for studying the origin of bilaterian genes.
We also found nine ancestral bilaterian PSs (Fig. 6, families 14–22) that had only been characterized in either deuterostomes or protostomes, which we propose to be of bilaterian origin. Among these nine bilaterian PSs, four vertebrate-type receptors, TRHR, Kiss1R, QRFPR, and (PTH + glucagon + PACAP)-R, have orthologous counterparts in lophotrochozoans, but only one is present in insects (PTH + glucagon + PACAP) and only one in nematodes (TRHR), and none is present in the genome of the best-studied animal model, Drosophila, which may explain why these have been overlooked as ancestral systems. Likewise, the five arthropod-type receptors, LK-R, ETH-R, luqin-R, (AstB + proctolin)-R, and PDF-R, all have orthologous sequences in ambulacrarian or amphioxus genomes, but only one of these has orthologs in vertebrate genomes, and these are still orphan receptors (GPR139 and GPR142). This again highlights that lophotrochozoans and ambulacrarians were necessary for this study; without them it would not have been possible to conclude that any of the Kiss1, QRFP, LK, luqin, or PDF systems were already present in the urbilaterian.
Our analysis led us to define four ancestral bilaterian groups (Fig. 6, families 21 and 23–25) that contain uncharacterized human receptors. The analysis of the trees gives us clues about where to look for their unknown ligands. In one instance (GPR139 and GPR142, family 21), orthologs have already been characterized (AstBR and proctolin-R) in protostomes. Given their position in the global rhodopsin tree we speculate that GPR39 is a neurotensin-like receptor, that the ligand of GPR150 is evolutionarily related to vasopressin, and that GPR83 is likely to be an RF-amide receptor, because it is present in a cluster with several others (PrlhR, NPYR, and luqin).
In one case, our peptide and receptor analysis gave irreconcilable results. Prokineticin (Prok) is a vertebrate cysteine-rich ligand that is known to occur in protostomes as astakine; in contrast, we found strong statistical support indicating that Prok receptors are restricted to deuterostomes (Fig. 2A). In most other cases, discrepancies between peptide and receptor data could be explained by the difficulty in finding peptide genes when no member from a related species is known. We predict that several peptides remain to be discovered in ambulacrarians and lophotrochozoans, including tachykinin, NPFF, galanin, Kiss1, QRFP, (PTH + glucagon + PACAP), LK, ETH, and RYa in ambulacrarians, and TRH, Kiss1, QRFP, and (PTH + glucagon + PACAP) in lophotrochozoans (Fig. 6). This study provides a rational framework for their search. Future expressed sequence tag (EST) sequencing projects using neural tissue from these animals should help to fill these knowledge gaps.
Comparison with Previous Studies.
Similar efforts to systematically annotate GPCRs and/or their neuropeptide ligands in nonvertebrate animals and relate them to known vertebrate or arthropod-type PS receptors have been confined to chordates (38, 39) or insects (9, 40, 41). In one important study (42), researchers used HMMs to annotate eukaryotic GPCRs and place them into one of the GRAFS (Glutamate, Rhodopsin, Adhesion, Frizzled, Secretin) classes. However, this classification strategy did not provide the level of detail necessary to specify the PS subclass to which neuropeptide receptors belong.
Another seminal study describing Drosophila peptides GPCRs and their ligand genes (11) was among the first to use whole genome data to demonstrate a large-scale orthology between receptor systems of insects and mammals. In their analysis, Hewes and Taghert propose phylogenetic relationships between human, C. elegans, and Drosophila peptide receptors, most of which were unannotated at that time. They used the neighbor-joining method to produces evolutionary trees of topology comparable with ours and identified 15 associations between vertebrate and ecdysozoan groups of receptors, corresponding to our PSs 1–9 and 11–13. However, the fact that some receptors are absent from the genome of Drosophila, including those of Ox, AVP, TRH, and Kiss1, hindered the correct interpretation of these proto/deutero–stomian PS relationships. Also the limited set of genomes that were investigated did not allow for a thorough picture of bilaterian PS diversity to emerge. Our work now complements this study and identifies bilaterian PSs 10 (NPS/CCAP), 14–17 (TRH, Kiss1, QRFP, PTH + glucagon) that are absent in Drosophila and bilaterian PSs 18–22 (LK, ETH, NepYR/luqin, AstB + proctolin, and PDF) that needed the inclusion of human orphan receptors and/or deuterostomian nonvertebrate sequences to be revealed.
In a third study (33) the author suggests a link between NPS peptide genes in human and in ambulacrarian, by making the observation that some peptide precursor genes in the urchin, acorn worm, and amphioxus genomes code for a C-terminal neurophysin domain and that the amphioxus peptide displays a high similarity to mammalian NPS. However, our interpretation differs in two aspects. First, the author suggested, based on the evidence that deuterostomian NPS-like peptides show no similarity with vasopressin, that neurophysin became associated with NPS peptides in a deuterostomian ancestor, whereas we favor an explanation whereby neurophysin was already associated with an ancestral NPS + AVP peptide before duplication of the two systems. This hypothesis is founded on our receptor analysis showing a close evolutionary relationship between the bilaterian NPS and AVP systems, and the tandem position of AVP and NPS in the amphioxus genome (Dataset S1). Second, the assumption that deuterostomian NPS-like peptides may be orthologous to SIFa based on a shared motif (NG) contrasts with our hypothesis of an NPS/CCAP and NPFF/SIFa orthology, which rests on our receptor analysis. These differences in interpretation can be explained by the emphasis we put on our receptor analysis to guide our peptide orthology hypotheses.
Annotations of Peptides.
PSs 14–17 deserve special attention, because their peptides are not known outside vertebrates, and although we did not find convincing peptide candidates in protostomes, we found TRH, Kiss1, QRFP, PTH, and glucagon-like peptides in the amphioxus genome (Dataset S1). It will be particularly interesting in the future to study the function of these genes in the amphioxus and lophotrochozoan species to learn more about the conserved features of these vertebrate PSs. Recently, several PSs have been characterized in the nematode, including vasopressin (43, 44). We give predictions for C. elegans peptides that had not been characterized to date, which include TK, DH31/Calc, SIFa, LK, luqin, and AstA, B, and C (Dataset S1).
Most ecdysozoan and vertebrate PSs have been already characterized, but we have only limited knowledge of ambulacrarian and lophotrochozoan PSs. Our study provides reliable annotations for neuropeptides and their GPCR receptors in an echinoderm, Strongylocentrotus purpuratus, a hemichordate, Saccoglossus kowalevskii, a mollusk, Lottia gigantea, and an annelid, Capitella teleta. The TRH system has been well-characterized in mammals and has not been characterized outside vertebrates, although a TRH-like peptide has recently been found in the urchin genome (45). The two lophotrochozoan genomes of L. gigantea and C. teleta have recently been probed in silico for the presence of arthropod-type peptides, yielding a surprisingly large number of candidates (46, 47). Here we confirm these findings and extend the searches to include crustacean and nematode peptides; and in all cases when a protostome peptide was found as part of a bilaterian-conserved system (Fig. 6, families 1–13 and 18–22), full protostomian alignments of peptides or their precursors could be built, including for CCAP, AT, SIFa, CCHa, LK, ETH, AstA, AstB, and luqin (Dataset S1). It proved more difficult to do so for deuterostomian peptides, owing to the larger evolutionary gap between chordate and ambulacrarian species. In most cases it was nonetheless possible to extend our peptide orthology characterization to chordates (Fig. 6), including for six peptide families that had not been described outside vertebrates (NPFF, QRFP, Ox, GRP/bombesin, Kiss1, glucagon, and PTH). For the best characterized bilaterian families (Fig. 6, families 1–13), most ambulacrarian peptides were found, including AVP, GnRH, CCK, NPY, CRH, Calc, Ox, and NPS peptides.
Coevolution.
GPCRs binding to the same monoamine neurotransmitters can be found in different regions of the rhodopsin phylogenetic tree (48) and peptides with recognizable motifs, such as RF-amides (e.g., NPY and Kiss1), bind receptors that are phylogenetically distant, suggesting that novel ligands may outcompete existing ones for a given receptor. A recent study (49) showed that a dendrogram constructed on human rhodopsin α receptors using a similarity measure on their ligands, showed significant differences with the traditional phylogenetic tree built on receptor sequences, going against the notion of a coevolution between receptors and their ligands. However, recently GnRH/AKH, CCK/SK, and NMU/PK peptides and receptors have been recognized as having coevolved in lineages leading to human, nematodes, and arthropods (50). Our analysis brings weight to this latter theory because we confirm the widespread presence of these PSs in all five major phylogenetic groups that we have scrutinized, and extend the hypothesis of coevolution of these systems to bilaterians (Fig. S3).
Ancestral Bilaterian Neuronal Types and Neuronal Circuits.
Behavioral processes rely in part on controlled brain expression of peptides and receptors in distinct groups of neurons. The existence of orthologous PSs poses the question of how conserved this wiring is in evolution and how conserved the functions of the different PSs are. Previous studies have shown that molecular markers defining ancestral RF-amide and vasopressin-like expressing neurons were conserved between fish and annelids, suggesting that these peptidergic cell types had been established before the deuterostome–protostome split (26). Our work provides markers to test whether sets of orthologous peptides define ancestral cell types with similarly conserved molecular coordinates (e.g., transcription factors and miRNA molecules). For instance, no TRH-like system has been studied in invertebrates, and we can speculate that, because the main function of TRH is to act on pituitary cells to release thyroid-stimulating hormone, TRH-like receptors may well be interesting markers to study ancient hormone-producing cells. We anticipate the existence of cells coexpressing TRHR and orthologs of mammalian glycoprotein hormones, which are known to occur in invertebrates.
Such orthology markers could also be used to compare neuronal microcircuits in distant animals. One salient feature of neuropeptide modulation, common to both vertebrates and invertebrates, is their role in gating and controlling the gain of sensory inputs (51, 52). Stress can trigger analgesia in mammals, a state whereby opioids signal to suppress the response of nociceptive neurons to aversive stimuli, and starvation induces an internal hunger state in flies, where increased dopamine signaling affects the sensitivity of taste neurons to sugar (52). One fascinating question will be to ask whether orthologous PSs perform gating on the same types of sensory neurons, such as mechanosensory, photo-, gustatory, or olfactory receptors. We found several examples of orthologous neuromodulatory systems in mammals and ecdysozoans that could be involved in orthologous circuits. Neuromodulation by noradrenaline in the mammalian brain and of tyramine and octopamine in the insect brain is responsible for the specification of an arousal state which sets off “flight or fight” behavioral responses (52), whereas signaling of dopamine and NPY/NPF in mammals and ecdysozoans participates in defining robust hunger states that qualitatively affect the response to food stimuli (53–55). Other examples of functional analogy between orthologous protostome and deuterostome PSs include the cholecystokinin/sulfakinin that are involved in satiety (22) in humans and worms, and GnRH and AKH that have analogous functions in reproduction in both humans and worms (20). Two recent studies (43, 44) showed that a bona fide vasopressin system was present in C. elegans and that it might participate in ancient circuits that control adaptive reproductive behaviors.
Peptidergic neuromodulation may also influence the activity of other neuromodulatory centers. This hierarchical nature of neuronal circuits seems to be a common feature of both vertebrate and invertebrate neuronal circuits. Cholecystokinin and vasopressin are known to activate orexinergic neurons (56), forming a neural circuit that is essential to maintain sleep and energy homeostasis. It will be interesting to see whether orexin neurons in protostome models interact in the same way with protostome homologs of these neuropeptides. Also, Kiss1 peptide is a hypothalamic peptide that is thought to regulate the activity of GnRH neurons (57). With our annotations we can interrogate whether GnRH neurons coexpress KissR to see whether this Kiss1–GnRH interaction is a conserved feature of peptidergic neuronal circuits of the bilaterian brain. Recognition of these PS homologies sets the stage for future studies on the conservation of peptidergic neuronal circuits’ architecture coordinating complex behavior in the bilaterian brain.
Taken together our results lend further support to the theory that the urbilaterian was an animal with a sophisticated physiology and nervous system, capable of integrating complex sensory information. We believe that some of these newly established homologies will provide the scientific community with markers to study ancestral cell types, yield insights into the fundamental functions of vertebrate peptidergic systems, and offer training data for computational biologists interested in the interaction between peptides and their receptors.
Methods
Genomes Investigated.
Rhodopsin-like receptors and their peptides were searched for in the genomes (Fig. S4) of the red flour beetle Tribolium castaneum (58), the fruitfly D. melanogaster (23, 59), one crustacean, Daphnia pulex (60), one nematode, C. elegans (61), two lophotrochozoans, the pond snail L.gigantea and C. teleta (62), two ambulacrarians, the sea urchin S. purpuratus (63) and the acorn worm S. kowalevskii, four chordates, the tunicate Ciona intestinalis (64), the amphioxus Branchiostoma floridae (65), the fish Takifugu rubripes (66), and Homo sapiens (67, 68). To better support the analysis, we partially included some genomic sequences from another nematode, Pristionchus pacificus (69), a tunicate, Ciona savignyi (70), and an insect, Acyrthosiphon pisum (71). We were also interested in annotating the neuropeptide GPCRs from the Lamprey Petromyzon marinus, whose genome was recently sequenced (72). The results of our receptor analysis that included lamprey and zebrafish gene models can be downloaded from http://neuroevo.org/phylogenetic_trees/receptors/.
Phylogenetic Analysis of Bilaterian pGPCRs.
GPCRs are seven transmembrane proteins that form a recognizable set of proteins that can be readily aligned (3). The first step of our study consisted of drawing a complete list of potential peptide GPCRs (pGPCRs) from each of the species considered. We first collected known human pGPCRs protein sequences from the UniProt database (73), including orphan GPCRs predicted to have peptide ligands (117 proteins). For each of the species sampled, we BLASTed these pGPCR sequences against full proteome sets from the Ensembl, JGI, Ghost (C. intestinalis), and BCM databases (Fig. 1). Reciprocal Basic Local Alignment SearchTool (BLAST) scores of human pGPCRs versus each proteome were then used to cluster pGPCR sequences through a single-linkage clustering algorithm. This procedure was applied for all species considered (Fig. 1) and resulted each time in three lists of about 50–60, 5–20, and 10–15 sequences respectively corresponding to separate groupings of β and γ rhodopsin-like receptors (rGPCRs) and secretin GPCRs (sGPCRs), consistent with the standard classification of human GPCRs (3).
We next built phylogenetic trees (74) for each of the three lists and identified sequences that formed clusters with vertebrate pGPCRs using Dendroscope (75). Selected sequences were then recursively added to the list of human pGPCRs (Fig. 1) to form three large lists of 582 β rGPCRs, 165 γ rGPCRs, and 148 sGPCRs. Pan-bilaterian alignments were then created using Muscle (76) and curated with a custom-made script, which filters out highly variable sites. Phylogenetic trees of β rGPCRs, γ rGPCRs, and sGPCRs were produced using both maximum likelihood (30) and Bayesian methods (29).
Phylogenetic Tree Inference.
For obtaining maximum likelihood trees with PhyML, the following parameters were used: LG as the substitution matrix (77), both Subtree Pruning and Regrafting and Nearest Neighbor Interchange for topological moves, and a number of discrete gamma rate categories equal to 4. Bayesian analysis were conducted with mixed amino acid (aamodelpr = mixed) and discrete gamma rates (rates = gamma) models. Two separate chains were launched starting from the maximum likelihood tree output from PhyML perturbed by 100 operations, and were stopped after 1 million generations. The 50% majority rule was used to produce final consensus trees. BSVs were generated using likelihood ratio tests (Shimodaira–Hasegawa-like procedure) implemented in PhyML (78), nonparametric bootstrapping (500 replicates) (79), and Bayesian posterior probability inference (29).
Strategy for Discovery of Ancestral Bilaterian Peptide Precursors.
Conservation between homologous peptide precursor sequences from different phyla (e.g., nematodes versus arthropods) is usually restricted to very few amino acids in the peptide region that are buried inside larger precursors. As a result, standard phylogenomics approaches are not applicable to study the evolution of peptide hormone genes. Instead we used a different approach. To obtain peptide precursor sequences candidates in bilaterian animals we screened lists of predicted genes and EST using a modified version of the PPH1 algorithm described in (31) where the scores used to rank precursor candidates are the expected density of cleavage inside a precursor (80). For each species we screened the top 500 candidate sequences for the presence of short conserved motifs often found at the C-terminal end of known vertebrate and arthropod-type peptides such as PRxG[KR]R for pyrokinins and neuromedins and R[FY]G[KR]R for RF-amides. Cleavage sites predicted by the HMM were checked with NeuroPred (81), a tool dedicated to prohormone cleavage site prediction. Alignments of homologous precursors were then built in a multistep trial-and-error process, gradually integrating or discarding these motif-containing candidate precursor sequences. Often the general structure of orthologous peptide precursor genes was found to be conserved in bilaterian sequences, such as the position of the peptide inside the precursor and the overall length of the precursor. In several interesting cases, including for bilaterian AVPs and ATs, protostomian luqin and SIF-amides, for chordate ghrelin/motilin and d-NPS, we noted regions of similarity outside the peptide region, suggesting unique conserved domains that reinforced our orthology hypotheses (Dataset S1). To measure the similarity between peptides we used a distance derived from the String alignment kernel from ref. 82.
Let 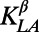 the kernel as defined in ref. 83 with parameters Blosum62 for the similarity matrix and gap penalties d = 12 (penalty for opening a gap) and e = 2 (penalty for extending a gap). We defined the distance between two peptides as
the kernel as defined in ref. 83 with parameters Blosum62 for the similarity matrix and gap penalties d = 12 (penalty for opening a gap) and e = 2 (penalty for extending a gap). We defined the distance between two peptides as
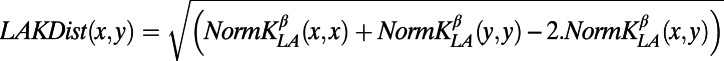 |
with the following normalization: 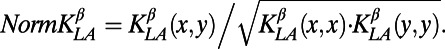
This normalized distance was used to build three alignment-free neighbor-joining trees of 575 rhodopsin β peptides, 129 rhodopsin γ peptides, and 165 secretin peptides.
Motif logos were drawn using the sequence logo generator tool (83), trees were visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/), and alignments with Jalview (84). Figures showing alignments were prepared with GeneDoc (www.psc.edu/biomed/genedoc).
Supplementary Material
Acknowledgments
We thank Dr. Christopher Lowe and Dr. Robert Freeman, who kindly provided us with the Saccoglossus kowalevskii gene models generated through an Augustus pipeline (85); and the BCM, the Department of Energy Joint Genome Institute, Ensembl, and the National Center for Biotechnology Information for providing free access to the genome and transcript databases. We thank Vincent Lefort, who generated initial GPCR tree bootstraps on the Laboratoire d'Informatique, de Robotique et de Microélectronique de Montpellier cluster; Samuel Blanquart for help in using Bayesian tree reconstruction programs; Vincent Laudet and Liliane Schoofs for insightful discussions; and Volker Hartenstein and Cornelius Gross for critical reading of the manuscript. This study was supported by a postdoctoral fellowship from the Fondation pour la Recherche Médicale and grants from the France Parkinson Association and the Agence Nationale de la Recherche.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1219956110/-/DCSupplemental.
References
- 1.Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: An evolutionary success. EMBO J. 1999;18(7):1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnan A, Almén MS, Fredriksson R, Schiöth HB. The origin of GPCRs: Identification of mammalian like Rhodopsin, Adhesion, Glutamate and Frizzled GPCRs in fungi. PLoS ONE. 2012;7(1):e29817. doi: 10.1371/journal.pone.0029817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63(6):1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 4.Douglass J, Civelli O, Herbert E. Polyprotein gene expression: Generation of diversity of neuroendocrine peptides. Annu Rev Biochem. 1984;53:665–715. doi: 10.1146/annurev.bi.53.070184.003313. [DOI] [PubMed] [Google Scholar]
- 5.Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2(1):31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 6.Eipper BA, Stoffers DA, Mains RE. The biosynthesis of neuropeptides: Peptide alpha-amidation. Annu Rev Neurosci. 1992;15:57–85. doi: 10.1146/annurev.ne.15.030192.000421. [DOI] [PubMed] [Google Scholar]
- 7.Tager HS, Steiner DF. Peptide hormones. Annu Rev Biochem. 1974;43(0):509–538. doi: 10.1146/annurev.bi.43.070174.002453. [DOI] [PubMed] [Google Scholar]
- 8.Hökfelt T, et al. Neuropeptides—an overview. Neuropharmacology. 2000;39(8):1337–1356. doi: 10.1016/s0028-3908(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 9.Hauser F, Cazzamali G, Williamson M, Blenau W, Grimmelikhuijzen CJ. A review of neurohormone GPCRs present in the fruitfly Drosophila melanogaster and the honey bee Apis mellifera. Prog Neurobiol. 2006;80(1):1–19. doi: 10.1016/j.pneurobio.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Civelli O, Saito Y, Wang Z, Nothacker HP, Reinscheid RK. Orphan GPCRs and their ligands. Pharmacol Ther. 2006;110(3):525–532. doi: 10.1016/j.pharmthera.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Hewes RS, Taghert PH. Neuropeptides and neuropeptide receptors in the Drosophila melanogaster genome. Genome Res. 2001;11(6):1126–1142. doi: 10.1101/gr.169901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nathoo AN, Moeller RA, Westlund BA, Hart AC. Identification of neuropeptide-like protein gene families in Caenorhabditiselegans and other species. Proc Natl Acad Sci USA. 2001;98(24):14000–14005. doi: 10.1073/pnas.241231298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riehle MA, Garczynski SF, Crim JW, Hill CA, Brown MR. 2002. Neuropeptides and peptide hormones in Anopheles gambiae. Science 298(5591):172–175.
- 14.Bargmann CI. 1998. Neurobiology of the Caenorhabditis elegans genome. Science 282(5396):2028–2033.
- 15.De Loof A, Schoofs L. Homologies between the amino acid sequences of some vertebrate peptide hormones and peptides isolated from invertebrate sources. Comp Biochem Physiol B. 1990;95(3):459–468. doi: 10.1016/0305-0491(90)90003-c. [DOI] [PubMed] [Google Scholar]
- 16.Tager HS, Markese J, Kramer KJ, Speirs RD, Childs CN. Glucagon-like and insulin-like hormones of the insect neurosecretory system. Biochem J. 1976;156(3):515–520. doi: 10.1042/bj1560515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritsch HA, Van Noorden S, Pearse AG. Gastro-intestinal and neurohormonal peptides in the alimentary tract and cerebral complex of Ciona intestinalis (Ascidiaceae) Cell Tissue Res. 1982;223(2):369–402. doi: 10.1007/BF01258496. [DOI] [PubMed] [Google Scholar]
- 18.Hoyle CH. Neuropeptide families: Evolutionary perspectives. Regul Pept. 1998;73(1):1–33. doi: 10.1016/s0167-0115(97)01073-2. [DOI] [PubMed] [Google Scholar]
- 19.Park Y, Kim YJ, Adams ME. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc Natl Acad Sci USA. 2002;99(17):11423–11428. doi: 10.1073/pnas.162276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindemans M, et al. Adipokinetic hormone signaling through the gonadotropin-releasing hormone receptor modulates egg-laying in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2009;106(5):1642–1647. doi: 10.1073/pnas.0809881106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindemans M, et al. A neuromedin-pyrokinin-like neuropeptide signaling system in Caenorhabditis elegans. Biochem Biophys Res Commun. 2009;379(3):760–764. doi: 10.1016/j.bbrc.2008.12.121. [DOI] [PubMed] [Google Scholar]
- 22.Janssen T, et al. Discovery of a cholecystokinin-gastrin-like signaling system in nematodes. Endocrinology. 2008;149(6):2826–2839. doi: 10.1210/en.2007-1772. [DOI] [PubMed] [Google Scholar]
- 23.Flicek P, et al. Ensembl 2012. Nucleic Acids Res. 2012;40(Database issue):D84–D90. doi: 10.1093/nar/gkr991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grigoriev IV, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40(Database issue):D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satou Y, Kawashima T, Shoguchi E, Nakayama A, Satoh N. An integrated database of the ascidian, Ciona intestinalis: Towards functional genomics. Zoolog Sci. 2005;22(8):837–843. doi: 10.2108/zsj.22.837. [DOI] [PubMed] [Google Scholar]
- 26.Tessmar-Raible K, et al. Conserved sensory-neurosecretory cell types in annelid and fish forebrain: Insights into hypothalamus evolution. Cell. 2007;129(7):1389–1400. doi: 10.1016/j.cell.2007.04.041. [DOI] [PubMed] [Google Scholar]
- 27.Pani AM, et al. Ancient deuterostome origins of vertebrate brain signalling centres. Nature. 2012;483(7389):289–294. doi: 10.1038/nature10838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomer R, Denes AS, Tessmar-Raible K, Arendt D. Profiling by image registration reveals common origin of annelid mushroom bodies and vertebrate pallium. Cell. 2010;142(5):800–809. doi: 10.1016/j.cell.2010.07.043. [DOI] [PubMed] [Google Scholar]
- 29.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 30.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 31.Mirabeau O, et al. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 2007;17(3):320–327. doi: 10.1101/gr.5755407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmel L, Rogozin IB, Wolf YI, Koonin EV. Patterns of intron gain and conservation in eukaryotic genes. BMC Evol Biol. 2007;7:192–207. doi: 10.1186/1471-2148-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elphick MR. NG peptides: A novel family of neurophysin-associated neuropeptides. Gene. 2010;458(1-2):20–26. doi: 10.1016/j.gene.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Lovejoy DA, Jahan S. Phylogeny of the corticotropin-releasing factor family of peptides in the metazoa. Gen Comp Endocrinol. 2006;146(1):1–8. doi: 10.1016/j.ygcen.2005.11.019. [DOI] [PubMed] [Google Scholar]
- 35.Furuya K, et al. Cockroach diuretic hormones: Characterization of a calcitonin-like peptide in insects. Proc Natl Acad Sci USA. 2000;97(12):6469–6474. doi: 10.1073/pnas.97.12.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitti T, Manoj N. Molecular evolution of the neuropeptide S receptor. PLoS ONE. 2012;7(3):e34046. doi: 10.1371/journal.pone.0034046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horodyski FM, et al. Isolation and functional characterization of an allatotropin receptor from Manduca sexta. Insect Biochem Mol Biol. 2011;41(10):804–814. doi: 10.1016/j.ibmb.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Kamesh N, Aradhyam GK, Manoj N. The repertoire of G protein-coupled receptors in the sea squirt Ciona intestinalis. BMC Evol Biol. 2008;8:129–147. doi: 10.1186/1471-2148-8-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nordström KJ, Fredriksson R, Schiöth HB. The amphioxus (Branchiostoma floridae) genome contains a highly diversified set of G protein-coupled receptors. BMC Evol Biol. 2008;8:9–17. doi: 10.1186/1471-2148-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hauser F, et al. A genome-wide inventory of neurohormone GPCRs in the red flour beetle Tribolium castaneum. Front Neuroendocrinol. 2008;29(1):142–165. doi: 10.1016/j.yfrne.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Fan Y, et al. The G protein-coupled receptors in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2010;40(8):581–591. doi: 10.1016/j.ibmb.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Fredriksson R, Schiöth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67(5):1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 43.Beets I, et al. 2012. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. 338(6106):543–545.
- 44.Garrison JL, et al. 2012. Oxytocin/vasopressin-related peptides have an ancient role in reproductive behavior. Science 338(6106):540–543.
- 45.Rowe ML, Elphick MR. The neuropeptide transcriptome of a model echinoderm, the sea urchin Strongylocentrotus purpuratus. Gen Comp Endocrinol. 2012;179(3):331–344. doi: 10.1016/j.ygcen.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Veenstra JA. Neurohormones and neuropeptides encoded by the genome of Lottia gigantea, with reference to other mollusks and insects. Gen Comp Endocrinol. 2010;167(1):86–103. doi: 10.1016/j.ygcen.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Veenstra JA. Neuropeptide evolution: Neurohormones and neuropeptides predicted from the genomes of Capitella teleta and Helobdella robusta. Gen Comp Endocrinol. 2011;171(2):160–175. doi: 10.1016/j.ygcen.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto K, et al. Evolution of dopamine receptor genes of the d1 class in vertebrates. Mol Biol Evol. 2013;30(4):833–843. doi: 10.1093/molbev/mss268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin H, Sassano MF, Roth BL, Shoichet BK. A pharmacological organization of G protein-coupled receptors. Nat Methods. 2013;10(2):140–146. doi: 10.1038/nmeth.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janssen T, Lindemans M, Meelkop E, Temmerman L, Schoofs L. Coevolution of neuropeptidergic signaling systems: From worm to man. Ann N Y Acad Sci. 2010;1200:1–14. doi: 10.1111/j.1749-6632.2010.05506.x. [DOI] [PubMed] [Google Scholar]
- 51.Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76(1):82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bargmann CI. Beyond the connectome: How neuromodulators shape neural circuits. Bioessays. 2012;34(6):458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- 53.Yang Y, Atasoy D, Su HH, Sternson SM. Hunger states switch a flip-flop memory circuit via a synaptic AMPK-dependent positive feedback loop. Cell. 2011;146(6):992–1003. doi: 10.1016/j.cell.2011.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krashes MJ, et al. A neural circuit mechanism integrating motivational state with memory expression in Drosophila. Cell. 2009;139(2):416–427. doi: 10.1016/j.cell.2009.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Milward K, Busch KE, Murphy RJ, de Bono M, Olofsson B. Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2011;108(51):20672–20677. doi: 10.1073/pnas.1106134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sakurai T. The neural circuit of orexin (hypocretin): Maintaining sleep and wakefulness. Nat Rev Neurosci. 2007;8(3):171–181. doi: 10.1038/nrn2092. [DOI] [PubMed] [Google Scholar]
- 57.Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30(6):713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Richards S, et al. Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452(7190):949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 59.Adams MD, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287(5461):2185–2195.
- 60.Colbourne JK, et al. 2011. The ecoresponsive genome of Daphnia pulex. Science 331(6017):555–561.
- 61.Consortium CeG 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282(5396):2012–2018.
- 62.Simakov O, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493(7433):526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sodergren E, et al. 2006. The genome of the sea urchin Strongylocentrotus purpuratus. Science 314(5801):941–952.
- 64.Dehal P, et al. 2002. The draft genome of Ciona intestinalis: Insights into chordate and vertebrate origins. Science 298(5601):2157–2167.
- 65.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453(7198):1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 66.Aparicio S, et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297(5585):1301–1310.
- 67.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 68.Bairoch A, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33(Database issue):D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dieterich C, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 2008;40(10):1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Small KS, Brudno M, Hill MM, Sidow A. A haplome alignment and reference sequence of the highly polymorphic Ciona savignyi genome. Genome Biol. 2007;8(3):R41–R54. doi: 10.1186/gb-2007-8-3-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. The International Aphid Genomics onsortium (2010) Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol 8(2):e1000313. [DOI] [PMC free article] [PubMed]
- 72.Smith JJ, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet. 2013;45(4):415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. UniProt Consortium (2009) The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res 37(Database issue):D169–174. [DOI] [PMC free article] [PubMed]
- 74.Dereeper A, et al. 2008. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36(Web Server issue):W465–469.
- 75.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460–465. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113–131. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 78.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55(4):539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 79.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985 doi: 10.1111/j.1558-5646.1985.tb00420.x. 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 80. Mirabeau O (2008) Searching for novel peptide hormones in the human genome. PhD thesis (Université De Montpellier, Montpellier, France)
- 81.Southey BR, Amare A, Zimmerman TA, Rodriguez-Zas SL, Sweedler JV. 2006. NeuroPred: A tool to predict cleavage sites in neuropeptide precursors and provide the masses of the resulting peptides. Nucleic Acids Res 34(Web Server issue):W267–272. [DOI] [PMC free article] [PubMed]
- 82.Saigo H, Vert JP, Ueda N, Akutsu T. Protein homology detection using string alignment kernels. Bioinformatics. 2004;20(11):1682–1689. doi: 10.1093/bioinformatics/bth141. [DOI] [PubMed] [Google Scholar]
- 83.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25(9):1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stanke M, Steinkamp R, Waack S, Morgenstern B. 2004. AUGUSTUS: A web server for gene finding in eukaryotes. Nucleic Acids Res 32(Web Server issue):W309–312. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.