Abstract
Huntington’s disease (HD) is an autosomal dominant genetic disorder that specifically causes neurodegeneration of striatal neurons, resulting in a triad of symptoms that includes emotional, cognitive, and motor disturbances. The HD mutation causes a polyglutamine repeat expansion within the N-terminal of the huntingtin (Htt) protein. This expansion causes aggregate formation within the cytosol and nucleus due to the presence of misfolded mutant Htt, as well as altered interactions with Htt’s multiple binding partners, and changes in post-translational Htt modifications. The present review charts efforts toward a therapy that delays age of onset or slows symptom progression in patients affected by HD, as there is currently no effective treatment. Although silencing Htt expression appears promising as a disease modifying treatment, it should be attempted with caution in light of Htt’s essential roles in neural maintenance and development. Other therapeutic targets include those that boost aggregate dissolution, target excitotoxicity and metabolic issues, and supplement growth factors.
Keywords: Huntington’s disease, HD, Htt, huntingtin, polyQ, triplet repeat disorders, polyglutamine expansions, neurodegenerative disease, autosomal dominant, protein misfolding, excitotoxicity, small molecule therapies, loss of function, gain of function, developmental disorder, gene therapy, RNAi
The Clinical Presentation of Huntington’s Disease
Huntington’s disease (HD) is a fatal neurodegenerative disorder affecting five to eight per 100,000 persons of European descent [1]. In 1872, a 22-year-old American neurologist published the first complete description of the disease [2]. George Huntington accurately characterized HD as a genetic condition and described the clinical presentation of HD as a triad of motor, emotional, and cognitive disturbances. The hallmark symptom of HD is the presence of involuntary movements, called chorea [3]. Symptom onset typically occurs in midlife and the disease progresses over the next 15 to 20 years [4].
In George Huntington’s day, diagnosis of HD could be tricky, relying heavily upon family history, but also upon postmortem brain analysis, where several pathological features are observed in Huntington’s diseased brains. There is overall atrophy, with marked cell loss in the striatum (caudate/putamen), and globus pallidus, with corresponding ventricular enlargement and gliosis [4] (Figure 1). Cortical pyramidal neuron degeneration also occurs, particularly in association areas of the temporal, frontal, and parietal lobes [5]. Within the HD striatum, the selective progressive loss of GABA-ergic projection neurons (called medium spiny neurons) results in choreic symptoms [6]. The medium spiny neurons receive robust cortical glutamatergic inputs [7], implicating excitotoxicity — a calcium-mediated pathological process that damages or kills cells by overstimulation of glutamate receptors — in the selective neuronal death seen in HD.
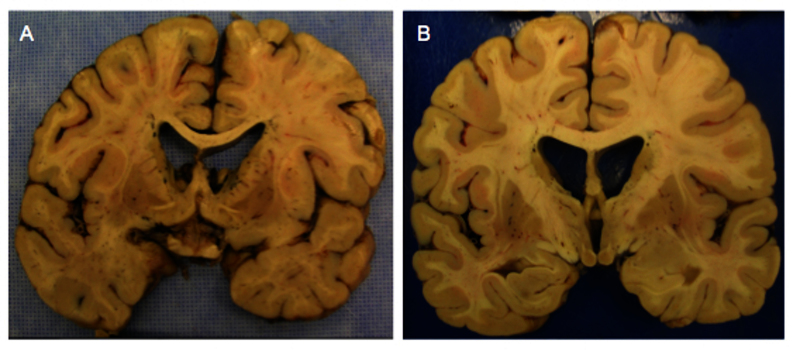
Figure 1.
Striatal pathology in Huntington’s disease. Huntington’s disease (HD) causes overall cortical atrophy and selective cell death of the medium spiny neurons in the striatum, with corresponding enlargement of the lateral ventrical, as evidenced by coronal autopsy sections from a 54-year-old male HD patient (a) and a 47-year-old female HD patient (b).
Back in 1872, George Huntington was able to categorize HD as a genetic condition. We now know that HD is an autosomal dominant disease, meaning that each offspring of an affected individual has a 50 percent chance of inheriting the disease, the disease does not skip a generation, and males and females are equally at risk [8]. One mutated gene is sufficient to cause the disease, regardless of the presence of a normal gene inherited from the other parent [9], and, in fact, homozygous individuals do not appear to differ significantly from heterozygotes in terms of age of onset or symptom severity [10].
The Molecular Biology of the HD Gene
The effort to find the HD gene is a remarkable story of collaboration between many researchers amid the earliest efforts of gene sequencing and cloning. Using genetic markers to probe specific American and Venezuelan kindreds, the gene responsible for the HD mutation was mapped to the tip of chromosome 4 [11]. By 1993, the Huntington’s Disease Collaborative Research Group isolated the HD mutation to a large gene (IT15, also called the HD gene) that encoded a 348 kDa novel protein [12]. The protein product, termed huntingtin (Htt), is the sole product expressed by this gene sequence. Expansion of a normally occurring glutamine (CAG) repeat within the Htt protein results in an extended N-terminal domain [13]. The average size of CAG repeats is 16 to 20 in the normal population and >36 in the affected population [14]. Htt glutamine repeat lengths between 27-35 are in the high normal range, but may elongate in future generations due to the unstable nature of the expansion; polyglutamine (polyQ) repeat lengths between 36 and 39 result in reduced penetrance, with delayed or no symptom onset [15].
The fact that HD is an inherited mutation with an expanded CAG repeat in the coding region of a gene lumps it into a category with eight more otherwise unrelated disorders, including dentatorubropallidoluysian atrophy (DRPLA), spinobulbar muscular atrophy (Kennedy’s disease), and several spinocerebellar ataxias, including type-1 (SCA1) and type-3 (SCA3 or Machado-Joseph disease) [16]. In these disorders, mutant alleles encode a protein with a corresponding number of polyQ repeats [13]. Each of these triplet repeat disorders demonstrate a progressive neurological phenotype in specific brain regions. The age of disease onset is inversely proportional to the number of CAG repeats — the longer the polyQ stretch, the earlier the individual will experience symptoms.
The dominant pattern of heredity displayed by HD focused immediate research efforts on a gain-of-function model. Researchers thought that Htt would be preferentially expressed in the areas most severely affected in HD, namely the striatum and cortex. But Htt is highly expressed in the entire brain and testis, predominately in neurons, as well as in glial cells [17]. Within the cell, Htt is a mostly cytoplasmic protein that is also found at low levels in the nucleus [18].
Once the HD gene was isolated, researchers were able to clone it and insert a mutated form into animals. Animal models catapulted the HD field forward. Htt is a highly conserved protein, and models of HD have been constructed in animals as diverse as C. elegans, D. melanogaster, and zebrafish [19] (Table 1). In 1995, targeted Htt disruption confirmed a gain-of-function model in HD [20-22], and in 1996, researchers in Gillian Bates’ lab showed that expression of an expanded Htt exon 1 alone was sufficient to induce a progressive neurological phenotype in mice [23].
Table 1. Animal Models of HD.
| Model | Construct/Promoter | PolyQ Length | Age of Onset | Pathology | Behavioral Phenotype | Year |
| Mouse R6/1 | Human Htt promoter; ~1.9kb fragment of 5´ human HD gene | 115 | 5 m | Intranuclear and neuropil aggregates throughout the brain; global brain atrophy; minimal cell death | Tremors and gait abnormalities; rotarod deficit; clasping behavior; learning deficit | 1996 [23] |
| R6/2 | 144 | 2 m | ||||
| Drosophila | GAL4-UAS system- using eye-specific P element expression vector pGMR; human HD exon 1 | 75 or 120 | 2 or 10 days | Late-onset progressive neurodegeneration dependent on repeat length; nuclear accumulation but no inclusions | Expression restricted to eyes | 1998 [19] |
| Zebrafish | Expanded N-terminal fragment of Htt protein fused with GFP | 102 | 24 h post fertilization | Increase in apoptotic cells, inclusions in non-apoptotic cells | Increase in embryonic lethality or in embryos with abnormal morphology | 1998 [134] |
| Mouse N171-82Q | N-terminal 171 amino acids of human Htt; mouse prion promoter | 82 | 5 m | Inclusions in striatum, cortex, hippocampus and amygdala; striatal degeneration | Tremors and gait abnormalities; rotarod deficit; loss of coordination; hypokinesis | 1999 [135] |
| Conditional mouse | TetO regulatable; Chimeric mouse/human exon 1. Replace the endogenous | 94 | 4.5 m | Nuclear/ cytoplasmic aggregates in striatum, cortex, and hippocampus; striatal degeneration; gliosis | Clasping behavior, tremor, decreased grooming | 2000 [60] |
| Mouse Hdh Q150 | polyQ with expanded polyQ; mouse Hdh promoter | 150 | 4 m | Nuclear inclusions in striatum; striatal gliosis | Clasping behavior; gait abnormalities; rotarod deficit; hypoactivity | 2001 [136] |
| Mouse YAC 128 | YAC expressing full-length human Htt; human HD promoter | 128 | 3 m | Inclusions in striatum; neuron loss in striatum | Rotarod deficit; clasping; gait abnormalities; circling behavior | 2003 [137] |
| Mouse Hdh Q140 | Replace mouse Htt exon 1 with expanded chimeric mouse/ human exon 1; mouse Hdh promoter | 140 | 12 m | Nuclear and neuropil inclusions in striatum, cortex, nucleus accumbens, and olfactory tubercle | Increased locomotor activity and rearing at 1 month, followed by hypoactivity and gait abnormalities | 2003 [138] |
| Transgenic Rat | A truncated Htt fragment; endogenous rat promoter | 51 | Adult onset | Neurological phenotypes, intracellular inclusions, striatal shrinkage | Progressive motor dysfunction | 2003 [139] |
| Mouse BAC-HD | Full-length human Htt; human HD promoter | 97 | 3 m | Synaptic dysfunction; cortical and striatal atrophy | Rotarod deficit | 2008 [140] |
| Rhesus Macaque Monkey | Human HD exon 1 fused to GFP; Human polyubiquitin-C promoter | 84 | Birth to 1 week | Neuronal inclusions | Dystonia, chorea | 2008 [141] |
| Rat BACHD | Human full-length HD genomic sequence; human HD promoter | 97 | Early onset | Cortical and striatal aggregates; neuropil aggregates appear earlier than inclusions; reduced dopamine receptor binding was detectable by in vivo imaging | Robust, early onset and progressive motor deficits and anxiety-related symptoms | 2012 [142] |
HD, human huntingtin gene; Hdh, mouse huntingtin gene; m, months of age; GAL4-UAS system, Transgenic flies expressing GAL4, a yeast transcriptional activator, are crossed with UAS-transgenic flies, carrying a gene of interest inserted downstream of the UAS (upstream activating sequence); YAC, yeast artificial chromosome; BAC, bacteria artificial chromosome; GFP, green fluorescent protein
Expression of mutant Htt in the animal models revealed a distinctive cellular phenotype — intranuclear inclusions and cytoplasmic aggregates that were mirrored in HD human patients [24] (Figure 2A). Aggregation was found in many brain areas and therefore could not explain the vulnerability of the striatum. The aggregates in dystrophic neurites were found in presymptomatic patients; however, the presence of intranuclear inclusions appeared to coincide with the onset of HD symptoms [25].
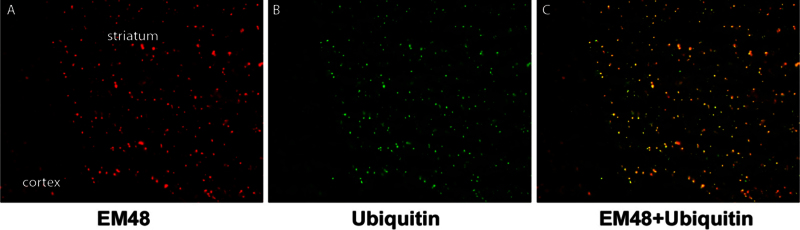
Figure 2.
Aggregate formation in a 140 polyQ knock-in mouse model of HD. In a model of HD made by inserting a chimeric mouse/human exon 1 with 140 CAG repeats into mice, inclusions and aggregates can be seen in the striatum and cortex using the EM48 antibody (a), which is specific for the N-terminal region of the mutant huntingtin protein. These bodies also contain ubiquitin (b), with colocalization of the two proteins (c).
A theory about HD pathogenesis emerged, strongly based on the data gleaned from the animal models. Functional subunits of the proteosome, ubiquitin, and heat shock proteins [24,26,27] are localized to polyQ disease inclusions, suggesting a cellular clearance effort (Figure 2B and 2C). If mutant Htt is resistant to proteolysis, then protein turnover is delayed. The concentration of protein increases with time, leading to aggregation. The aggregates draw other proteins in (including normal Htt), sequestering them and rendering them useless [28]. Key cellular components, such as neurofilaments, are disrupted by aggregate formation [29]. The cell then becomes dysfunctional, dies, and the patient becomes symptomatic.
Physiological Modifiers
Due to its large size, the tertiary structure of Htt remains unknown, but the structure of several of its protein domains has been described. Analysis of Htt protein composition revealed a glutamine-rich region followed by a proline-rich domain, several caspase cleavage sites, and three sets of HEAT repeats (Htt, elongation factor 3, the PR65/A subunit of protein phosphatase 2A and the lipid kinase Tor) (Figure 3) [30-32]. The polyQ stretch, being the subject of intense speculation, is not found in all organisms that express an Htt homolog. The polyQ stretch is absent in the N-terminal in Drosophila and Ciona (sea squirt), maintained at 4 glutamines in fish, birds, and amphibians, and expanded to its longest stretch in humans [33]. Deletion of Htt’s polyQ stretch in mice causes neurological consequences and alterations in energy homeostasis in adults, but its absence does not appear to overtly impact development [34]. Loss of the entire Htt protein results in embryonic lethality in mice due to organization defects in the extra-embryonic tissues [22,35].
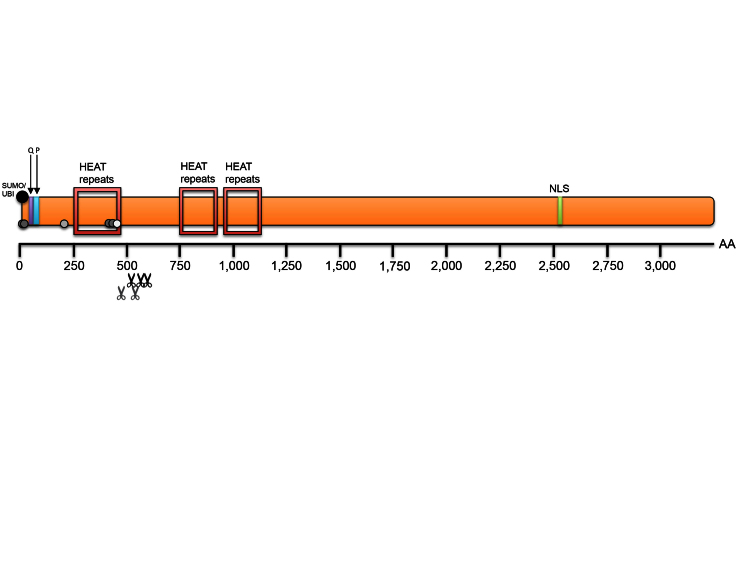
Figure 3.
Huntingtin Protein Domains (adapted from Zuccato 2010). Huntingtin (Htt) is a large protein (352 KDa) with several highly conserved protein domains. The N-terminus contains the polyglutamine (polyQ) stretch that causes Huntington’s disease when elongated, as well as a proline rich region (PRR). There are three sets of HEAT repeats (Htt, elongation factor 3, the PR65/A subunit of protein phosphatase 2A and the lipid kinase Tor) that stretch throughout a large portion of the protein. Several cleavage sites have been identified in Htt, concentrated between amino acids 400-600, as well as a nuclear localization signal (NLS) towards the C-terminus. Htt also is subject to multiple forms of post-translational modifications, including acetylation, phosphorylation, palmitoylation, sumoylation and ubiquitination, which can be altered in the presence of the expanded allele.
The Htt proline-rich region is found only in mammals [33], and although it may contribute to the solubility of the protein [36], deletion of the proline-rich domain in mice does not appear to significantly affect Htt’s normal function [37]. In contrast, the large majority of the HEAT repeats are present throughout all the homologs, including insects [33]. HEAT repeat proteins are typically very large, function as part of protein complexes, and are often involved in cytoplasmic transport processes [30]. The conserved nature of the HEAT repeats in Htt is perhaps our best clue to Htt’s normal function, as their presence indicates a propensity to interact with other proteins, and suggests Htt is a type of scaffold on which other proteins can assemble.
The presence of accumulated misfolded proteins classifies HD as a conformational disease and groups it with a diverse brain disorders such as prion encephalopathies, Alzheimer’s disease, and Parkinson’s disease, although the aggregation sites differ [38]. Direct evidence for misfolded mutant Htt lies in the fact that certain antibodies are able to distinguish between the mutant and normal forms of the protein [28].
A misfolding of the mutant Htt protein due to the extended N-terminal could alter protein function, as the polyQ stretch may present itself or nearby Htt domains in a more or less provocative way to potential binding partners. Indeed, an assessment of the changes in mutant Htt’s protein interactions shows that HD has elements of loss-of-function occurring at the same time as gain-of-function, both perturbing normal Htt functions and gaining deleterious new cellular activities [39]. These altered binding partner relationships are all potential therapeutic targets.
Being a large protein, Htt has numerous binding partners, including transcription co-activators [40], co-repressors [41], and apoptosis-related kinases [42]. Htt’s normal function may impact many cellular processes, including signal transduction, endocytosis, cytoskeletal structure, transcription and axonal transport [43-45], and in the presence of the expanded polyQ stretch, the interactions with binding partners can be increased or decreased (Table 2). Interestingly, expression of the N-terminal section of mutant Htt is enough to cause neuronal degeneration, but is not sufficient to maintain Htt’s axonal transport functions [46].
Table 2. Huntingtin Interacting Proteins [39,143-145].
| Name | Protein function | Htt binging region | Influence of mu HTT |
| Transcription | |||
| CA150 | Transcription activator | Unknown | None |
| CBP | Transcription activator | Amino acids 1-588 | Enhances |
| CtBP | Transcription repressor | Unknown | Decreases |
| HYP-A, B | RNA splicing factors | Polyproline | Enhances |
| HYP-C | Transcription factor | Polyproline | Enhances |
| NCOR | Transcription repressor | Amino acids 1-171 | Enhances |
| NF-ĸB | Transcription factor | HEAT repeats | Unknown |
| SP1 | Transcription activator | Amino acids 1-171 | Enhances |
| TAFII130 | Transcription activator | Amino acids 1-480 | None |
| TBP | Basal transcription factor | Unknown | Unknown |
| P53 | Transcription factor | Polyproline | None |
| REST-NRSE | Trascription suppressor | Amino acids 1-548 | Decreases |
| Trafficking and endocytosis | |||
| HAP1 | Trafficking, endocytosis | Amino acids 1-230 | Enhances |
| HIP1 | Endocytosis, pro-apoptotic | Amino acids 1-540 | Decreases |
| HIP14 | Trafficking, endocytosis | Amino acids 1-550 | Decreases |
| PACSIN1 | Endocytosis | Polyproline | Enhances |
| Phosphatidylethanolamine | Phospholipids | Amino acids 171-287 | Enhances |
| PI(3,4,5)P3 | |||
| PSD-905 | Synaptic scaffolding | Unknown | Decreases |
| Signaling | |||
| Calmodulin | Calcium-binding regulatory protein | Unknown | Enhances |
| CIP-4 | Cdc42-related signaling | Amino acids 1-152 | Enhances |
| FIP2 (HYP-L) | GTPase Rab8 interactor | Amino acids 1-550 | Unknown |
| GRb2 | Growth factor signaling | Polyproline | Unknown |
| IP31 | Calcium release channel | Amino acids 1-158 | Enhances |
| SH3GL3 | Endocytosis and vesicle recycling | Polyproline | Enhances |
| RasGAP | Ras GTPase-activating protein | Polyproline | Unknown |
| Metabolism | |||
| Cystathionine b-synthase | Generation of cysteine | Amino acids 1-171 | None |
| GAPDH | Glycolitic enzyme | Polyproline | Enhances |
| gp78 | ER membrane-anchored ubiquitin ligase | HEAT repeats 2/3 | Enhances |
| HIP2 | Ubiquitin-conjugated enzyme | Amino acids 1-540 | None |
| Protein Synthesis | |||
| Gnb211 | Translation (indirect,) ribosomal protein | Unknown | Decreases |
| Myo5a | RNA transport to spines | Unknown | Decreases |
| Rps6 | Translation (direct), Ribosomal protein | Unknown | Decreases |
| Prkra | Translation (indirect), PKR regulation | Unknown | Decreases |
Abbreviations: CA150, co-activator 150; CBP, (cAMP-response element binding protein) binding protein; CIP-4, cdc42-interacting protein 4; Co-IP, co-immunoprecipitation; CtBP, C-terminal-binding protein; FIP2, for 14.7K interacting protein; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Gnb2l1, guanine nucleotide-binding protein (G protein) polypeptide 2-like 1; gp78, glycoprotein 78; GRB2, growth factor receptor-binding protein 2; GST, glutathione S-transferase; HAP1, htt-associated protein 1; HIP, htt-interacting protein; HYP, htt-yeast partner; IP31, inositol (1,4,5)-trisphosphate receptor type 1; Myo5a, myosin V;A NCOR, nuclear receptor co-repressor; NF-kB, nuclear factor-kB transcription factor; PACSIN1, protein kinase C and casein kinase substrate in neurons 1; [PI(3,4,5)P3] phosphoinositol, (PI) 3,4-bisphosphate, PI 3,5-bisphosphate, and PI 3,4,5-triphosphate; PKR, double-stranded RNA-activated protein kinase; Prkra, interferon inducible double-stranded RNA-dependent protein kinase activator A; PSD- 95, postsynaptic density 95; RasGAP, Ras GTPase-activating protein; REST–NRSE, the repressor element-1 transcription factor–the neuron restrictive silencer element; Rps6, ribosomal protein S6; SH3GL3, SH3-containing GRB2-like protein 3; SP1, specificity protein-1; TAFII130, TBP-associated factor; TBP, TATA box binding protein.
Htt was the first neurodegenerative disease protein to be identified as a caspase substrate [47]. Htt can be cleaved by caspases (including caspase-2 [48] and caspase-3 [47]), calpain [49], and the matrix metalloproteinase MMP-10 [50] at a regional “hot spot” within Htt between 400 and 600 amino acids, resulting in N-terminal Htt fragments that are small enough to passively translocate into the nucleus. Once inside the nucleus, the mutant Htt cleavage product can form nuclear inclusions that recruit transcription factors, and soluble mutant Htt can aberrantly repress transcription itself [18].
Proteolytic processing is likely an important initial step in pathogenesis, since expressing the smaller Htt truncation product results in greater cell toxicity than expressing the entire mutant htt protein [51,52] and inhibition of cleavage can lessen neurotoxicity in animal models [53]. Htt contains a strictly conserved nuclear export signal that is cleaved away in HD [54], and nuclear targeting of mutant Htt increases toxicity [55], so devising a way to keep mutant Htt out of the nucleus could be beneficially therapeutic.
In addition to cleavage, Htt is normally subject to several types of post-translational modifications, including acetylation, phosphorylation, methylation, sumoylation, and ubiquitination, which can be altered in the presence of the expanded allele [56]. For example, Htt phosphorylation at serine 421 promotes anterograde transport within the neuron, but this function is impaired in the presence of the mutant allele [57]. Htt phosphorylation at serines 13 and 16 can protect against expanded polyQ toxicity [58], and increasing or mimicking phosphorylation at these sites is currently an area of therapeutic investigation.
Biochemical Modifiers
Although the certainty of a genetic diagnosis can be daunting for affected patients and their families, early genetic testing of individuals at risk for an autosomal dominant disorder would allow ample time for a potential therapy to be administered [59]. However, with no available treatment, and with more than 10 potential disease-modifying drugs showing no significant difference in clinical trials [58], many at-risk individuals choose not to find out their genetic status.
In 2000, the first regulatable mouse model of HD showed that it is possible to reverse aggregate formation and disease symptoms after they have manifested [60], which gave researchers hope that a potential therapy could even be effective in symptomatic patients. The cumulative damage hypothesis states that in neurodegenerative disease, neurons are slowly overwhelmed by accumulated damage (such as that caused by oxidative stress or toxic protein accumulation), and those neurons become increasingly committed to an apoptotic future. On the contrary, the “one-hit” biochemical model of several inherited diseases, including HD, uses statistical analysis to argue against the cumulative damage hypothesis [61]. The “one-hit” model proposes that a single catastrophic intracellular event results in neuronal death. At any one time, each neuron is at constant risk of cell death, which implies that treatment administered at any disease stage should be beneficial.
In animal models, we can delay the disease process and even reverse it — we can dismantle aggregates and rescue phenotypes [60,62-65]. Theoretically, if we can do it in animals, in humans we should be able to administer a small molecule drug to keep pre-symptomatic patients healthy or to restore proper neuronal function at any stage of the disease, as long as the neurons are still present. However, the post-mitotic nature of neurons, combined with difficulties crossing the blood brain barrier, pose substantial hurdles to effectively reaching the striatal target cells. Recent advances in transforming adipose-derived stem cells derived from HD patient into pluripotent stem cells (that can be transformed into neurons) should assist with elucidating disease mechanisms and hasten the testing of small molecule therapies [66].
One such small molecule under investigation aims to inhibit histone deacetylases (HDACs). Truncated mutant Htt can inhibit, mislocalize, and degrade acetyltransferases, which are enzymes that normally modify proteins to increase gene activity [67]. This interaction is mediated through the proline rich domain, as well as the polyQ stretch, and results in reduced levels of acetylated histones [68]. HDACs are able to reverse this reduction and reduce lethality in animal models of HD, even after symptom onset, so HDAC inhibitors are another potential treatment, as they can influence not only gene transcription but also potentially alleviate endoplasmic reticulum stress or modulate chaperone activity [67].
The HD phenotype could conceivably be delayed by preventing aggregate formation and/or increasing aggregate clearance by targeting proteosome function, increasing ubiquitination, or increasing autophagy. Eukaryotic cells have two pathways for clearance — under normal circumstances, the ubiquitin/proteosome system functions at high levels, whereas the autophagy/lysosome system maintains low activity levels [69]. If mutant Htt overwhelms the ubiquitin/proteosome system in HD, the neuron will then induce autophagy for protein clearance [70].
Indeed, the autophagic response is one of the first neuronal responses to mutant Htt [71] and is predominately responsible for clearing the cytoplasmic aggregates [72]. Polymorphisms in autophagy-related genes contribute to the age of onset in HD [73]. Remarkably, Htt may normally regulate mechanisms of protein degradation that are ultimately involved in its own clearance [74], and in the disease process, expression of Htt with a deleted polyQ tract in a 140Q/+ knock-in mouse model can upregulate autophagic markers and increase lifespan [65]. Unlike the cytoplasmic aggregates, HD nuclear inclusions appear to be cleared using the ubiquitin/proteosome system instead [69], making both the autophagic and the ubiquitin/proteosome systems attractive therapeutic targets.
Modulating Excitotoxicity and Metabolism
Excitotoxicity has been implicated in the selective neuronal death seen in HD [75]. There is an intimate relationship between cellular metabolism and excitotoxicity. Although implicated in HD several ways, the strongest evidence that mitochondria are involved in HD pathogenesis is the fact that administration of the mitochondrial toxin 3-nitropropionic acid (3-NP) can mimic HD characteristics [76], including selective cell death in the striatum, cognitive impairment, and the development of motor symptoms in a non-human primate model [77].
Both creatine and coenzyme Q10 administration reduce reactive oxygen species to address the metabolic defects in HD. Creatine is involved with energy buffering and the connection between energy production and consumption within the cell. When orally administered in a HD mouse model, creatine improves survival and delays atrophy and aggregate formation [78]. Coenzyme Q10 is an antioxidant, as well as an essential part of the electron transport chain. Coenzyme Q10 can alleviate symptoms, extend survival time, and slow striatal atrophy in a mouse model of HD [76]. Unfortunately, limited efficacy of these agents has been observed in HD patients; however, the optimal therapeutic dose may have been underestimated and higher dose administration is under investigation [73]. Another metabolic therapeutic target is the P2X receptor, part of the signaling machinery mediating ATP responses to neurodegenerative stressors [79].
Growth factors and cytokines play a role in HD pathology and may particularly modulate the effects of excitotoxicity. Transforming growth factor β1 (TGF-β1) is reduced in cortical neurons of HD patients and mouse models [80], so supplementation is a possibility, or perhaps TGF-β1 could be a useful biomarker for disease progression.
Retroviral administration of ciliary neurotrophic factor (CNTF) can alter the neuronal degeneration and prevent deficits in an excitotoxic HD rat model [81,82]. Bilateral striatal implantations of CNTF releasing cells into a primate model of HD at symptom onset protects neurons from further degeneration, as well as offering cognitive and motor improvement [83]. Peripheral administration of CNTF is not well tolerated due to side effects, but implantation of CNTF-releasing cells remains a possibility [84]. A phase I study implanting a device with a semi-permeable membrane encapsulating a cell line engineered to synthesize CNTF in HD patients showed that administration within the ventricle is safe and feasible; however, the technique needs improvement, as CNTF levels were low in many patients and there were varying cell survival numbers within capsules after removal [85].
Brain-derived growth factor (BDNF) is a neurotrophic factor speculated to play a role in neuronal development and survival [86], and BDNF can prevent cell death in excitotoxic models of HD [87]. One of normal Htt’s regular jobs in the cell is to bind up transcriptional repressors (such as REST–NSFR [the repressor element-1 transcription factor–the neuron restrictive silencer element]) in the cytoplasm. Mutant Htt does a poor job of binding to the repressors, resulting inhibition of target genes, including brain-derived neurotrophic factor (BDNF) [73]. Not surprisingly, a 53 percent to 82 percent reduction in BDNF expression was found in the striatum of HD patients upon autopsy [88]. Enrollment was recently completed for a phase 2/3 clinical trial investigating cysteamine bitartrate delayed-release capsules (RP103) for HD in France, following results showing that cysteamine increases BDNF levels in rodents and primate models of HD [89].
Glial cells play a major role in local trophin availability, and astrocytes are able to both respond to and produce BDNF [90]. Efforts can be made to increase gene products like BDNF, for example, astrocytes engineered to overexpress BDNF are being explored as a potential gene therapy in a rodent model of HD [91].
A Role for Astrocytes in HD
Recently, the research focus on HD neuronal dysfunction has expanded to include a possible glial role in pathogenesis. Glial cells do express Htt [17] and original pathology work showed marked gliosis as a disease marker, becoming more widespread as the disease progresses [92]. Mouse models expressing mutant Htt show glial nuclear aggregates [93], and specific astrocytic expression of 160Q N-terminal mutant Htt fragments can induce neurological symptoms in mice [94]. Interestingly, mouse stem cells expressing no Htt are much more likely to differentiate into glial cells than cells expressing Htt with 20, 50, 111 or 140 polyQ repeats, even when treated with the same in vitro neural differentiation protocol [95].
Neurons are dependent on astrocytes metabolically and cooperate very closely with astrocytes when it comes to circumventing glutamate-mediated excitotoxicity [96], so investigating this relationship in HD seems reasonable. Nearly 80 percent of glutamate is removed from the synapse by the astrocytic transporters glutamate transporter 1 (GLT1) and glutamate-aspartate transporter (GLAST) [97]. Interestingly, knockouts of either of these glial glutamate transporters results in excess extracellular glutamate levels and a progressive motor phenotype [98]. If the GLT1 transporters are nonfunctional or missing, neuronal damage due to glutamate over-stimulation is likely to occur [99,100], and indeed, decreased mRNA levels for GLT1 are found in HD [101].
Astrocytes are also responsible for supplying adult neurons with cholesterol [102], a key ingredient for normal synaptogenesis and neurotransmitter release [103,104]. Cholesterol biosynthesis is reduced in astrocytes isolated from HD mouse models [105], though Htt’s involvement in cholesterol homeostasis remains to be fully elucidated. Since astrocytes are very sensitive to cues in the environment surrounding the neurons, they may also be affected by increased ciliogenesis caused by mutant Htt [106]. The non-motile cilia have a sensory role in regulating signaling pathways, such as hedgehog and PDGF-α [106]. The restoration of normal ciliary function — though certainly not a complete treatment — could be a potential therapeutic target.
Genetic Modifiers
Early investigations showed that a gene closely linked to the HD gene may modify age of onset [107]. Targeting cis-regulatory elements to delay the appearance of symptoms is a strategy that remains to be elucidated, as the exact nature of these regulatory elements are still unknown. However, sequence variations in the PPARGC1A gene encoding PGC-1α (involved in mitochondrial function), as well as polymorphisms in PGC1α’s downstream targets, can exert modifying effects on the age of onset in HD [108]. Subtypes of N-methyl D-aspartate receptor genes (GRIN2A and GRIN2B) may also modify age of onset [109].
The length of Htt’s polyQ stretch in the normal allele does not influence when HD symptoms first appear [110], which suggests that strategies to decrease Htt expression itself may be effective. This proposal is more complicated than simply ridding the cell of a benign protein that has turned noxious. The development of knockout and conditional knockout mouse models demonstrate that Htt is essential for early embryogenesis [20-22] and spermatogenesis [111]. Rather than seeing a decrease in Htt expression following execution of its critical role in embryogenesis, postnatal Htt expression levels actually rise in the adult [112]. Htt plays a critical role in the development of proper neuronal connections and apoptosis [113], and it is not yet known if lack of Htt expression as an adult would be benign.
Existing literature suggests that Htt loss of function may comprise essential neurodevelopment programs, including neuronal organization through a pivotal role in mitotic spindle orientation [114] and neuronal maturation via its role in ciliogenesis [115]. Htt itself has antiapoptotic properties [116], and depletion of wild type Htt has been found not just in mouse models of HD, but also in models of neurodegeneration secondary to ischemia and traumatic brain injury [117]. Eliminating expression without a complete understanding of Htt’s normal cellular function could confound the plight of already sick neurons.
The HD gene can be silenced in vivo using RNA strategies (small interfering RNA [siRNA] or short hairpin RNA [shRNA]) or by antisense oligonucleotides (ASO). Temporally sensitive administration of RNA therapy could reduce Htt production in HD gene carriers and potentially eliminate Htt protein in adult tissues. However, practical applications of these therapies struggle to find the best routes of administration, due to the blood brain barrier, and the appropriate cells to target, as we know that HD is not solely a striatal specific disease [118]. In addition, gene suppression strategies must be carefully designed to avoid off-target effects and dosage control.
Small-scale siRNA knockdown experiments in monkeys [119] and mice using siRNA [120] appear promising; however, in the process of eliminating Htt protein expression, wild-type Htt protein expression is often decreased as well. Continuous partial suppression of both forms of Htt expression in rodent models decreases neuropathology, reduces symptoms, and prolongs survival, even when wild-type Htt was also eliminated [121,122]. To selectively target just the mutant Htt allele, RNA strategies can capitalize on the presence of the expanded polyQ [123] or on the presence of single nucleotide polymorphisms (SNPs) associated with the presence of the expanded polyQ allele [124] as targets. The recent achievement of an allele-selective siRNA in an HD mouse model may make the siRNA technique the most effective way forward for Htt silencing efforts [125].
But can these knockdown techniques be translated effectively from mice to humans? Recently, a convection-enhanced delivery system delivered 7 days of a siRNA treatment to the much larger non-human primate brain and was able to decrease Htt expression effectively throughout the striatum, removing a technical delivery hurdle for human therapy translation [126]. Still more promising, favorable results from a phase 1 trial that used siRNA to block the expression of SOD1 in familial forms of Amyotrophic Lateral Sclerosis (ALS) were presented at the 2012 American Academy of Neurology Annual Meeting. In humans affected with HD, neurons are able to sustain the expression of mutant Htt for many years before aggregates form and neurodegeneration begins. It may be that a brief elimination of mutant Htt synthesis is all a neuron needs to get a better handle on clearing the mutant Htt from the cell and to keep symptoms at bay.
Huntingtin as a Developmental Disorder
Although HD patients are usually not symptomatic until mid-life, abnormal brain development may contribute to HD pathophysiology. Affected patients often experience weight loss that pre-dates motor abnormalities [127], and brain scans reveal enlarged cortex size [128] and decreased intracranial volume [127] in presymptomatic patients, which suggests abnormalities in neural development. Presymptomatic children carrying the HD expansion have lower body mass index (BMI) and head circumference than controls, suggesting defects with energy regulation and brain growth are present 30+ years before overt symptoms would normally appear [129].
These findings group HD into yet another disease category (with disorders such as schizophrenia, familial Alzheimer’s disease, and SCA-1), where abnormal development sets the stage for a later stressor, resulting in cell death [130]. The evidence for very early disease manifestation, in combination with Htt’s essential roles in early development, brings HD full circle from a neurodegenerative disease to a developmental disease with cellular homeostasis defects that predispose neurons to die in mid-life [131].
Htt, as a fairly social protein within the cell, may have multiple roles to play during development and during adulthood, and different protein domains may be essential at distinct time points [132]. Even normal variations in the size of the Htt polyQ stretch are associated with differences in brain measurements in the pallidum, with longer repeat lengths associated with more grey matter in normal individuals [133]. More research is needed to discover the normal functions of Htt at multiple stages in development. Thinking about HD as a type of developmental disorder opens the door for very early intervention targets, and yet pointedly introduces the issue of responsible genetic testing of juveniles with no available cure.
Conclusion
It has been more than a century since the first description of the disease was published. With the advent of molecular genetics techniques, we now are able to view a blueprint of each individual’s DNA. In a few short decades, HD diagnosis became as easy as a blood test. But Htt turns out to have multiple roles within the cell, and the HD story has been far more complex than most imagined. Many of the original pioneering HD researchers are still working on this disease, and the hope is that a physician will be able to hand patients an effective treatment along with an HD diagnosis. The HD gene was the first to be mapped; perhaps it will also be the first neurodegenerative disease to be cured.
Acknowledgments
The author would like to thank Jim Mandell and Joy Vetter in the Department of Pathology and Scott Zeitlin in the Department of Neuroscience at the University of Virginia for contributing photos for Figures 1 and 2, respectively, and Talbot Weston and Courtney Lowers, undergraduate students at Randolph-Macon College, for their assistance with Figure 3 and referencing.
Abbreviations
- HD
Huntington’s disease
- Htt
huntingtin
- CAG
glutamine
- polyQ
polyglutamine
- HDAC
histone deacetylase
- TGF-β1
transforming growth factor β1
- CNTF
ciliary neurotrophic factor
- BDNF
brain-derived neurotrophic factor
- siRNA
small interfering RNA
- shRNA
short hairpin RNA
- ASO
antisense oligonucleotides
- GLT1
glutamate transporter 1
- SOD1
superoxide dismutase 1
- 3-NP
3-nitropropionic acid
- SNP
single- nucleotide polymorphism
References
- Conneally PM. Huntington disease: Genetics and epidemiology. Am J Hum Genet. 1984;36:506–526. [PMC free article] [PubMed] [Google Scholar]
- Huntington G. On chorea. Medical and Surgical Reporter. 1872;26:317–321. [Google Scholar]
- Koller RL, Watts WC. Movement Disorders: Neurologic Principles and Practice. New York: McGraw Hill Professional Publishing, Inc.; 1997. [Google Scholar]
- Harper PS. Huntington’s Disease. London: Saunders Company Ltd.; 1996. [Google Scholar]
- Heinsen H, Strik M, Bauer M, Luther K, Ulmas G, Gangus D. et al. Cortical and striatal neurone number in Huntington's disease. Acta Neuropathol (Berl) 1994;88:320–333. doi: 10.1007/BF00310376. [DOI] [PubMed] [Google Scholar]
- Graveland GA, Williams RS, DiFiglia M. Evidence for degenerative and regenerative changes in neostriatal spiny neurons in Huntington's disease. Science. 1985;227(4688):770–773. doi: 10.1126/science.3155875. [DOI] [PubMed] [Google Scholar]
- Zigmond MJ, Bloom FE, Landis SC, Roberts JL, Squire LR, editors. Fundamental Neuroscience. San Diego: Academic Press; 1999. [Google Scholar]
- Folstein SE. Huntington’s Disease: A disorder of families. Baltimore, MD: Johns Hopkins University Press; 1989. [Google Scholar]
- Ho LW, Carmichael J, Swartz J, Wyttenbach A, Rankin J, Rubinsztein DC. et al. The molecular biology of Huntington's disease. Psychol Med. 2001;31(1):3–14. doi: 10.1017/s0033291799002871. [DOI] [PubMed] [Google Scholar]
- Wexler NS, Young AB, Tanzi RE, Travers H, Starosta-Rubinstein S, Penney JB. et al. Homozygotes for Huntington's disease. Nature. 1987;326(6109):194–197. doi: 10.1038/326194a0. [DOI] [PubMed] [Google Scholar]
- Gusella JS, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE. et al. A polymorphic DNA marker genetically linked to Huntington’s disease. Nature. 1983;306(5940):234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- HD Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Jou Y, Myers RM. Evidence from antibody studies that the CAG repeat is the Huntington’s disease gene is expressed in the protein. Hum Mol Genet. 1995;4(3):465–469. doi: 10.1093/hmg/4.3.465. [DOI] [PubMed] [Google Scholar]
- Warby SC, Visscher H, Collins JA, Doty CN, Carter C, Butland SL. et al. HTT haplotypes contribute to differences in Huntington disease prevalence between Europe and East Asia. Eur J Hum Genet. 2011;19(5):561–566. doi: 10.1038/ejhg.2010.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey ER. Characterization of a large group of individuals with Huntington disease and their relatives enrolled in the COHORT study. PLoS One. 2012;7(2):e29522. doi: 10.1371/journal.pone.0029522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Fischbeck KH. Trinucleotide repeats in neurogenetic disorders. Annu Rev Neurosci. 1996;19:79–107. doi: 10.1146/annurev.ne.19.030196.000455. [DOI] [PubMed] [Google Scholar]
- Sharp AH, Love SJ, Schiling G, Li S, Li X, Bao J. et al. Widespread expression of Huntington's disease gene (IT15) protein product. Neuron. 1995;14:1065–1074. doi: 10.1016/0896-6273(95)90345-3. [DOI] [PubMed] [Google Scholar]
- Kegel KB, Meloni AR, Yi Y, Kim YJ, Doyle E, Cuiffo BG. et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J Biol Chem. 2002;277(9):7466–7476. doi: 10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- Jackson GR, Salecker I, Dong X, Yao X, Arnheim N, Faber PW. et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron. 1998;21(3):633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- Nasir J, Floresco SB, O’Kusky JR, Diewert VM, Richman JM, Zeisler J. et al. Targeted disruption of the Huntingto’s disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- Duyao MP, Auerbach AB, Ryan A, Persichetti F, Barnes GT, McNeil SM. et al. Inactivation of the mouse Huntington's Disease homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- Zeitlin S, Liu JP, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homolog. Nat Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- Mangiarini L, Sathasivam K, Seller M, Cozens B, Harper A, Hetherington C. et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 1996;87(3):493–506. doi: 10.1016/s0092-8674(00)81369-0. [DOI] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA. et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90(3):537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP. et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277(5334):1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Cummings JC, Mancini MA, Antalffy B, DeFranco DB, Orr HT, Zoghbi HY. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19:148–154. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- Stenoien DL, Cummings CJ, Adams HP, Mancini MG, Patel K, Demartino GN. et al. Polyglutamine-expanded androgen receptors form aggregates that sequester heat shock proteins, proteasome components and SRC-1, and are suppressed by the HJD-2 chaperone. Hum Mol Genet. 1999;8(5):731–741. doi: 10.1093/hmg/8.5.731. [DOI] [PubMed] [Google Scholar]
- Dyer RB, McMurray CT. Mutant protein in Huntington disease is resistant to proteolysis in affected brain. Nat Genet. 2001;29(3):270–278. doi: 10.1038/ng745. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Onodera O, Chun J, Strittmatter WJ, Burke JR. Expanded polyglutamine domain proteins bind neurofilament and alter the neurofilament network. Exp Neurol. 1999;155(2):195–203. doi: 10.1006/exnr.1998.6991. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11:115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- Wellington CL, Ellerby LM, Hackam AS, Margolis RL, Trifiro MA, Singaraja R. et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273(15):9158–9167. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- Maddox J. Triplet repeat genes raise questions. Nature. 1994;368(6473):685. doi: 10.1038/368685a0. [DOI] [PubMed] [Google Scholar]
- Tartari M, Gissi C, Lo Sardo V, Zuccato C, Picardi E, Pesole G. et al. Phylogenetic comparison of huntingtin homologues reveals the appearance of a primitive polyQ in sea urchin. Mol Biol Evol. 2008;25(2):330–338. doi: 10.1093/molbev/msm258. [DOI] [PubMed] [Google Scholar]
- Clabough EB, Zeitlin SO. Deletion of the triplet repeat encoding polyglutamine within the mouse Huntington's disease gene results in subtle behavioral/motor phenotypes in vivo and elevated levels of ATP with cellular senescence in vitro. Hum Mol Genet. 2006;15(4):607–623. doi: 10.1093/hmg/ddi477. [DOI] [PubMed] [Google Scholar]
- Dragatsis I, Efstratiadis A, Zeitlin S. Mouse mutant embryos lacking huntingtin are rescued from lethality by wild-type extraembryonic tissues. Development. 1998;125(8):1529–1539. doi: 10.1242/dev.125.8.1529. [DOI] [PubMed] [Google Scholar]
- Ignatova Z, Gierasch LM. Inhibition of protein aggregation in vitro and in vivo by a natural osmoprotectant. Proc Natl Acad Sci USA. 2006;103(36):13357–13361. doi: 10.1073/pnas.0603772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveklovska M, Clabough EB, Zeitlin S. Deletion of the huntingtin proline-rich region does not significantly affect normal huntingtin function in mice. J Huntingtons Dis. 2012;1(1):71–87. doi: 10.3233/JHD-2012-120016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Barrow CJ. Protein conformational misfolding and amyloid formation: characteristics of a new class of disorders that include Alzheimer's and Prion diseases. Curr Med Chem. 2002;9(19):1751–1762. doi: 10.2174/0929867023369123. [DOI] [PubMed] [Google Scholar]
- Culver BP, Savas JN, Park SK, Choi JH, Zheng S, Zeitlin SO. et al. Proteomic Analysis of Wild-type and Mutant Huntingtin-associated Proteins in Mouse Brains Identifies Unique Interactions and Involvement in Protein Synthesis. J Biol Chem. 2012;287(26):21599–21614. doi: 10.1074/jbc.M112.359307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SH, Cheng AL, Zhou H, Lam S, Rao M, Li H. et al. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol Cell Biol. 2002;22(5):1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutell JM, Thomas P, Neal JW, Weston VJ, Duce J, Harper PS. et al. Aberrant interactions of transcriptional repressor proteins with the Huntington's disease gene product, huntingtin. Hum Mol Genet. 1999;8(9):1647–1655. doi: 10.1093/hmg/8.9.1647. [DOI] [PubMed] [Google Scholar]
- Peel AL, Rao RV, Cottrell BA, Hayden MR, Ellerby LM, Bredesen DE. Double-stranded RNA-dependent protein kinase, PKR, binds preferentially to Huntington’s disease (HD) transcripts and is activated in HD tissue. Hum Mol Genet. 2001;10(15):1531–1538. doi: 10.1093/hmg/10.15.1531. [DOI] [PubMed] [Google Scholar]
- Gunawardena S, Her LS, Brusch RG, Laymon RA, Niesman IR, Gordesky-Gold B. et al. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40(1):25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- Gusella JF, MacDonald ME. Huntingtin: a single bait hooks many species. Curr Opin Neurobiol. 1998;8(3):425–430. doi: 10.1016/s0959-4388(98)80071-8. [DOI] [PubMed] [Google Scholar]
- Trushina E, Dyer RB, Badger JD 2nd, Ure D, Eide L, Tran DD. et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24(18):8195–8209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pages M, Dompierre JP, Rangone H, Cordelieres FP. et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118(1):127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Goldberg YP, Nicholson DW, Rasper DM, Kalchman MA, Koide HB, Graham RK. et al. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996;13(4):442–449. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- Hermel E, Gafni J, Propp SS, Leavitt BR, Wellington CL, Young JE. et al. Specific caspase interactions and amplification are involved in selective neuronal vulnerability in Huntington's disease. Cell Death Differ. 2004;11(4):424–438. doi: 10.1038/sj.cdd.4401358. [DOI] [PubMed] [Google Scholar]
- Gafni J, Ellerby LM. Calpain activation in Huntington’s disease. J Neurosci. 2002;22(12):4842–4849. doi: 10.1523/JNEUROSCI.22-12-04842.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JP, Holcomb J, Al-Ramahi I, de Haro M, Gafni J, Zhang N. et al. Matrix metalloproteinases are modifiers of huntingtin proteolysis and toxicity in Huntington’s disease. Neuron. 2010;67(2):199–212. doi: 10.1016/j.neuron.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackam AS, Singaraja R, Wellington CL, Metzler M, McCutcheon K, Zhang T. et al. The influence of huntingtin protein size on nuclear localization and cellular toxicity. J Cell Biol. 1998;141(5):1097–1105. doi: 10.1083/jcb.141.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindale D, Hackam A, Wieczorek A, Ellerby L, Wellington C, McCutcheon K. et al. Length of huntingtin and its polyglutamine tract influences localization and frequency of intracellular aggregates. Nat Genet. 1998;18(2):150–154. doi: 10.1038/ng0298-150. [DOI] [PubMed] [Google Scholar]
- Gafni J, Hermel E, Young JE, Wellington CL, Hayden MR, Ellerby LM. Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J Biol Chem. 2004;279(19):20211–20220. doi: 10.1074/jbc.M401267200. [DOI] [PubMed] [Google Scholar]
- Xia J, Lee DH, Taylor J, Vandelft M, Truant R. Huntingtin contains a highly conserved nuclear export signal. Hum Mol Genet. 2003;12(12):1393–1403. doi: 10.1093/hmg/ddg156. [DOI] [PubMed] [Google Scholar]
- Peters MF, Nucifora FC Jr., Kushi J, Seaman HC, Cooper JK, Herring WJ. et al. Nuclear targeting of mutant Huntingtin increases toxicity. Mol Cell Neurosci. 1999;14(2):121–128. doi: 10.1006/mcne.1999.0773. [DOI] [PubMed] [Google Scholar]
- Ross CA, Tabrizi SJ. Huntington’s disease: from molecular pathogenesis to clinical treatment. Lancet Neurol. 2010;10(1):83–98. doi: 10.1016/S1474-4422(10)70245-3. [DOI] [PubMed] [Google Scholar]
- Colin E, Zala D, Liot G, Rangone H, Borrell-Pages M, Li XJ. et al. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27(15):2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S. et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron. 2009;64(6):828–840. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman RJ, Aisen PS, De Strooper B, Fox NC, Lemere CA, Ringman JM. et al. Autosomal-dominant Alzheimer’s disease: a review and proposal for the prevention of Alzheimer’s disease. Alzheimers Res Ther. 2011;3(1):1. doi: 10.1186/alzrt59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Lucas JJ, Hen R. Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell. 2000;101(1):57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- Clarke G, Collins RA, Leavitt BR, Andrews DF, Hayden MR, Lumsden CJ. et al. A one-hit model of cell death in inherited neuronal degenerations. Nature. 2000;406(6792):195–199. doi: 10.1038/35018098. [DOI] [PubMed] [Google Scholar]
- Arregui L, Benitez JA, Razgado LF, Vergara P, Segovia J. Adenoviral astrocyte-specific expression of BDNF in the striata of mice transgenic for Huntington’s disease delays the onset of the motor phenotype. Cell Mol Neurobiol. 2011;31(8):1229–1243. doi: 10.1007/s10571-011-9725-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang NK, Lin JH, Lin JT, Lin CI, Liu Em, Lin CJ. et al. A new drug design targeting the adenosinergic system for Huntington’s disease. PLoS One. 2011;6(6):e20934. doi: 10.1371/journal.pone.0020934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder-Keller A, McLear JA, Hathorn T, Messer A. Early or late-stage anti-N-terminal Huntingtin intrabody gene therapy reduces pathological features in B6.HDR6/1 mice. J Neuropathol Exp Neurol. 2010;69(10):1078–1085. doi: 10.1097/NEN.0b013e3181f530ec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Clabough EB, Sarkar S, Futter M, Rubinsztein DC, Zeitlin SO. Deletion of the huntingtin polyglutamine stretch enhances neuronal autophagy and longevity in mice. PLoS Genet. 2010;6(2):e1000838. doi: 10.1371/journal.pgen.1000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The HD iPSC Consortium. Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11(2):264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SG. Targeting histone deacetylases for the treatment of Huntington’s disease. CNS Neurosci Ther. 2010;16(6):348–361. doi: 10.1111/j.1755-5949.2010.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan JS, Bodai L, Pallos J, Poelman M, McCampbell A, Apostol BL. et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413(6857):739–743. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- Li XJ, Li H, Li S. Clearance of mutant huntingtin. Autophagy. 2010;6(5) doi: 10.4161/auto.6.5.12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280(48):40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- Heng MY, Duong DK, Albin RL, Tallaksen-Greene SJ, Hunter JM, Lesort MJ. et al. Early autophagic response in a novel knock-in model of Huntington disease. Hum Mol Genet. 2010;19(19):3702–3720. doi: 10.1093/hmg/ddq285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS. et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc Natl Acad Sci USA. 2005;102(37):13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C, Tartari M, Crotti A, Goffredo D, Valenza M, Conti L. et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat Genet. 2003;35(1):76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- Steffan JS. Does Huntingtin play a role in selective macroautophagy? Cell Cycle. 2010;9(17):3401–3413. doi: 10.4161/cc.9.17.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman SM, Olney JW. Excitotoxicity and the NMDA receptor―still lethal after eight years. Trends Neurosci. 1995;18(2):57–58. doi: 10.1016/0166-2236(95)93869-y. [DOI] [PubMed] [Google Scholar]
- Beal MF. Coenzyme Q10 as a possible treatment for neurodegenerative diseases. Free Radic Res. 2002;36(4):455–460. doi: 10.1080/10715760290021315. [DOI] [PubMed] [Google Scholar]
- Palfi S, Ferrante RJ, Brouillet E, Beal MF, Dolan R, Guyot MC. et al. Chronic 3-nitropropionic acid treatment in baboons replicates the cognitive and motor deficits of Huntington's disease. J Neurosci. 1996;16(9):3019–3025. doi: 10.1523/JNEUROSCI.16-09-03019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante RJ, Andreassen OA, Jenkins BG, Dedeoglu A, Kuemmerle S, Kubilus JK. et al. Neuroprotective effects of creatine in a transgenic mouse model of Huntington's disease. J Neurosci. 2000;20(12):4389–4397. doi: 10.1523/JNEUROSCI.20-12-04389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apolloni S, Montilli C, Finocchi P, Amadio S. Membrane compartments and purinergic signalling: P2X receptors in neurodegenerative and neuroinflammatory events. FEBS J. 2009;276(2):354–364. doi: 10.1111/j.1742-4658.2008.06796.x. [DOI] [PubMed] [Google Scholar]
- Battaglia G, Cannella M, Riozzi B, Orobello S, Maat-Schieman ML, Aronica E. et al. Early defect of transforming growth factor beta1 formation in Huntington’s disease. J Cell Mol Med. 2010;15(3):555–571. doi: 10.1111/j.1582-4934.2010.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordower JH, Isacson O, Emerich DF. Cellular delivery of trophic factors for the treatment of Huntington’s disease: is neuroprotection possible? Exp Neurol. 1999;159:4–20. doi: 10.1006/exnr.1999.7156. [DOI] [PubMed] [Google Scholar]
- Pereira de Almeida L, Zala D, Aebischer P, Deglon N. Neuroprotective effect of a CNTF expressing lentiviral vector in the quinolinic acid rat model of Huntington’s disease. Neurobiol Dis. 2001;8(3):433–446. doi: 10.1006/nbdi.2001.0388. [DOI] [PubMed] [Google Scholar]
- Mittoux V, Joseph JM, Conde F, Palfi S, Dautry C, Poyot T. et al. Restoration of cognitive and motor functions by ciliary neurotrophic factor in a primate model of Huntington’s disease. Hum Gene Ther. 2000;11(8):1177–1187. doi: 10.1089/10430340050015220. [DOI] [PubMed] [Google Scholar]
- Bachoud-Levi AC, Deglon N, Nguyen JP, Bloch J, Bourdet C, Winkel L. et al. Neuroprotective gene therapy for Huntington’s disease using a polymer encapsulated BHK cell line engineered to secrete human CNTF. Hum Gene Ther. 2000;11(12):1723–1729. doi: 10.1089/10430340050111377. [DOI] [PubMed] [Google Scholar]
- Bloch J, Bachoud-Levi AC, Deglon N, Lefaucheur JP, Winkel L, Palfi S. et al. Neuroprotective gene therapy for Huntington's disease, using polymer-encapsulated cells engineered to secrete human ciliary neurotrophic factor: results of a phase I study. Hum Gene Ther. 2004;15(10):968–975. doi: 10.1089/hum.2004.15.968. [DOI] [PubMed] [Google Scholar]
- Hofer M, Pagliusi SR, Hohn A, Leibrock J, Barde Y-A. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990;9(8):2459–2464. doi: 10.1002/j.1460-2075.1990.tb07423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Navarro E, Canudas AM, Akerud P, Alberch J, Arenas E. Brain-derived neurotrophic factor, neurotrophin-3, and neurotrophin-4/5 prevent the death of striatal projection neurons in a rodent model of Huntington’s disease. J Neurochem. 2000;75(5):2190–2199. doi: 10.1046/j.1471-4159.2000.0752190.x. [DOI] [PubMed] [Google Scholar]
- Zuccato C, Ciammola A, Rigamonti D, Leavitt BR, Goffredo D, Conti L. et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293(5529):493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- Borrell-Pages M, Canals JM, Cordelieres FP, Parker JA, Pineda JR, Grange G. et al. Cystamine and cysteamine increase brain levels of BDNF in Huntington disease via HSJ1b and transglutaminase. J Clin Invest. 2006;116(5):1410–1424. doi: 10.1172/JCI27607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty KD, Dreyfus CF, Black IB. Brain-derived neurotrophic factor in astrocytes, oligodendrocytes, and microglia/macrophages after spinal cord injury. Neurobiol Dis. 2000;7(6 Pt B):574–585. doi: 10.1006/nbdi.2000.0318. [DOI] [PubMed] [Google Scholar]
- Giralt A, Rodrigo T, Martin ED, Gonzalez JR, Mila M, Cena V. et al. Brain-derived neurotrophic factor modulates the severity of cognitive alterations induced by mutant huntingtin: involvement of phospholipaseCgamma activity and glutamate receptor expression. Neuroscience. 2009;158(4):1234–1250. doi: 10.1016/j.neuroscience.2008.11.024. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, Myers RH, Stevens TJ, Ferrante RJ, Bird ED, Richardson EP Jr.. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44(6):559–577. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- Wang CE, Tydlacka S, Orr AL, Yang SH, Graham RK, Hayden MR. et al. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington’s disease. Hum Mol Genet. 2008;17(17):2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford J, Shin JY, Roberts M, Wang CE, Li XJ, Li S. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc Natl Acad Sci USA. 2009;106(52):22480–22485. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti P, Camnasio S, Mutti C, Valenza M, Thompson M, Fossale E. et al. Lack of huntingtin promotes neural stem cells differentiation into glial cells while neurons expressing huntingtin with expanded polyglutamine tracts undergo cell death. Neurobiol Dis. 2013;50:160–170. doi: 10.1016/j.nbd.2012.10.015. [DOI] [PubMed] [Google Scholar]
- Palmada M, Centelles JJ. Excitatory amino acid neurotransmission. Pathways for metabolism, storage and reuptake of glutamate in brain. Front Biosci. 1998;3:D701–D718. doi: 10.2741/a314. [DOI] [PubMed] [Google Scholar]
- Hertz L, Dringen R, Schousboe A, Robinson SR. Astrocytes: glutamate producers for neurons. J Neurosci Res. 1999;57:417–428. [PubMed] [Google Scholar]
- Rothstein JD, Dykes-Hoberg M, Pardo CA, Bristol LA, Jin L, Kuncl RW. et al. Knockout of glutamate transporters reveals a major role for astroglial transport in excitotoxicity and clearance of glutamate. Neuron. 1996;16:675–686. doi: 10.1016/s0896-6273(00)80086-0. [DOI] [PubMed] [Google Scholar]
- Shin JY, Fang ZH, Yu ZX, Wang CE, Li SH, Li XJ. Expression of mutant huntingtin in glial cells contributes to neuronal excitotoxicity. J Cell Biol. 2005;171(6):1001–1012. doi: 10.1083/jcb.200508072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faideau M, Kim J, Cormier K, Gilmore R, Welch M, Auregan G. et al. In vivo expression of polyglutamine-expanded huntingtin by mouse striatal astrocytes impairs glutamate transport: a correlation with Huntington’s disease subjects. Hum Mol Genet. 2010;19(15):3053–3067. doi: 10.1093/hmg/ddq212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens JC, Woodman B, Mahal A, Spasic-Boscovic O, Samuel D, Kerkerian-Le Goff L. et al. Impaired glutamate uptake in the R6 Huntington’s disease transgenic mice. Neurobiol Dis. 2001;8(5):807–821. doi: 10.1006/nbdi.2001.0430. [DOI] [PubMed] [Google Scholar]
- Bjorkhem I, Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;24(5):806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- Mauch DH, Nagler K, Schumacher S, Goritz C, Muller EC, Otto A. et al. CNS synaptogenesis promoted by glia-derived cholesterol. Science. 2001;294(5545):1354–1357. doi: 10.1126/science.294.5545.1354. [DOI] [PubMed] [Google Scholar]
- Thiele C, Hannah MJ, Fahrenholz F, Huttner WB. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat Cell Biol. 2000;2(1):42–49. doi: 10.1038/71366. [DOI] [PubMed] [Google Scholar]
- Valenza M, Cattaneo E. Emerging roles for cholesterol in Huntington's disease. Trends Neurosci. 2011;34(9):474–486. doi: 10.1016/j.tins.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Keryer G, Pineda JR, Liot G, Kim J, Dietrich P, Benstaali C. et al. Ciliogenesis is regulated by a huntingtin-HAP1-PCM1 pathway and is altered in Huntington disease. J Clin Invest. 2011;121(11):4372–4382. doi: 10.1172/JCI57552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Wiater P, Conneally PM, Gusella JF, Myers RH. The normal Huntington disease (HD) allele, or a closely linked gene, influences age at onset of HD. Am J Hum Genet. 1993;53(1):125–130. [PMC free article] [PubMed] [Google Scholar]
- Taherzadeh-Fard E, Saft C, Akkad DA, Wieczorek S, Haghikia A, Chan A. et al. PGC-1alpha downstream transcription factors NRF-1 and TFAM are genetic modifiers of Huntington disease. Mol Neurodegener. 2011;6(1):32. doi: 10.1186/1750-1326-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saft C, Epplen JT, Wieczorek S, Landwehrmeyer GB, Roos RA, de Yebenes JG. et al. NMDA receptor gene variations as modifiers in Huntington disease: a replication study. PLoS Curr. 2011;3:RRN1247. doi: 10.1371/currents.RRN1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JM, Ramos EM, Lee JH, Gillis T, Mysore JS, Hayden MR. et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78(10):690–695. doi: 10.1212/WNL.0b013e318249f683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragatsis I, Levine MS, Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat Genet. 2000;26(3):300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- Bhide PG, Day M, Sapp E, Schwarz C, Sheth A, Kim J. et al. Expression of normal and mutant huntingtin in the developing brain. J Neurosci. 1996;16(17):5523–5535. doi: 10.1523/JNEUROSCI.16-17-05523.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JK, Auerbach W, Duyao MP, Vonsattel JP, Gusella JF, Joyner AL. et al. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat Genet. 1997;17(4):404–410. doi: 10.1038/ng1297-404. [DOI] [PubMed] [Google Scholar]
- Godin JD, Colombo K, Molina-Calavita M, Keryer G, Zala D, Charrin BC. et al. Huntingtin is required for mitotic spindle orientation and mammalian neurogenesis. Neuron. 2010;67(3):392–406. doi: 10.1016/j.neuron.2010.06.027. [DOI] [PubMed] [Google Scholar]
- Liu JP, Zeitlin SO. The long and the short of aberrant ciliogenesis in Huntington disease. J Clin Invest. 2011;121(11):4237–4241. doi: 10.1172/JCI60243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigamonti D, Bauer JH, De-Fraja C, Conti L, Sipione S, Sciorati C. et al. Wild-type huntingtin protects from apoptosis upstream of caspase-3. J Neurosci. 2000;20(10):3705–3713. doi: 10.1523/JNEUROSCI.20-10-03705.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li M, Drozda M, Chen M, Ren S, Mejia Sanchez RO. et al. Depletion of wild-type huntingtin in mouse models of neurologic diseases. J Neurochem. 2003;87(1):101–106. doi: 10.1046/j.1471-4159.2003.01980.x. [DOI] [PubMed] [Google Scholar]
- Gu X, Andre VM, Cepeda C, Li SH, Li XJ, Levine MS. et al. Pathological cell-cell interactions are necessary for striatal pathogenesis in a conditional mouse model of Huntington's disease. Mol Neurodegener. 2007;2:8. doi: 10.1186/1750-1326-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grondin R, Kaytor MD, Ai Y, Nelson PT, Thakker DR, Heisel J. et al. Six-month partial suppression of huntingtin is well tolerated in the adult rhesus striatum. Brain. 2012;135(Pt 4):1197–1209. doi: 10.1093/brain/awr333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M. et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci USA. 2007;104(43):17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ. et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol Ther. 2009;17(6):1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet V, Perrin V, Hassig R, Dufour N, Auregan G, Alves S. et al. Sustained effects of nonallele-specific Huntingtin silencing. Ann Neurol. 2009;65(3):276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- Fiszer A, Mykowska A, Krzyzosiak WJ. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011;39(13):5578–5585. doi: 10.1093/nar/gkr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H. et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet. 2009;84(3):351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze EE. et al. Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant huntingtin expression. Cell. 2012;150(5):895–908. doi: 10.1016/j.cell.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles DK, Zhang Z, Ge P, Nelson B, Grondin R, Ai Y. et al. Widespread suppression of huntingtin with convection-enhanced delivery of siRNA. Exp Neurol. 2012;233(1):463–471. doi: 10.1016/j.expneurol.2011.11.020. [DOI] [PubMed] [Google Scholar]
- Marder K, Zhao H, Eberly S, Tanner CM, Oakes D, Shoulson I. et al. Dietary intake in adults at risk for Huntington disease: analysis of PHAROS research participants. Neurology. 2009;73(5):385–392. doi: 10.1212/WNL.0b013e3181b04aa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen JS, Magnotta VA, Mikos AE, Paulson HL, Penziner E, Andreasen NC. et al. Brain structure in preclinical Huntington's disease. Biol Psychiatry. 2006;59(1):57–63. doi: 10.1016/j.biopsych.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Lee JK, Mathews K, Schlaggar B, Perlmutter J, Paulsen JS, Epping E. et al. Measures of growth in children at risk for Huntington disease. Neurology. 2012;79(7):668–674. doi: 10.1212/WNL.0b013e3182648b65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopoulos PC, Aylward EH, Ross CA, Mills JA, Langbehn DR, Johnson HJ. et al. Smaller intracranial volume in prodromal Huntington's disease: evidence for abnormal neurodevelopment. Brain. 2011;134(Pt 1):137–142. doi: 10.1093/brain/awq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehler MF, Gokhan S. Mechanisms underlying neural cell death in neurodegenerative diseases: alterations of a developmentally-mediated cellular rheostat. Trends Neurosci. 2000;23(12):599–605. doi: 10.1016/s0166-2236(00)01705-7. [DOI] [PubMed] [Google Scholar]
- Cattaneo E, Zuccato C, Tartari M. Normal huntingtin function: an alternative approach to Huntington’s disease. Nat Rev Neurosci. 2005;6(12):919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- Muhlau M, Winkelmann J, Rujescu D, Giegling I, Koutsouleris N, Gaser C. et al. Variation within the Huntington’s disease gene influences normal brain structure. PLoS One. 2012;7(1):e29809. doi: 10.1371/journal.pone.0029809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiffer NW, Broadley SA, Hirschberger T, Tavan P, Kretzschmar HA, Giese A. et al. Identification of anti-prion compounds as efficient inhibitors of polyglutamine protein aggregation in a zebrafish model. J Biol Chem. 2007;282(12):9195–9203. doi: 10.1074/jbc.M607865200. [DOI] [PubMed] [Google Scholar]
- Schilling G, Becher MW, Sharp AH, Jinnah HA, Duan K, Kotzuk JA. et al. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum Mol Genet. 1999;8(3):397–407. doi: 10.1093/hmg/8.3.397. [DOI] [PubMed] [Google Scholar]
- Lin CH, Tallaksen-Greeene S, Chien WM, Cearley JA, Jackson WS, Crouse AB. et al. Neurological abnormalities in a knock-in mouse model of Huntington’s disease. Hum Mol Genet. 2001;10(2):137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y. et al. Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet. 2003;12(13):1555–1567. doi: 10.1093/hmg/ddg169. [DOI] [PubMed] [Google Scholar]
- Menalled LB, Sison JD, Dragatsis I, Zeitlin S, Chesselet MF. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington's disease with 140 CAG repeats. J Comp Neurol. 2003;465(1):11–26. doi: 10.1002/cne.10776. [DOI] [PubMed] [Google Scholar]
- von Horsten S, Schmitt I, Nguyen HP, Holzmann C, Schmidt T, Walther T. et al. Transgenic rat model of Huntington’s disease. Hum Mol Genet. 2003;12(6):617–624. doi: 10.1093/hmg/ddg075. [DOI] [PubMed] [Google Scholar]
- Gray M, Shirasaki DI, Cepeda C, Andre VM, Wilburn B, Lu XH. et al. Full-length human mutant huntingtin with a stable polyglutamine repeat can elicit progressive and selective neuropathogenesis in BACHD mice. J Neurosci. 2008;28(24):6182–6195. doi: 10.1523/JNEUROSCI.0857-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Cheng PH, Banta H, Piotrowska-Nitsche K, Yang JJ, Cheng EC. et al. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453(7197):921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu-Taeger L, Petrasch-Parwez E, Osmand AP, Redensek A, Metzger S, Clemens LE. et al. A Novel BACHD Transgenic Rat Exhibits Characteristic Neuropathological Features of Huntington Disease. J Neurosci. 2012;32(44):15426–15438. doi: 10.1523/JNEUROSCI.1148-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel KB, Sapp E, Alexander J, Valencia A, Reeves P, Li X. et al. Polyglutamine expansion in huntingtin alters its interaction with phospholipids. J Neurochem. 2009;110(5):1585–1597. doi: 10.1111/j.1471-4159.2009.06255.x. [DOI] [PubMed] [Google Scholar]
- Li SH, Li XJ. Huntingtin-protein interactions and the pathogenesis of Huntington’s disease. Trends Genet. 2004;20(3):146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu C, Zhong Y, Luo S, Monteiro MJ, Fang S. Huntingtin interacts with the cue domain of gp78 and inhibits gp78 binding to ubiquitin and p97/VCP. PLoS One. 2010;5(1):e8905. doi: 10.1371/journal.pone.0008905. [DOI] [PMC free article] [PubMed] [Google Scholar]