Abstract
A recent study by Massirer et al. in the nematode C. elegans has shown that a family of microRNAs (miRNAs), miR-35-41, regulates the efficiency of RNA interference (RNAi), revealing a new connection between these small RNA pathways. In this commentary, we discuss the potential mechanisms for cross regulation in the miRNA and RNAi pathways and the implications for gene expression. While miRNAs are genetically encoded, the small interfering RNAs (siRNAs) that function in RNAi can originate from processing of exogenous dsRNA (exo-RNAi) or from the production of siRNAs from endogenous transcripts (endo-RNAi). These small RNA pathways involve Dicer and Argonaute proteins and typically use antisense base pairing to target mRNAs for downregulated expression. The discovery that loss of miR-35–41 results in enhanced exo-RNAi sensitivity and reduced endo-RNAi effectiveness suggests that these miRNAs normally help balance the RNAi pathways. The effect of mir-35–41 on RNAi is largely through lin-35, the C. elegans homolog of the tumor suppressor Retinoblastoma (Rb) gene. lin-35/Rb previously has been shown to regulate RNAi sensitivity through unclear mechanisms and the new finding that accumulation of LIN-35/Rb protein is dependent on miR-35–41 adds another layer of complexity to this process. The utilization of miRNAs to control the responsiveness of RNAi exemplifies the cross-regulation embedded in small RNA-directed pathways.
Keywords: C. elegans, RNAi, lin-35, miR-35-41, miRNA, retinoblastoma (Rb)
Gene Silencing By Small RNAs
The first miRNAs were discovered in C. elegans as temporal regulators of development.1,2 Worms deficient in lin-4 or let-7 activity repeat larval cell fates and fail to produce adult characteristics in several tissues. A crucial role for specific miRNAs in human cells has also been established with the realization that aberrant expression of certain miRNAs contributes to diseases, such as cancer, heart ailments and neurodegeneration.3 Consequently, miRNAs are also being developed as both therapeutic molecules and targets for inactivation for medical interventions.4 An encouraging example of this approach is the recent demonstration that inhibition of miR-122 in chimpanzees suppressed hepatitis C virus and improved liver physiology.5 A significant challenge for utilizing miRNA-based therapeutics is to understand the biological effects of these molecules on not just their intended targets but also on other endogenous small RNA pathways that may share processing and effector proteins.
The biogenesis of most miRNAs begins with the synthesis of long primary RNA transcripts (pri-miRNAs) by RNA Pol II (Fig. 1).6 These pri-miRNAs are processed in the nucleus by the RNase III enzyme Drosha (DRSH-1), generating ~65 nt long hairpin precursor miRNAs (pre-miRNAs). The pre-miRNAs are exported to the cytoplasm, where they are processed by the RNase III enzyme Dicer (DCR-1) to form the mature ~22 nt miRNAs. Mature miRNAs are loaded onto Argonaute (ALG-1/-2) proteins to form the miRNA induced silencing complex (miRISC). Typically, miRNAs use imperfect base pairing to recognize target mRNAs and repress their expression. Cofactors associated with miRISC induce destabilization or translational repression of bound target mRNAs.7
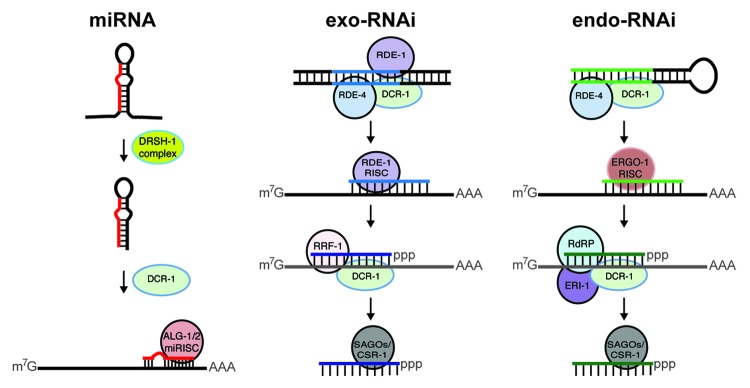
Figure 1. Small RNA pathways. MiRNAs are processed from long structured primary transcripts by the Drosha (DRSH-1) complex into precursor miRNAs, which undergo Dicer (DCR-1) processing to produce mature ~22 nt miRNAs. Mature miRNAs use imperfect complementarity to guide Argonaute-containing (ALG-1 or ALG-2) miRISC to inhibit target mRNA expression. In the exo-RNAi pathway, long exogenous dsRNAs are recognized by RDE-4 and cleaved by the Dicer complex into primary exo-siRNAs, which recruit the Argonaute RDE-1 and other RISC factors to bind complementary sequences in target mRNAs. Amplification of primary siRNAs can generate secondary exo-siRNAs by the RNA dependent RNA Polymerase (RdRP) RRF-1, which uses the previously targeted mRNA as a template. The newly synthesized siRNAs contain 5′ triphosphates and form RISC complexes with the Secondary Argonautes (SAGOs) or the Argonaute CSR-1. In the endo-RNAi pathway, endogenous dsRNAs derived from hairpin RNAs or sense-antisense dsRNAs undergo Dicer processing to form some types of primary endo-siRNAs that are bound by Argonautes, such as ERGO-1. Targeted mRNAs serve as templates for the generation of secondary exo-siRNAs by the RdRP RRF-3 and associated factors. In some cases, mRNAs are directly chosen by an unknown mechanism as templates for the synthesis of 26 nt endo-siRNAs by the RRF-3 complex. An additional phase involving RRF-1 results in 22 nt endo-siRNAs. In the germline, 22G-RNAs are produced by RRF-1/EGO-1 activity that is independent of the DCR-1/ERI-1 complex and are incorporated into Worm-specific Argonautes (WAGOs). The exo- and endo-siRNAs typically base-pair perfectly with target sites and can induce mRNA cleavage if they recruit catalytically active Argonaute proteins.
The ability of exogenous double-stranded RNA (dsRNA) to induce silencing of endogenous genes was also discovered in C. elegans.8 Similar to the miRNA pathway, dsRNA introduced into worms is processed by Dicer into siRNAs that are then incorporated into Argonaute-containing complexes, generally referred to as RISC (Fig. 1).6 By design, the siRNAs are capable of pairing perfectly to mRNA sequences and, if a catalytically competent Argonaute protein is recruited, endonucleolytic cleavage of the target proceeds. This is considered the exo-RNAi pathway to distinguish it from the endo-RNAi pathway, where the siRNAs are generated from endogenous sources of RNA. In worms, some primary endo-siRNAs are thought to derive from dsRNAs formed from intramolecular hairpins or intermolecular duplexes that are cleaved by Dicer (Fig. 1). Secondary endo-siRNAs, which are the more abundant class, are synthesized by RNA dependent RNA polymerases (RdRPs) that use RNAs targeted by primary siRNAs as templates.9-11 In some cases, RdRP activity directly produces endo-siRNAs by copying mRNAs chosen as templates through an unknown mechanism.9,12,13 Endo-siRNAs have also been detected in mammals and flies, although RdRPs capable of amplifying the RNAi pathway have not been found in these organisms.
Forward and reverse genetic screens have identified numerous genes required for an effective RNAi response in C. elegans. At the core, Dicer is required to convert long dsRNA into the siRNA guides.14-16 The dsRNA binding protein, RDE-4, aids in this process and the Argonaute RDE-1 utilizes the siRNAs to initiate exo-RNAi, while other Argonautes with redundant functions mediate endo-RNAi.17-19 The production of secondary siRNAs by the RdRPs RRF-1 and RRF-3 amplify the exo- and endo-RNAi pathways, respectively.9-12,20,21 Some endo-siRNAs are directly synthesized by the RdRPs, RRF-1, RRF-3 and EGO-1, which select mRNA templates through a yet to be defined mechanism.9,12,13 Additionally, worms have a dedicated nuclear RNAi pathway capable of co-transcriptionally regulating gene expression.22
Enhanced RNAi
Since the endo- and exo-RNAi mechanisms share some factors, inactivation of one of these pathways can result in enhanced gene silencing by the other (Table 1). The first indication that this can occur came with the discovery that loss of rrf-3 causes exo-RNAi hypersensitivity in C. elegans.23 Later it was shown that endo-siRNA levels are strongly reduced in rrf-3 mutants, suggesting that depletion of the endogenous pathway might release limiting factors for exo-RNAi.24,25 Even though the endo-RNAi pathway is compromised in rrf-3 mutants, the worms appear phenotypically normal at 20°C but become sterile at 25°C.23 This feature makes rrf-3 mutants a valuable strain for performing RNAi screens with heightened sensitivity.20
Table 1. Genes with enhanced RNAi phenotypes. t.s., temperature sensitive.
| Gene | Gene product | Other phenotypes | References |
|---|---|---|---|
|
eri-1 |
Exonuclease |
t.s. sterile, X-chromosome non-disjunction |
26
|
|
eri-3 |
Novel |
t.s. sterile, X-chromosome non-disjunction |
24
|
|
eri-5 |
Tudor domain protein |
t.s. sterile, X-chromosome non-disjunction |
24
|
|
eri-6/ eri-7 |
Helicase |
none reported |
28
|
|
eri-9 |
Novel |
none reported |
27
|
|
rrf-3 |
RNA-directed RNA polymerase |
t.s. sterile, X-chromosome non-disjunction |
23
|
|
ergo-1 |
Argonaute |
none reported |
19
|
| Transgenic overexpression of sago-1/-2 |
Argonaute |
none reported |
19
|
|
lin-35 |
Retinoblastoma homolog |
t.s. sterile and embryonic lethal, t.s. arrested development, synthetic multivulva |
31
-
33
,
45
|
|
lin-15B, dpl-1, lin-53, lin-9, lin-13, hpl-2 |
Syn muv B genes |
synthetic multivulva |
30
-
33
,
45
|
| mir-35–41 | miRNAs | t.s. embryonic lethal | 38 , 39 |
A screen aimed at finding additional enhanced RNAi (eri) mutants uncovered several genes with diverse functions (Table 1). The eri-1 gene encodes an evolutionarily conserved exonuclease.26 ERI-1 interacts with Dicer and the endo-RNAi pathway depends on this factor for accumulation of some endo-siRNAs.24,25 Although ERI-1 has been shown to degrade siRNAs in vitro,26 its role in the endo-siRNA pathway remains to be fully understood. Like ERI-1, ERI-3 and ERI-5 associate with Dicer and loss of these factors results in temperature sensitive sterility.24,26,27 The helicase ERI-6/7 and novel protein ERI-9 are also required for the accumulation of some endo-siRNAs, but mutations in these genes do not result in obvious phenotypes, indicating that sterility is not always associated with defects in the endo-RNAi pathway.27,28
Another class of RNAi sensitive mutants includes several members of the lin-35/Rb pathway (Table 1). In worms, lin-35 functions in many cellular processes and developmental steps, usually in concert with other genes in the class B SynMuv family.29 Genes categorized as class B SynMuv display the multiple vulva phenotype only when a class A SynMuv gene is also mutated.30 However, single mutants for lin-35/Rb, lin-53 (homolog of the mammalian chromatin modifying complex subunit, RbAp48) and dpl-1 (homolog of the mammalian transcription factor, DP), to name a few, all display enhanced RNAi.31-33 Consistent with other enhanced RNAi mutants, loss of lin-35/Rb results in broad mis-regulation of endo-siRNA targets.34 Some Argonaute genes that function in exo-RNAi are among these upregulated targets. Thus, the RNAi hypersensitivity of lin-35/Rb mutants could arise from increased expression of RNAi factors and reduced competition with the endogenous pathway. However, the mechanistic role of lin-35/Rb and other SynMuv B genes in the endo-RNAi pathway is yet to be elucidated.
miRNA Regulation of the RNAi Pathway
A new gene that enhances RNAi sensitivity was discovered while trying to determine the targets of a family of related miRNAs. Since animal miRNAs typically use partial base-pairing to recognize mRNAs, identifying direct targets and pathways regulated by specific miRNAs has proven challenging.7 Numerous computational approaches, using motifs such as perfect pairing of the 5′ end or seed region of the miRNA, have provided lists of candidate targets for individual miRNAs. Molecular and biochemical methods that detect mRNAs upregulated in the absence of miRNA activity or isolate sequences associated with Argonaute proteins have also been used to successfully match miRNAs to target genes. Additionally, genetic studies have been invaluable for both finding targets as well as validating candidates from other approaches as direct miRNA targets. For example, targets of the first miRNAs, lin-4 and let-7, emerged as suppressors of the lineage and lethality phenotypes exhibited by strains with mutations in these miRNA genes35,36
The availability of deletion strains for most C. elegans miRNAs provides a rich resource for attempting to identify miRNA targets through biochemical and genetic methods.37 While the majority of C. elegans miRNA mutants fail to exhibit obvious phenotypes under normal growth conditions, deletion of the miR-35–41 cluster of miRNAs results in temperature sensitive embryonic lethality.38,39 Presumably, targets of miR-35–41 fail to be repressed in the mutant strain, causing the lethality. Thus, reduction of target gene expression by RNAi is predicted to rescue this phenotype. When Massirer et al. attempted to identify genes that suppressed the lethality of mir-35–41(gk262) mutants, they fortuitously discovered that this strain was generally hypersensitive to RNAi.39 For example, RNAi of unc-22 resulted in paralysis in mir-35–41(gk262) but only the weaker twitching phenotype in wild-type worms. Loss of the miR-35–41 miRNAs produced RNAi hypersensitivity in multiple tissues and stages of development that was similar or greater than that observed in rrf-3 mutants. The detection of enhanced RNAi phenotypes in larval and adult stages is surprising given the fact that expression of miR-35–41 appears to be restricted to embryos.38 Thus, the absence of these miRNAs in early development affects how future cells will respond to RNAi in the developing worm.
Like all of the other enhanced RNAi mutants, deletion of mir-35–41 also results in defects in the endo-RNAi pathway. Endo-siRNA targets were found to be significantly enriched in the list of genes upregulated in mir-35–41(gk262) compared with wild-type embryos by microarray analysis.39 Initially, the scarcity of predicted targets in the mir-35–41(gk262) upregulated gene set was puzzling. The Duchaine lab found that targets of miR-35–41 undergo deadenylation but remain otherwise stable and translationally repressed in embryos,40 thus, providing an explanation for the inability to detect direct targets of these miRNAs through transcriptome profiling.
Several similarities between mir-35–41 and lin-35/Rb mutants, such as extensive mis-regulation of endo-RNAi targets, exceptional RNAi hypersensitivity and temperature sensitive embryonic lethal phenotypes, led to the hunch that these genes might regulate each other.39 While expression of miR-35 miRNA proved independent of lin-35/Rb, protein levels of LIN-35/Rb were found to be reduced almost 5-fold in mir-35–41(gk262) embryos.39 The significance of this deficiency was demonstrated by showing that transgenic overexpression of lin-35/Rb largely rescued the RNAi hypersensitivity of mir-35–41 mutants. Since miRNAs typically repress the expression of direct targets and lin-35/Rb lacks obvious complementary sites to miR-35–41, positive regulation of LIN-35/Rb protein levels by these miRNAs is likely indirect. While an effect on protein but not mRNA levels points to post-transcriptional control of lin-35/Rb by mir-35–41, the mechanism is yet to be unraveled.
Another curious feature of the effect of mir-35–41 and lin-35/Rb on RNAi effectiveness is that it can be inherited. Massirer et al. showed that the enhanced sensitivity to unc-22(RNAi) can be maternally rescued.39 If mother worms contain one wild-type copy of mir-35–41 or lin-35/Rb, this is sufficient to abolish RNAi hypersensitivity in progeny carrying homozygous mutations for either of the genes. These results suggest that mir-35–41 and lin-35/Rb have a far-reaching influence on the function of other small RNA pathways and this effect seems to be established very early in development.
Cross-Regulation Among Small RNA Pathways
Cross-regulation between the exo- and endo-RNAi pathways first became apparent when representative endo-siRNAs failed to accumulate in enhanced RNAi strains, such as eri-1 and rrf-3 mutants.24,25 Conversely, increased levels of exo-siRNAs were detected in these same mutants and an active RNAi response was found to cause reduced endo-siRNAs.24 These observations suggest that factors limiting for siRNA production or stabilization are shared between the exo- and endo-RNAi pathways. The secondary siRNAs produced by RdRP activity require specific Argonautes, which seem to be limiting since overexpression of SAGO-1, for example, results in increased levels of exo- and endo-siRNAs.19 Additionally, mRNAs for Argonaute proteins involved in both pathways, such as SAGO-2, are upregulated in lin-35 mutant larvae, providing a possible explanation for the enhanced RNAi sensitivity of these worms.34 However, this increased expression of Argonaute genes is not observed in mir-35–41 mutant embryos, which express decreased levels of LIN-35/Rb.39 Dicer is another factor utilized by the general siRNA, as well as the miRNA, pathway. In fact, Dicer is the only factor described so far to be broadly required for siRNA and miRNA biogenesis. Thus, the absence of the abundant miR-35–41 miRNAs in embryogenesis could liberate Dicer to allow for more effective RNAi. However, the endo-RNAi pathway is not enhanced and, to the contrary, appears defective in mir-35–41 mutants, where many endo-siRNA targets are misregulated.39 Thus, it remains a mystery as to how loss of the miR-35–41 miRNAs disrupts the endo-RNAi and enhances the exo-RNAi pathways.
Another surprising connection between the RNAi and miRNA pathways is the finding that a miRNA directs the production of endo-siRNAs against a specific target gene. While rde-1 is required for the initiation of exo-RNAi (Fig. 1), its endogenous function has been unclear, as no phenotypes have been attributed to mutations in this gene. Surprisingly, this Argonaute binds representatives of most classes of small RNAs, including miRNAs.41 In particular it associates with miR-243, which guides RDE-1 to a perfect complementary site in the 3′ UTR of Y47H10A.5, which encodes a protein of unknown function. This interaction triggers silencing by a mechanism similar to the exo-RNAi pathway with the utilization of RRF-1 to synthesize endo-siRNAs and the requirement of secondary Argonautes to bind the siRNAs and repress the expression of Y47H10A.5.12,41 While plants also use miRNAs to guide the synthesis of endo-siRNAs, whether the miR-243 example is unique or represents a more prevalent connection between small RNA pathways in animals is yet to be determined.
Concluding Remarks
MiRNAs are predicted to regulate well over half of the transcriptome and, given their extensive complexity, endo-siRNAs are likely to also target a large fraction of the worm genome. Thus, small RNAs potentially influence directly the expression of most genes in this animal. While there are proteins dedicated to the miRNA, exo- and endo-RNAi pathways, there also appears to be substantial cross-regulation. Dicer is at the hub of most small RNA pathways, but it forms distinct complexes for miRNA, endo- and exo-RNAi functions.42 How the small RNA guides and their cofactors sort into the appropriate complexes is not entirely clear. Although loss of specific endo- or exo-RNAi factors results in enhanced activity of the competing pathway, miRNA expression and function is generally unaffected. This separation of miRNA and RNAi pathways has been clouded by the recent discovery that loss of the miR-35–41 miRNAs leads to exo-RNAi hypersensitivity and endo-RNAi deficiency.39 These miRNAs positively regulate the expression of LIN-35/Rb, a factor previously shown to impact RNAi efficiency.31-33 The connection between mir-35–41 and lin-35/Rb is likely indirect but points to a new mechanism for the regulation of LIN-35/Rb protein levels.
Endo-siRNAs have been identified by small RNA cloning not only in C. elegans, but also in Drosophila and mammalian cells and are predicted to target thousands of endogenous genes, particularly mRNAs present in germline and embryos.6 In mouse oocytes, the endo-siRNA pathway is essential for meiosis, while miRNA function is suppressed in this cell type.43 The specific role of endo-siRNAs in oogenesis and the mechanism for blocking miRNA activity in oocytes are currently unknown. In worms, the endo-RNAi pathway plays an important function in sperm development, raising the possibility of a conserved role for endogenous RNAi in gametes and reproduction.13,27,44 It is yet to be shown if disruption of the miRNA or endo-RNAi pathway influences RNAi initiated from exogenous sources in other organisms. While in worms it is well established that perturbations in endo-RNAi can result in enhanced silencing efficiency by exo-siRNAs, the discovery of a family of miRNAs that regulates the effectiveness of these RNAi pathways further interconnects small RNA regulation. Additionally, the work by Massirer et al. demonstrates that miRNAs can broadly regulate other small RNA pathways and, thus, have far reaching effects on gene expression beyond directly targeting specific mRNAs.
Acknowledgments
We thank S. Kennedy (UW Madison) and P. Van Wynsberghe (Colgate U) for critical reading of the manuscript. Funding was provided by the US National Institutes of Health (GM071654), Keck, and Peter Gruber Foundations.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/21835
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–87. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 4.Jackson A, Linsley PS. The therapeutic potential of microRNA modulation. Discov Med. 2010;9:311–8. [PubMed] [Google Scholar]
- 5.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aalto AP, Pasquinelli AE. Small non-coding RNAs mount a silent revolution in gene expression. Curr Opin Cell Biol. 2012;24:333–40. doi: 10.1016/j.ceb.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–82. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 8.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 9.Gent JI, Lamm AT, Pavelec DM, Maniar JM, Parameswaran P, Tao L, et al. Distinct phases of siRNA synthesis in an endogenous RNAi pathway in C. elegans soma. Mol Cell. 2010;37:679–89. doi: 10.1016/j.molcel.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–4. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- 11.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–7. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- 12.Gu W, Shirayama M, Conte D, Jr., Vasale J, Batista PJ, Claycomb JM, et al. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell. 2009;36:231–44. doi: 10.1016/j.molcel.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han T, Manoharan AP, Harkins TT, Bouffard P, Fitzpatrick C, Chu DS, et al. 26G endo-siRNAs regulate spermatogenic and zygotic gene expression in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2009;106:18674–9. doi: 10.1073/pnas.0906378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, et al. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/S0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 15.Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–9. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–71. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–32. doi: 10.1016/S0092-8674(00)81644-X. [DOI] [PubMed] [Google Scholar]
- 18.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–71. doi: 10.1016/S0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 19.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, et al. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–57. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 20.Simmer F, Moorman C, van der Linden AM, Kuijk E, van den Berghe PV, Kamath RS, et al. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 2003;1:E12. doi: 10.1371/journal.pbio.0000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasale JJ, Gu W, Thivierge C, Batista PJ, Claycomb JM, Youngman EM, et al. Sequential rounds of RNA-dependent RNA transcription drive endogenous small-RNA biogenesis in the ERGO-1/Argonaute pathway. Proc Natl Acad Sci U S A. 2010;107:3582–7. doi: 10.1073/pnas.0911908107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guang S, Bochner AF, Burkhart KB, Burton N, Pavelec DM, Kennedy S. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010;465:1097–101. doi: 10.1038/nature09095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–9. doi: 10.1016/S0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 24.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr., Pang K, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–54. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 25.Lee RC, Hammell CM, Ambros V. Interacting endogenous and exogenous RNAi pathways in Caenorhabditis elegans. RNA. 2006;12:589–97. doi: 10.1261/rna.2231506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy S, Wang D, Ruvkun G. A conserved siRNA-degrading RNase negatively regulates RNA interference in C. elegans. Nature. 2004;427:645–9. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 27.Pavelec DM, Lachowiec J, Duchaine TF, Smith HE, Kennedy S. Requirement for the ERI/DICER complex in endogenous RNA interference and sperm development in Caenorhabditis elegans. Genetics. 2009;183:1283–95. doi: 10.1534/genetics.109.108134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischer SE, Butler MD, Pan Q, Ruvkun G. Trans-splicing in C. elegans generates the negative RNAi regulator ERI-6/7. Nature. 2008;455:491–6. doi: 10.1038/nature07274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–91. doi: 10.1016/S0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson EL, Horvitz HR. The multivulva phenotype of certain Caenorhabditis elegans mutants results from defects in two functionally redundant pathways. Genetics. 1989;123:109–21. doi: 10.1093/genetics/123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ceron J, Rual JF, Chandra A, Dupuy D, Vidal M, van den Heuvel S. Large-scale RNAi screens identify novel genes that interact with the C. elegans retinoblastoma pathway as well as splicing-related components with synMuv B activity. BMC Dev Biol. 2007;7:30. doi: 10.1186/1471-213X-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, et al. Loss of LIN-35, the Caenorhabditis elegans ortholog of the tumor suppressor p105Rb, results in enhanced RNA interference. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang D, Kennedy S, Conte D, Jr., Kim JK, Gabel HW, Kamath RS, et al. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–7. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- 34.Grishok A, Hoersch S, Sharp PA. RNA interference and retinoblastoma-related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2008;105:20386–91. doi: 10.1073/pnas.0810589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/S1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 36.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 37.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–73. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Massirer KB, Perez SG, Mondol V, Pasquinelli AE. The miR-35-41 family of microRNAs regulates RNAi sensitivity in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002536. doi: 10.1371/journal.pgen.1002536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu E, Thivierge C, Flamand M, Mathonnet G, Vashisht AA, Wohlschlegel J, et al. Pervasive and cooperative deadenylation of 3’UTRs by embryonic microRNA families. Mol Cell. 2010;40:558–70. doi: 10.1016/j.molcel.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corrêa RL, Steiner FA, Berezikov E, Ketting RF. MicroRNA-directed siRNA biogenesis in Caenorhabditis elegans. PLoS Genet. 2010;6:e1000903. doi: 10.1371/journal.pgen.1000903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thivierge C, Makil N, Flamand M, Vasale JJ, Mello CC, Wohlschlegel J, et al. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nat Struct Mol Biol. 2012;19:90–7. doi: 10.1038/nsmb.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suh N, Blelloch R. Small RNAs in early mammalian development: from gametes to gastrulation. Development. 2011;138:1653–61. doi: 10.1242/dev.056234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gent JI, Schvarzstein M, Villeneuve AM, Gu SG, Jantsch V, Fire AZ, et al. A Caenorhabditis elegans RNA-directed RNA polymerase in sperm development and endogenous RNA interference. Genetics. 2009;183:1297–314. doi: 10.1534/genetics.109.109686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fay DS, Keenan S, Han M. fzr-1 and lin-35/Rb function redundantly to control cell proliferation in C. elegans as revealed by a nonbiased synthetic screen. Genes Dev. 2002;16:503–17. doi: 10.1101/gad.952302. [DOI] [PMC free article] [PubMed] [Google Scholar]


