Abstract
We recently found that genes involved in mitochondrial dynamics and autophagy are required for removal of UVC-induced mitochondrial DNA damage. However, drp-1 and pink-1, unlike the autophagy and fusion genes tested, were not necessary for larval development after exposure. We hypothesized that increased fusion resulting from mutations in these genes facilitated recovery of mitochondrial function. In this work, we investigated this hypothesis by studying the effects of fis-1, fis-2, drp-1 and pink-1 mutations on mitochondrial responses to UVC exposure including ATP levels, mitochondrial DNA copy number, larval development and mitochondrial morphology. Our results suggest that mutations that promote highly networked mitochondria have the capacity to lessen the effects of mitochondrial genotoxicants on the function of this organelle.
Keywords: ultraviolet C radiation, Caenorhabditis elegans, mitochondrial DNA, autophagy, mitochondrial dynamics, pink-1, drp-1, fis-1, fis-2
Introduction
Mitochondrial dynamics (i.e., fusion and fission) play a critical role in maintaining mitochondrial function and are essential for normal development,1 maintenance of mitochondrial DNA (mtDNA),2 exchange of mitochondrial proteins, lipids and nucleoids,3,4 mtDNA replication5,6 and repair,5-7 and more. Mutations in mitochondrial fusion genes OPA1 and MFN2 cause neurodegenerative diseases,8,9 further emphasizing the importance of mitochondrial dynamics particularly in post-mitotic cells. Furthermore, mitochondrial dynamics serve as a compensatory mechanism for mitochondrial dysfunction and stress. Mixing of mitochondrial components through fusion allows for complementation of damaged and undamaged components, a process referred to as functional complementation.3,4 Additionally, dysfunctional mitochondria can be isolated by fission and selective fusion and targeted for autophagic degradation;10 the selective degradation of mitochondria by autophagy is referred to as mitophagy.11
Mitophagy has received significant attention recently due to its potential role in the pathology of Parkinson disease. PTEN-induced kinase 1 (PINK1) and Parkin, both of which are commonly mutated in those with early–onset familial Parkinson disease,12,13 have been shown to mediate mitophagy of depolarized mitochondria under certain in vitro conditions.14,15 Abnormal mitochondrial autophagy and accumulation of dysfunctional mitochondria had been well documented in several neurodegenerative disorders including Parkinson,16,17 and these findings suggest that defective mitochondrial degradation may drive pathogenesis rather than occur as a secondary effect. Furthermore, autophagy and mitophagy have a protective role against mitochondrial dysfunction, apoptosis and cell death following toxicant exposure or cell stress.18-20
mtDNA can be more susceptible than nuclear DNA (nDNA) to damage resulting from environmental contaminant exposure, particularly those genotoxicants that cause “bulky” or helix-distorting lesions including PAHs,21 mycotoxins (e.g., aflatoxin B1),22 UV C radiation (UVC),23 and cisplatin.24 These lesions are irreparable in mtDNA due to a lack of nucleotide excision repair (NER), the repair mechanism utilized in the nuclear genome to remove most “bulky” or helix-distorting DNA damage. We recently reported that UVC-induced lesions in mtDNA are slowly removed in human cells25 and Caenorhabditis elegans.26 In C. elegans, this process was dependent upon fusion, fission and autophagy genes including pink-1 and drp-1. We also found that serial UVC exposure, which resulted in persistent mtDNA damage, resulted in decreases in many mitochondria-mediated functions including oxygen consumption, maintenance of ATP levels and mtDNA copy number and development from the L3 to L4 stage26,27 in wild-type (N2 Bristol strain) nematodes. Interestingly, genetic or pharmaceutical inhibition of autophagy slowed recovery from UVC-induced damage, while recovery was completely inhibited by fusion gene knockout. However, knockout of pink-1 and drp-1 did not affect recovery despite their role in mtDNA damage removal. These data suggested that while removal of mtDNA damage aids in recovery from mitochondrial dysfunction, tolerance of such damage is primarily driven by fusion.
We hypothesized that mutations in drp-1 and pink-1 enhance fusion, thus promoting fusion-induced functional complementation and enhanced mtDNA replication which compensate for lack of removal of damaged mtDNAs. In this work, we investigated this hypothesis by studying the effects of fis-1, fis-2, drp-1 and pink-1 mutations on the response to UVC exposure including ATP levels, mitochondrial DNA copy number, larval development and mitochondrial morphology. The results highlighted the importance of fusion in short-term buffering against mtDNA damage.
Results
We recently found that although pink-1 and drp-1 are involved in removal of irreparable mtDNA damage,26 mutations in these genes had little effect on recovery from mtDNA-damage induced developmental delay in C. elegans. When autophagy was inhibited, recovery was delayed suggesting that removal of damaged mtDNAs does aid in the recovery process. To further probe a potential role for fission in these processes, we tested the effect of mutations in fis-1 and fis-2, apparent homologs to mammalian FIS1. These experiments were performed identically to the damage removal and larval development experiments reported by Bess et al.26 While knockout of these genes alone does not affect mitochondrial morphology or development,28 RNAi knockdown of fis-1 inhibited mtDNA damage removal similarly to drp-1 (Fig. 1). fis-1 knockout, like drp-1 knockout, did not affect recovery from UVC-induced larval arrest (Fig. 1). Knockdown or knockout of fis-2 had no effect (Fig. 1).
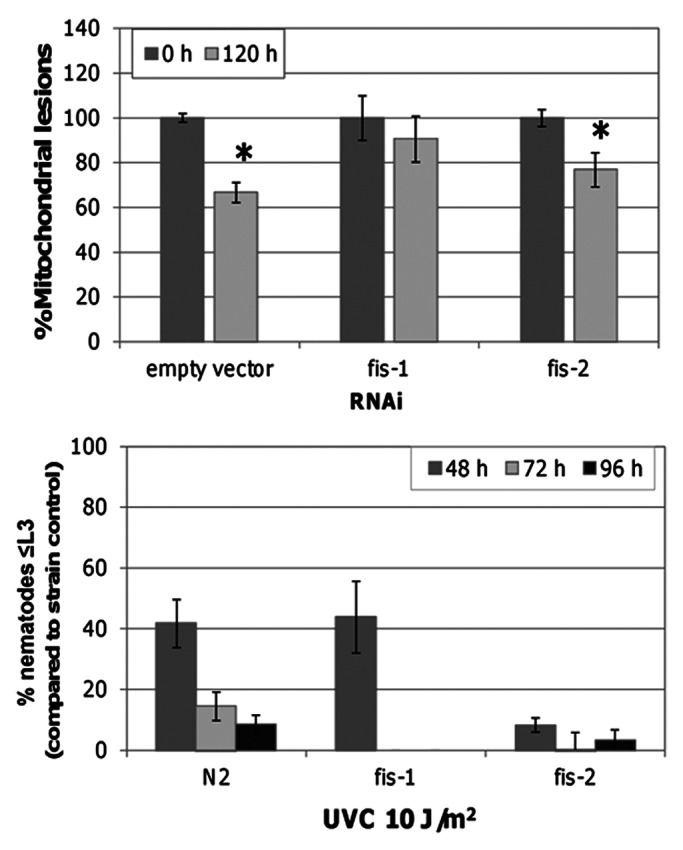
Figure 1.fis-1 is required for removal of UVC-induced mtDNA damage but not recovery from larval arrest. mtDNA lesions were removed in empty vector control and fis-2 120 h post a single UVC exposure in post-mitotic adult C. elegans (glp-1). RNAi knockdown of fis-1 inhibited mtDNA damage removal as indicated by a lack of a significant difference between 0 and 120 h lesion frequency. fis-2 did not significantly inhibit removal of mtDNA damage. fis-1 or fis-2 knockout did not exacerbate larval arrest. Two-way ANOVA indicated a significant interaction between RNAi and recovery (p < 0.05). Asterisks denote a significant difference between 0 h and 120 h mtDNA lesions within RNAi treatment (Fisher’s PLSD, p < 0.05). Percent mtDNA lesions remaining after 120 h was calculated based on 0 h lesion frequency within each RNAi treatment. Asterisks denote p < 0.05.
In combination with our previously-reported results for drp-1 and pink-1,26 the effect of the fis-1 mutation led us to hypothesize that increased fusion, a process critical for recovery from mitochondrial damage, was compensating for lack of removal of damaged mtDNAs in these mutants. We therefore performed additional experiments to test whether knockout of pink-1 and drp-1 would protect against UVC-induced reduction in mtDNA copy number and ATP levels.
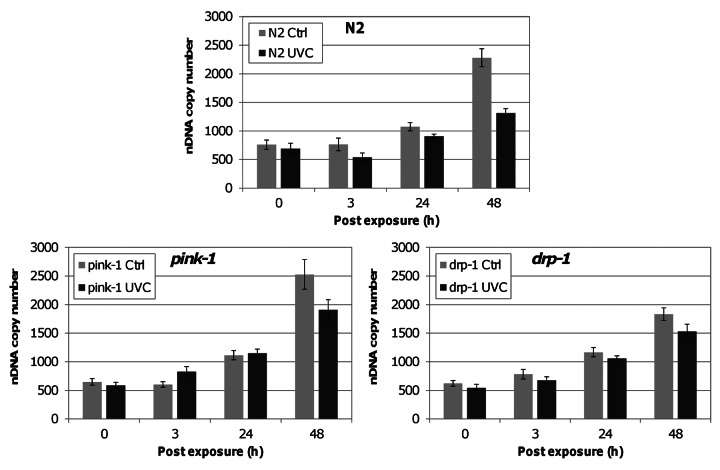
We exposed wild-type (N2), drp-1 and pink-1 nematodes (all in a PE255 background) to serial UVC as described in Bess et al.26 This exposure protocol results in accumulated mtDNA damage while permitting repair of nDNA damage.26 Following the last exposure, we placed L1 nematodes on food and measured DNA copy number and ATP at 0, 3, 24 and 48 h post-exposure. In non-exposed nematodes, we observed a significant increase in mtDNA:nDNA with time (development) in all strains, consistent with previous work.27,29,30 Since we also measured nDNA copy number (below), and since nDNA copy number is closely related to cell number, measurement of both allows us to infer the mtDNA content per cell. UVC exposure significantly decreased the mtDNA copy number per cell in N2 and mutant strains (Fig. 2) at 24 h and 48 h post exposure. The fact that there was little effect of UVC exposure on mtDNA copy number at earlier timepoints likely reflects the biology of the organism. mtDNA apparently undergoes very little replication early in development,27,29,30 and mtDNA replication is in fact dispensable for early larval development.29 Interestingly, pink-1 and drp-1 had significantly higher mtDNA:nDNA compared with N2 in the absence of UVC exposure (Fig. 2) but similar copy number after UVC exposure, such that the percent reduction in mtDNA copy number per cell was greater in these mutant strains (p < 0.05 for strain × treatment interaction).
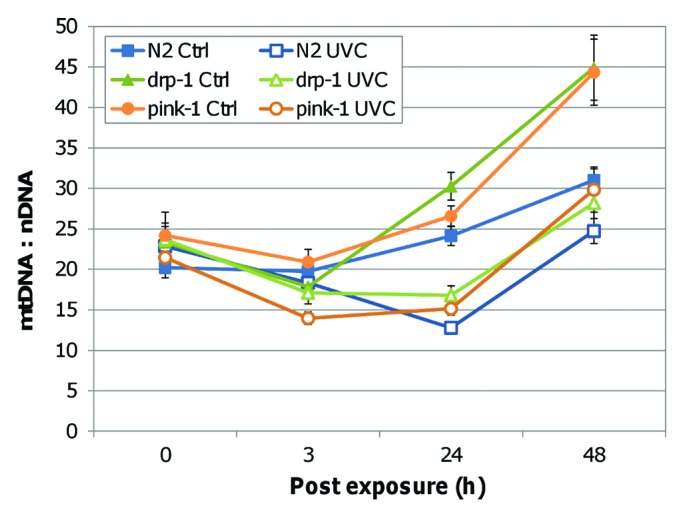
Figure 2. Serial UVC treatment resulted in a greater reduction of mtDNA copy number per cell in pink-1 and drp-1 when compared with the N2 strain. UVC exposure resulted in significantly lower mtDNA:nDNA, by 24 h, in all strains (ANOVA, treatment x timepoint, p < 0.001; FPLSD p < 0.001). pink-1 and drp-1 have higher mtDNA:nDNA under control conditions but were similar to N2 when exposed to UVC (ANOVA, strain × treatment, p < 0.05; FPLSD, p < 0.05).
nDNA copy number was also affected by UVC exposure. Exposed strains had significantly lower nDNA copy number at 48 h (Fig. 3). This data are consistent with our previous observations that this dose of UVC leads to a mild L3 arrest at 48 h.26 The degree of decrease was no greater in pink-1 and drp-1 than N2, also consistent with our previous observation of no more developmental delay in these mutants than N2.26 nDNA copy number in pink-1 and drp-1 nematodes did not differ significantly from N2 in the absence of UVC, but was higher in pink-1 than N2 and drp-1 after UVC exposure (FPLSD).
Figure 3. UVC reduced nDNA copy number. UVC exposure resulted in significantly reduced nDNA copy number at 48 h (ANOVA, treatment x timepoint, p < 0.001; FPLSD p < 0.001). Exposed pink-1 had greater nDNA copy number compared with drp-1 and N2 (ANOVA, strain × treatment, p < 0.05; FPLSD, p < 0.05).
We observed a marked increase in steady-state ATP level between 24 and 48 h in unexposed N2 (PE255s) (Fig. 4), as observed previously.26,31 While this increase was also present in UVC-exposed N2s, ATP level was significantly lower at 48 h compared with untreated worms (Fig. 4). The higher mtDNA copy number that we observed in pink-1 and drp-1 did not translate into higher ATP levels under control conditions. Rather, in the untreated mutants, basal ATP levels were actually significantly lower than N2 and only a small increase in absolute ATP level occurred between 24 and 48 h (Fig. 4). UVC exposure had little to no effect on absolute ATP level at 48 h in the mutant strains (Fig. 4) and the percentage decrease in ATP level at 48 h was greater in N2s (~40%) compared with pink-1 (~20%) and drp-1 (no significant difference). It is important to point out that the slower increase in ATP levels in the mutant strains cannot be attributed to developmental delay. Based on our previous direct observation of larval stage,26 neither drp-1 nor pink-1 shows developmental delay compared with N2. Similarly, our measurement (Fig. 3) of nDNA copy number (which is a proxy for developmental stage, since C. elegans development is invariant with respect to cell division) does not indicate a statistically significant difference in developmental pace in either drp-1 or pink-1 compared with N2.
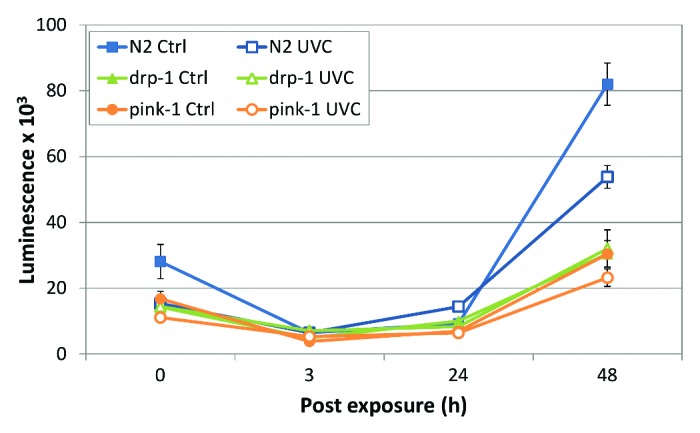
Figure 4.pink-1 and drp-1 have altered ATP levels and the normal activity of pink-1 and drp-1 mediates the effect of UVC exposure on ATP level. UVC exposure resulted in significantly reduced ATP level in N2 but had little to no effect on pink-1 and drp-1 compared with strain and time-matched controls (ANOVA, treatment × strain × timepoint, treatment × timepoint, p < 0.05; FPLSD p < 0.05). Absolute ATP levels in pink-1 and drp-1 were significantly lower than in N2 (ANOVA, strain, p < 0.05; FPLSD, p < 0.05) and ATP levels were not significantly altered by UVC exposure in pink-1 and drp-1 (FPLSD p > 0.05).
As described above, we hypothesized that pink-1 and drp-1 mutations facilitate recovery from UVC exposure by increasing mitochondrial fusion. Mutations in drp-1 have been shown in C. elegans to prevent fission resulting in excessively fused, elongated mitochondria.32 Mutations in pink-1, on the other hand, have been reported to result in reduced length of mitochondrial cristae, as well as increased paraquat sensitivity, in C. elegans.33 We made preliminary examinations of mitochondrial ultrastructure in body muscle cells by transmission electron microscopy (TEM) at 48 h in each strain, with and without exposure to UVC. We identified elongated mitochondria in 1 of 19 mitochondria in control wild type nematodes, 0 of 17 in UVC-exposed wild types, 8 of 12 in control drp-1s, 3 of 8 in UVC-exposed drp-1s, 3 of 7 control pink-1s and 3 of 8 UVC-exposed pink-1s. The TEM analysis allowed us to complement our previous examination of body wall muscle cells26 via fluorescence microscopy with a different methodology. While this was certainly not an exhaustive analysis, we again failed to discern an effect of UVC, despite being able to detect strain-related differences in mitochondrial morphology.
Discussion
The health of the mitochondrial population is maintained in part by mitochondrial dynamics and autophagy, which regulate mitochondrial turnover and selective removal of dysfunctional mitochondrial. Enhanced mitochondrial fusion is particularly important in the response to certain environmental stressors including DNA intercalators,34 UV exposure35 and chronic oxidant exposure.36 Mixing of mitochondrial components allowing for complementation of damaged and undamaged components (i.e., functional complementation) and enhanced mtDNA replication may account for the buffering effect of fusion. The data we present here complements our previous work26 in supporting a role for fused mitochondria in protecting against UVC-induced mitochondrial alterations.
In vitro, UVC-induced photodimers stall DNA polymerase γ, the only polymerase found in mitochondria,37 as well as the mitochondrial RNA polymerase.38 The lower mtDNA:nDNA ratio observed in all UVC-exposed strains, when compared with non-treated strains, likely reflects the in vivo inhibition of mtDNA replication in all strains during development. pink-1 and drp-1 are clearly required for the normal developmental increase in mtDNA:nDNA copy number in the context of UVC exposure. The greater mtDNA copy number per cell that we observed in mutant strains under control conditions may reflect enhanced mtDNA replication and/or lack of mitophagy resulting in accumulation of mtDNA that in the N2 background are degraded. The fact that UVC-exposed mutants have a very large percent decrease in mtDNA per cell supports the hypothesis that these mutations enhance mtDNA replication. If they merely reduced degradation, copy number would be expected to be reduced less by UVC than in N2, rather than more. Since mtDNA copy number per cell is similar among strains at the time of UVC exposure and is similar after 48 h in all strains (rather than being higher in the mutants), we propose that at this dose of UVC, undamaged template for mtDNA replication is limiting across all strains such that the replicative advantage of the mutants is lost. Alternatively, it is possible that the UVC damage somehow interferes with the mechanism that confers replicative advantage on the mutants.
It was interesting that the pink-1 mutants had a higher nDNA copy number than N2 after UVC. The drp-1 mutants did not have a higher nDNA copy number after UVC, but did show an apparently smaller decrease in nDNA copy number after UVC than N2 (this smaller proportional decrease could not be tested formally statistically due to the lack of a significant strain × treatment × timepoint interaction). This result suggests that pink-1 and possibly drp-1 mutants are in fact somewhat protected against the developmental delay caused by UVC exposure, since nDNA copy number is a proxy for development in C. elegans. This interpretation is supported by the fact that there was also a trend toward less inhibition of larval development by UVC in the pink-1 strain26 and the fis-1 strain (Fig. 1). These effects were not statistically significant, but our screen was designed to detect exacerbation, not rescue, of larval arrest.
UVC-exposed N2 nematodes had lower ATP levels particularly at 48 h post exposure, when ATP production and utilization is peaking. We speculate that this may result from a lower number of mtDNAs available to serve as template for transcription or from the presence of transcription blocking lesions that may decrease mtDNA-encoded mitochondrial protein production resulting in lower ATP levels. Additionally, mitochondrial dysfunction may trigger compensatory stress responses that consume ATP at a faster rate thus reducing ATP levels. In pink-1 and drp-1 untreated mutants, basal ATP level was significantly lower than N2 which is consistent with previous studies in Drosophila melanogaster and mammalian cell culture.39-41 We hypothesized that UVC would decrease ATP more in the mutants than in N2. To our surprise, we saw the opposite: ATP levels were decreased less in pink-1 and drp-1 mutants than in N2. One possible explanation for this result is that in these mutants, mtDNA template is not limiting in ATP production. In other words, through functional complementation, damaged and undamaged mtDNAs permit a full set of mitochondrial proteins to be produced. Despite lower basal ATP level and little effect of UVC on ATP, nDNA copy number was not decreased in untreated mutants and UVC similarly decreased nDNA copy number in all strains. Furthermore, Bess et al.26 demonstrated that UVC exposure in these strains resulted in similar developmental delay. Based on these results and those previously reported,26 it does not appear that ATP level directly controls developmental arrest. This conclusion is in keeping with previous evidence that signaling events mediate the L3-L4 larval arrest.42,43
Both enhanced mitochondrial elongation and fragmentation have been reported to result from knockdown or knockout of pink-1 in different species. The only prior analysis of mitochondrial ultrastructure in C. elegans reported shortened cristae in pink-1 mutants.33 While this may at first glance appear to conflict with our results, we note that studies in D. melanogaster and primary human neurons report enhanced elongation of mitochondria yet also fragmentation and disorganization of cristae,44,45 consistent with both sets of C. elegans observations. In addition, it is possible that the effect of pink-1 loss of function on mitochondrial morphology depends on cell type, environmental stressors such as those that promote mitophagy or developmental stage. This is further supported by the fact that in other model organisms genetic inhibition of pink-1 appears to have variable effects.46,47 Overall, these studies show that pink-1 interacts with fusion and fission machinery to alter mitochondrial morphology. Additionally, mitochondrial stress resulting from pink-1 inactivation could enhance mitochondrial fusion indirectly. We did not observe overt changes in mitochondrial morphology induced by UVC exposure. This is consistent with our previous findings in UVC exposed adult C. elegans26 as well as primary human fibroblasts25 using fluorescence microscopy.
In a broader context, these results highlight the importance of fusion in short-term buffering against mtDNA damage. In a genetic background where mitophagy is slowed or inhibited, mitochondrial fusion appears to minimize mitochondrial dysfunction resulting from toxicant exposure. However, this does not appear to be a long-term solution.48 Many wide-spread diseases including neurodegenerative disorders are marked by accumulation of dysfunctional mitochondria and damaged/mutated mtDNA and in some cases (e.g., cardiovascular disease) mitochondrial dysfunction persists despite extensive mitochondrial fusion.
Methods and Materials
C. elegans culture, larval exposures and larval arrest assay
Populations of C. elegans were maintained on K agar plates49 seeded with OP50 bacteria. fis-1(tm1867), VC801 fis-2(gk363) and drp-1 (tm1108) were provided by Ding Xue, University of Colorado, and pink-1(tm1779) was provided by Guy Caldwell, University of Alabama. drp-1 and pink-1 were crossed with the transgenic strain, PE255 (feIs5), provided by Christina Lagido, University of Aberdeen. Larval C. elegans UVC exposure and larval arrest quantification were conducted as previously described.26
Relative ATP assay
Steady-state ATP levels were determined in PE255 as described previously with some modifications.50 Briefly, luminescence (used to determine ATP level) was measured in a 96-well microplate reader (FLUOstar OPTIMA, BMG Labtech) with approximately 300 nematodes per well (in 100 μl). Luminescence buffer (50 μl) was automatically dispensed into each well. Two to three separated experiments with 3–5 technical replicates at each timepoint were conducted.
Real-time PCR quantification of DNA copy number
Absolute mitochondrial and nuclear copy number were measured by quantitative, real-time PCR as previously described.29 Primers used to measure nDNA copy number were designed with Primer3 to amplify a 164bp region of the cox-4 gene: forward 5′-GCC GAC TGG AAG AAC TTG TC-3′; reverse 5′-GCG GAG ATC ACC TTC CAG TA-3′. Two independent experiments with 6 biological replicates per experiment at each timepoint were conducted. For each biological replicate, three technical replicates were averaged from a single real-time PCR run.
TEM
Worms were fixed in 1.5 ml 2% osmium tetroxide in 0.1 M cacodylate buffer for 30 min 48 h after the last UVC exposure and placement on food. Fixed worms were washed twice in 0.1 M cacodylate buffer and transported to the Laboratory for Advanced Electron and Light Optical Methods (LAELOM), North Carolina State University College of Veterinary Medicine. Primarily fixed worms were placed in secondary fixative (1% osmium tetroxide/0.1 M sodium phosphate buffer, pH 7.2–7.4) for 1 h at room temperature then dehydrated in graded ethanol solutions and embedded in Spurr’s resin. Semithin sections (250–500 nm thick) were cut with glass knives, mounted on glass slides and stained with 1% toluidine blue O in 1% sodium borate. These sections were first viewed under a light microscope to verify the presence of worms in the block. Then, ultrathin sections (70–90 nm thick) were cut with a diamond knife, stained with uranyl acetate and lead citrate and examined using a FEI/Philips 208S transmission electron microscope (TEM) at 80 kV accelerating voltage. Analysis of ultrastructure of mitochondria included determination of length of mitochondrial profiles in transverse orientation in body wall muscle cells. Mitochondria were considered elongated if the length was greater than 1.5 microns. Evidence for overt mitochondrial structural alteration (clear mitochondrial swelling or membrane rupture) was not encountered in this study.
mtDNA damage removal
RNAi knockdown of fis-1 and fis-2, adult glp-1 exposure and mtDNA damage quantification via QPCR were performed as previously described.26
Statistical analysis
Multi-factorial ANOVA was used to determine main effects and interactions of experiment variables (e.g., UVC treatment, strain, time) followed by Fisher’s protected least significant difference (FPLSD) posthoc analysis if warranted.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences [1R01-ES017540–01A2], the Society of Toxicology [Colgate-Palmolive Awards for Student Research Training in Alternative Methods] and the American Foundation of Aging Research [GlaxoSmithKline Foundation Award].
Glossary
Abbreviations:
- UVC
ultraviolet C radiation
- NER
nucleotide excision repair
- mtDNA
mitochondrial DNA
- nDNA
nuclear DNA
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/23763
References
- 1.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim JY, Hwang JM, Ko HS, Seong MW, Park BJ, Park SS. Mitochondrial DNA content is decreased in autosomal dominant optic atrophy. Neurology. 2005;64:966–72. doi: 10.1212/01.WNL.0000157282.76715.B1. [DOI] [PubMed] [Google Scholar]
- 3.Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–96. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ono T, Isobe K, Nakada K, Hayashi JI. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat Genet. 2001;28:272–5. doi: 10.1038/90116. [DOI] [PubMed] [Google Scholar]
- 5.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, et al. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–9. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elachouri G, Vidoni S, Zanna C, Pattyn A, Boukhaddaoui H, Gaget K, et al. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011;21:12–20. doi: 10.1101/gr.108696.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rouzier C, Bannwarth S, Chaussenot A, Chevrollier A, Verschueren A, Bonello-Palot N, et al. The MFN2 gene is responsible for mitochondrial DNA instability and optic atrophy ‘plus’ phenotype. Brain. 2012;135:23–34. doi: 10.1093/brain/awr323. [DOI] [PubMed] [Google Scholar]
- 8.Züchner S, Mersiyanova IV, Muglia M, Bissar-Tadmouri N, Rochelle J, Dadali EL, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36:449–51. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 9.Delettre C, Lenaers G, Griffoin JM, Gigarel N, Lorenzo C, Belenguer P, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–10. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 10.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–46. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys. 2007;462:245–53. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 13.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 14.Geisler S, Holmström KM, Treis A, Skujat D, Weber SS, Fiesel FC, et al. The PINK1/Parkin-mediated mitophagy is compromised by PD-associated mutations. Autophagy. 2010;6:871–8. doi: 10.4161/auto.6.7.13286. [DOI] [PubMed] [Google Scholar]
- 15.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vila M, Ramonet D, Perier C. Mitochondrial alterations in Parkinson’s disease: new clues. J Neurochem. 2008;107:317–28. doi: 10.1111/j.1471-4159.2008.05604.x. [DOI] [PubMed] [Google Scholar]
- 17.Santos RX, Correia SC, Wang X, Perry G, Smith MA, Moreira PI, et al. A synergistic dysfunction of mitochondrial fission/fusion dynamics and mitophagy in Alzheimer’s disease. J Alzheimers Dis. 2010;20(Suppl 2):S401–12. doi: 10.3233/JAD-2010-100666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen LH, Chu PM, Lee YJ, Tu PH, Chi CW, Lee HC, et al. Targeting Protective Autophagy Exacerbates UV-Triggered Apoptotic Cell Death. Int J Mol Sci. 2012;13:1209–24. doi: 10.3390/ijms13011209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cotán D, Cordero MD, Garrido-Maraver J, Oropesa-Ávila M, Rodríguez-Hernández A, Gómez Izquierdo L, et al. Secondary coenzyme Q10 deficiency triggers mitochondria degradation by mitophagy in MELAS fibroblasts. FASEB J. 2011;25:2669–87. doi: 10.1096/fj.10-165340. [DOI] [PubMed] [Google Scholar]
- 20.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–6. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 21.Chen RH, Maher VM, Brouwer J, van de Putte P, McCormick JJ. Preferential repair and strand-specific repair of benzo[a]pyrene diol epoxide adducts in the HPRT gene of diploid human fibroblasts. Proc Natl Acad Sci U S A. 1992;89:5413–7. doi: 10.1073/pnas.89.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bedard LL, Massey TE. Aflatoxin B1-induced DNA damage and its repair. Cancer Lett. 2006;241:174–83. doi: 10.1016/j.canlet.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 23.Hanawalt PC. Subpathways of nucleotide excision repair and their regulation. Oncogene. 2002;21:8949–56. doi: 10.1038/sj.onc.1206096. [DOI] [PubMed] [Google Scholar]
- 24.LeDoux SP, Wilson GL, Beecham EJ, Stevnsner T, Wassermann K, Bohr VA. Repair of mitochondrial DNA after various types of DNA damage in Chinese hamster ovary cells. Carcinogenesis. 1992;13:1967–73. doi: 10.1093/carcin/13.11.1967. [DOI] [PubMed] [Google Scholar]
- 25.Bess AS, Ryde IT, Hinton DE, Meyer JN. UVC-induced mitochondrial degradation via autophagy correlates with mtDNA damage removal in primary human fibroblasts. J Biochem Mol Toxicol. 2013;27:28–41. doi: 10.1002/jbt.21440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bess AS, Crocker TL, Ryde IT, Meyer JN. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012;40:7916–31. doi: 10.1093/nar/gks532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leung M, Rooney J, Ryde I, Bernal A, Bess A, Crocker T, et al. Effects of early life exposure to ultraviolet C radiation on mitochondrial DNA content, transcription, ATP production, and oxygen consumption in developing Caenorhabditis elegans. BMC Pharmacology and Toxicology. 2013;14 doi: 10.1186/2050-6511-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breckenridge DG, Kang BH, Kokel D, Mitani S, Staehelin LA, Xue D. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Mol Cell. 2008;31:586–97. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bratic I, Hench J, Henriksson J, Antebi A, Bürglin TR, Trifunovic A. Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic Acids Res. 2009;37:1817–28. doi: 10.1093/nar/gkp018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang WY, Lemire BD. Mitochondrial genome content is regulated during nematode development. Biochem Biophys Res Commun. 2002;291:8–16. doi: 10.1006/bbrc.2002.6394. [DOI] [PubMed] [Google Scholar]
- 31.Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, et al. Ageing is reversed, and metabolism is reset to young levels in recovering dauer larvae of C. elegans. Exp Gerontol. 2002;37:1015–21. doi: 10.1016/S0531-5565(02)00063-3. [DOI] [PubMed] [Google Scholar]
- 32.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–26. doi: 10.1016/S1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 33.Sämann J, Hegermann J, von Gromoff E, Eimer S, Baumeister R, Schmidt E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J Biol Chem. 2009;284:16482–91. doi: 10.1074/jbc.M808255200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashley N, Poulton J. Anticancer DNA intercalators cause p53-dependent mitochondrial DNA nucleoid re-modelling. Oncogene. 2009;28:3880–91. doi: 10.1038/onc.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, et al. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28:1589–600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koopman WJH, Verkaart S, Visch H-J, van der Westhuizen FH, Murphy MP, van den Heuvel LWPJ, et al. Inhibition of complex I of the electron transport chain causes O2-. -mediated mitochondrial outgrowth. Am J Physiol Cell Physiol. 2005;288:C1440–50. doi: 10.1152/ajpcell.00607.2004. [DOI] [PubMed] [Google Scholar]
- 37.Kasiviswanathan R, Gustafson MA, Copeland WC, Meyer JN. Human mitochondrial DNA polymerase γ exhibits potential for bypass and mutagenesis at UV-induced cyclobutane thymine dimers. J Biol Chem. 2012;287:9222–9. doi: 10.1074/jbc.M111.306852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 2012; 1819:979-91. [DOI] [PMC free article] [PubMed]
- 39.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, et al. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gegg ME, Cooper JM, Schapira AH, Taanman JW. Silencing of PINK1 expression affects mitochondrial DNA and oxidative phosphorylation in dopaminergic cells. PLoS One. 2009;4:e4756. doi: 10.1371/journal.pone.0004756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morais VA, Verstreken P, Roethig A, Smet J, Snellinx A, Vanbrabant M, et al. Parkinson’s disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ndegwa S, Lemire BD. Caenorhabditis elegans development requires mitochondrial function in the nervous system. Biochem Biophys Res Commun. 2004;319:1307–13. doi: 10.1016/j.bbrc.2004.05.108. [DOI] [PubMed] [Google Scholar]
- 43.Ventura N, Rea SL, Schiavi A, Torgovnick A, Testi R, Johnson TE. p53/CEP-1 increases or decreases lifespan, depending on level of mitochondrial bioenergetic stress. Aging Cell. 2009;8:380–93. doi: 10.1111/j.1474-9726.2009.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W, Sun Y, Guo S, Lu B. The PINK1/Parkin pathway regulates mitochondrial dynamics and function in mammalian hippocampal and dopaminergic neurons. Hum Mol Genet. 2011;20:3227–40. doi: 10.1093/hmg/ddr235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poole AC, Thomas RE, Andrews LA, McBride HM, Whitworth AJ, Pallanck LJ. The PINK1/Parkin pathway regulates mitochondrial morphology. Proc Natl Acad Sci U S A. 2008;105:1638–43. doi: 10.1073/pnas.0709336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Ouyang Y, Yang L, Beal MF, McQuibban A, Vogel H, et al. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc Natl Acad Sci U S A. 2008;105:7070–5. doi: 10.1073/pnas.0711845105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dagda RK, Cherra SJ, 3rd, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–55. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer JN, Bess AS. Involvement of autophagy and mitochondrial dynamics in determining the fate and effects of irreparable mitochondrial DNA damage. Autophagy. 2012;8:1822–3. doi: 10.4161/auto.21741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis JA, Fleming JT. Basic culture methods. Methods Cell Biol. 1995;48:3–29. doi: 10.1016/S0091-679X(08)61381-3. [DOI] [PubMed] [Google Scholar]
- 50.Lagido C, Pettitt J, Flett A, Glover LA. Bridging the phenotypic gap: real-time assessment of mitochondrial function and metabolism of the nematode Caenorhabditis elegans. BMC Physiol. 2008;8:7. doi: 10.1186/1472-6793-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]