Abstract
Background:
Wnt-signalling has an important role in renal cancer and it is modulated by genistein in other cancers. Recently, microRNAs (miRNAs) have emerged as new regulators of gene expression. Thus, we focused on miRNAs to examine the regulatory mechanism of genistein on the Wnt-signalling pathway in renal cell carcinoma (RCC).
Methods:
Initially, we investigated the effect of genistein on Wnt-signalling (TOPflash reporter assay (TCF reporter assays)) in renal cancer cells, and using microarray identified candidate miRNAs whose expression was decreased by genistein. We performed functional analyses and investigated the relationship between miRNA expression and renal cancer patient outcomes. We also did 3′UTR luciferase assays to look at direct miRNA regulation of Wnt-signalling-related genes.
Results:
Genistein promoted apoptosis while inhibiting RCC cell proliferation and invasion. Genistein also decreased TCF reporter activity in RCC cells. We found that miR-1260b was highly expressed and significantly downregulated by genistein in RCC cells. The expression of miR-1260b was significantly higher in renal cancer tissues compared with normal, and significantly related to overall shorter survival. In addition, miR-1260b promoted renal cancer cell proliferation and invasion in RCC cells. The 3′UTR luciferase activity of target genes (sFRP1, Dkk2, Smad4) was significantly decreased and their protein expression significantly upregulated in miR-1260b inhibitor-transfected renal cancer cells.
Conclusion:
Our data suggest that genistein inhibited Wnt-signalling by regulating miR-1260b expression in renal cancer cells.
Keywords: Genistein, miR-1260b, RCC, Dkk2, sFRP1, Smad4, real-time RT-PCR, MTS assay, invasion assay, apoptosis, FACS, western blot, 3′ UTR luciferase assay, TOPflash
Renal cell carcinoma (RCC) is the third leading cause of death among urological tumours and accounts for 2% of adult malignancies (Jemal et al, 2008). Metastatic lesions are found at diagnosis in about 30% of RCC patients (Bukowski, 1997), and patients with advanced cancer (stage IV) have a significantly reduced 5-year survival rate (<30% Jemal et al, 2008). Compared with other cancers, there are very few tumour markers for renal cancer (Linehan et al, 2010). Also, renal cancer patients respond poorly to conventional chemotherapy because RCC is regarded as a multidrug-resistant cancer (Walsh et al, 2009). Thus, early detection and early treatment is very important for RCC patients as with other cancers.
Several nutritional compounds have been used for the treatment and prevention of renal cancer. Of the nutritional compounds, genistein is one of the most popular isoflavones found in soy products. Previous reports have shown the anticancer effects of genistein, including antiproliferation, anti-invasion, promotion of apoptosis and modulation of cell cycle in a number of cancers including renal cancer (Sasamura et al, 2002; Banerjee et al, 2008; Gullett et al, 2010).
Several cancer pathways have been identified that are modulated by genistein (Sarkar and Li, 2002). Wnt-signalling has an important role in several cancers including renal cancer, and there have been some reports showing the relationship between genistein and the Wnt-signalling pathway (Zhang and Chen 2011; Wang et al, 2012). We initially performed functional analyses of genistein on renal cancer cell lines and observed that genistein inhibited cell proliferation, invasion and promoted apoptosis in renal cancer cell lines. In order to understand the effect of genistein on Wnt-signalling pathway in renal cancer cell lines, we performed TCF reporter assay with genistein treatment and found that genistein inhibited TCF reporter activity in three renal cancer cell lines. We also focused on microRNAs (miRNAs) and their role in the effect of genistein on the Wnt-signalling, as several miRNAs have been identified and reported to have an important role in this pathway (Hirata et al, 2012; Hassan et al, 2012). MicroRNAs have emerged as important regulators of gene expression as they bind to the 3′UTR of target mRNA and repress translation from mRNA to protein or induce mRNA cleavage (Inui et al, 2010). Using a commercial microarray service, we identified several miRNAs that are modulated by genistein, and observed that miR-1260b was highly expressed and significantly downregulated by genistein in two renal cancer cell lines. MiRNA-1260b expression was also significantly higher in renal caner tissues compared with normal kidney tissues and miR-1260b overexpression increased TCF reporter activity in renal cancer cell lines. Thus, we focused on miR-1260b to examine the effect of genistein on Wnt-signalling pathways in renal cancer.
We searched for target genes of miR-1260b involved in the Wnt-signalling pathway with miRNA target prediction algorithms such as ‘http://www.microRNA.org'. In addition, we performed functional analysis with these target genes.
Materials and methods
Clinical samples
A total of 43 patients (29 male and 14 female) with pathologically confirmed clear cell RCC (cc-RCC) were enrolled in this study (Toho University Hospital, Tokyo, Japan). The mean age of the patients was 62-years (range 37–77). They were classified according to the WHO criteria and staged according to the tumour node metastasis (TNM) classification. Namely, T refers to the size of the renal cancer and whether or not it has invaded nearby tissue, N refers to whether regional lymph nodes are involved, and M whether there is distant metastasis or not. Samples were obtained from the patients after written informed consent was obtained at Toho University hospital.
Genistein and transfection
Genistein was purchased from (Sigma-Aldrich Corp., St Louis, MO, USA) and used at a concentration of 25 μℳ based on our laboratory's previous report (Zaman et al, 2012). Renal cancer cells were treated with DMSO or genistein (25 μℳ) for 4 days and used in experiments. Cell culture media was replaced every day.
Cell culture
Renal cancer cell lines (A-498; ATCC number: HTB-44, 786-O; ATCC number: CRL-1932, Caki-2; ATCC number: HTB-47) were purchased from the American Type Culture Collection (Manassas, VA, USA). The cell lines were cultured in RPMI 1640 medium supplemented with 10% foetal bovine serum.
Total RNA and protein extraction
RNA (miRNA and total RNA) was extracted from formalin-fixed, paraffin-embedded (FFPE) human renal cancer, and adjacent non-cancerous normal kidney tissues using a miRNeasy FFPE kit (Qiagen, Valencia, CA, USA) after microdissection based on a pathologist's reviews. To digest DNA, the Qiagen RNase-Free DNase kit was used. Total RNA was also extracted from renal cancer cell lines using a miRNeasy mini kit (Qiagen). Cells were lysed with RIPA buffer (Pierce, Brebieres, France) containing protease inhibitors (Sigma, St. Louis, MO, USA). Protein quantification was done using a BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA).
MicroRNA transfection
Pre-miR miRNA precursors (negative control (miR-NC) or hsa-miR-1260b (miR-1260b), Ambion, Foster City, CA, USA) were transiently transfected into renal cancer cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA), according to the manufacturer's instructions.
Anti-miR miRNA inhibitor (negative control (inh-NC) or miR-1260b inhibitor (miR-1260b inhibitor), Ambion) was transiently transfected into renal cancer cells by siPORT NeoFX Transfection Agent (Ambion), according to the manufacturer's instructions. After transfection, cells were incubated at 37 °C for 48 h until used.
TOPflash luciferase assay
TOPflash luciferase assays (TCF/LEF reporter assays) were performed to assess the effect of genistein effect on the Wnt-signalling pathway. After DMSO (4 days) or genistein (25 μℳ–4days) treatment, cells were trypsinised and resuspended in 48-well plates overnight. Cells were then transiently co-transfected with TOPflash (Upstate, Lake Placid, NY, USA) and pRL-TK Vector (Promega, Madison, WI, USA), encoding renilla luciferase as an internal control for transfection efficiency using lipofectamine LTX (Invitrogen).
To look at the effect of miR-1260b on the β-catenin-dependent pathway, we overexpressed miR-1260b with miR-1260b precursor (48 h transfection) or knocked down miR-1260b with miR-1260b inhibitor (48 h transfection). Cells were trypsinised and resuspended in 48-well plates overnight, and then were transiently co-transfected with TOPflash (Upstate) and pRL-TK Vector (Promega).
Cell viability and cell invasion
Cell viability was measured 3 days after transfection with MTS (CellTiter 96 Aqueous One Solution Cell Proliferation Assay, Promega). Data are the mean±s.d. of four independent experiments. Cell invasion assays were performed with the CytoSelect 24-well cell invasion assay kit (Cell BioLab, San Diego, CA, USA), according to the manufacturer's instructions. Transfected cells were resuspended in culture medium without FBS and placed in the upper chamber in triplicate. After 48 h incubation at 37 °C (5% CO2), cells migrating through the membrane were stained. The results were expressed as invaded cells quantified at OD 560 nm.
Apoptosis analyses
Cells (4 days after DMSO or genistein transfection, or 48 h after transfection with miRNA precursor or plasmid) were washed twice with 1 × PBS and trypsinised. After inactivating trypsin in complete medium, the cells were resuspended in ice-cold 1 × binding buffer (70 μl). Annexin V-FITC solution (10 μl) and 7-AAD viability dye (20 μl) (Beckman Coulter, Brea, CA, USA) were added to 70 μl of the cell suspensions. After incubation for 15 min in the dark, 400 μl of ice-cold 1 × binding buffer was added. The apoptotic distribution of the cells in each sample was then determined using a FACS (Cell Lab QUANTA SC, Beckman Coulter, Fullerton, CA, USA). Data are the mean±s.d. of four independent experiments.
Plasmid construction and 3′UTR-luciferase assay
Individual plasmids were constructed for each binding site in the 3′UTR of mRNA from potential target genes based on http://www.microRNA.org information. Then we confirmed miR-1260b binding to the target genes mRNA 3′UTR by luciferase assay with miR-1260b precursor. PmirGLO Dual-Luciferase miRNA Target Expression Vector was used to perform 3′UTR-luciferase assay (Promega). The primer sequences used for plasmid inserts are shown in Table 1. In a total volume of 20 μl, 5 μl each of 100 μℳ forward primer and reverse primer, 2 μl of 10 × annealing buffer (100 mℳ Tris-HCl, pH 7.5, 1 ℳ NaCl, 10 mℳ EDTA) and 8 μl water were added to a 200-μl PCR tube and incubated at 95 °C for 5 min, and then placed at room temperature for 1 h. The oligonucleotides were ligated into the PmeI-XbaI site of pmirGLO Dual-Luciferase miRNA Target Expression Vector. Colony direct PCR was performed for insert recognition using REDTaq (Sigma). The primers used for PCR were as follows: forward primer, 5′-CGTGCTGGAACACGGTAAAA-3′ reverse primer, 5′-GCAGCCAACTCAGCTTCCTT-3′. PCR parameters for cycling were as follows: 94 °C for 3 min, 30 cycles of PCR at 94 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s, 72 °C for 10 min and 4 °C for 10 min. The PCR product was digested with NotI (TaKaRa/Fisher Scientific, Pittsburgh, PA, USA). The sizes of vectors containing inserts were about 200 bp and 100 bp by electrophoresis, as the NotI recognition sequence was incorporated into the primers. For miR-1260b precursor transfection, renal cancer cells were co-transfected with miR-NC and pmirGLO or miR-1260b and pmirGLO Dual-Luciferase miRNA Target Expression Vectors, using Lipofectamine 2000 (Invitrogen). Luciferase activity was assessed using the Dual-Luciferase Reporter Assay System (Promega; 48 h after their transfection), and the ratio of firefly/renilla luciferase activity was determined.
Table 1. Primer sequences used for plasmid construction.
| Name | Sequence |
|---|---|
| Dkk2-1S |
5′-AAACTAGCGGCCGCTAGTtgtccaatatatGTGGGAaT-3′ |
| Dkk2-1AS |
5′-CTAGAtTCCCACatatattggacaACTAGCGGCCGCTAGTTT-3′ |
| sFRP1-1S |
5′-AAACTAGCGGCCGCTAGTcctcgGGTGGGGTGGGAgT-3′ |
| sFRP1-1AS |
5′-CTAGAcTCCCACCCCACCcgaggACTAGCGGCCGCTAGTTT-3′ |
| sFRP1-2S |
5′-AAACTAGCGGCCGCTAGTgctagGGCCAAGGTGGGAtT-3′ |
| sFRP1-2AS |
5′-CTAGAaTCCCACCTTGGCCctagcACTAGCGGCCGCTAGTTT-3′ |
| Smad4-1S |
5′-AAACTAGCGGCCGCTAGTccGGGAGTAGATCGTGGGAtT-3′ |
| Smad4-1AS |
5′-CTAGAaTCCCACGATCTACTCCCggACTAGCGGCCGCTAGTTT-3′ |
| sFRP1 NheI cloning forward primer |
5′-GCTAGCgaggtccctggaagtttgc-3′ |
| sFRP1 XhoI cloning reverse primer |
5′-CTCGAGcgactggattacaatgtccactac-3′ |
| Smad4 NheI cloning forward primer |
5′-GCTAGCttgcttcagaaattggagacatatt-3′ |
| Smad4 XhoI cloning reverse primer | 5′-CTCGAGattttgtagtccaccatcctgataa-3′ |
Overexpression plasmid of target genes (sFRP1, Dkk2, Smad4) and functional analyses
Plasmids containing the human full-length cDNA fragment of Dkk2 (Genbank accession number NM_014421) were purchased from Origene (Rockville, MD, USA). This clone (pCMV6-Dkk2) expresses the complete Dkk2 open reading frame with a tag (MYC/DDK) at the COOH-terminus.
In order to construct other target gene (sFRP1 and Smad4)-overexpressing plasmids, the gene was amplified with total RNA from human adult normal kidney tissues (catalogue number: R1234142-50, Biochain Institute, Newark, CA, USA) by reverse transcription–PCR (RT-PCR). The sequences of primers for cloning are shown in Table 1.
PCR products were cloned into the pTargeT-Mammalian Expression Vector System (Promega). Then, pCMV6-sFRP1 or -Smad4 was obtained by subcloning a NheI–XhoI fragment from pTargeT-sFRP1 or -Smad4 into the NheI–XhoI site of pCMV6-Entry Vector. Initially, we transfected pCMV6-empty and pCMV6-Dkk2 or -sFRP1, or -Smad4 into renal cancer cells, and RNA and protein were extracted. Overexpression of Dkk2, sFRP1 or Smad4 was validated by real-time PCR and western analysis, and then functional analyses were performed.
Quantitative real-time RT-PCR
Quantitative real-time RT-PCR was performed in triplicate with an Applied Biosystems Prism 7500 Fast Sequence Detection System, using TaqMan universal PCR master mix, according to the manufacture's protocol (Applied Biosystems Inc., Foster City, CA, USA). The TaqMan probes and primers were purchased from Applied Biosystems. Human GAPDH and RNU48 were used as an endogenous control. Levels of RNA expression were determined using the 7500 Fast System SDS software version 1.3.1 (Applied Biosystems).
Western analysis
Total cell protein (15–20 μg) was used for western blotting. Samples were resolved in 4–20% Precise Protein Gels (Thermo Scientific, Rockford, IL, USA) and transferred to PVDF membranes (Amersham Biosciences, Fairfield, CT, USA). The membranes were immersed in 0.3% skim milk in TBS containing 0.1% Tween 20 for 1 h, and probed overnight at 4 °C with primary polyclonal and monoclonal antibody against sFRP1(number 3534), Smad4 (number 9515) and β-tubulin (number 2128) from Cell Signaling Technology, Beverly, MA, USA. Dkk2 antibody was purchased from ABGENT (number AP1522a, San Diego, CA, USA). Blots were washed in TBS containing 0.1% Tween 20 and labelled with horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibody (Cell Signaling Technology). Proteins were enhanced by chemiluminescence (Amersham ECL plus Western Blotting detection system) for visualisation. The protein expression levels were expressed relative to β-tubulin levels.
Statistical analysis
All statistical analyses were performed using StatView (version 5; SAS Institute Inc., Cary, NC, USA). Error bars in all the figures represent s.d. Statistical significance was determined using the Students t-test or analysis of variance for functional analysis. A P-value of <0.05 was regarded as statistically significant.
Results
Effect of genistein on renal cancer cells
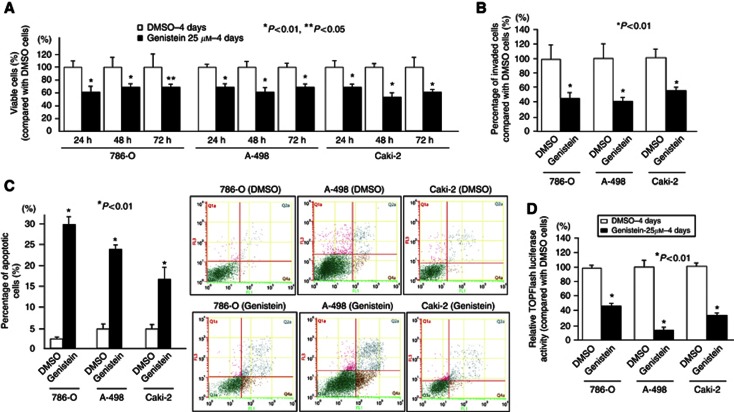
Cancer cell proliferation (Figure 1A), invasion ability (Figure 1B) was significantly decreased by 4 days of genistein treatment (25 μℳ) in renal cancer cell lines (786-O, A-498, Caki-2). The percentage of apoptotic cells was significantly higher in genistein-treated renal cancer cells (Figure 1C). TOPflash reporter assay activity was significantly decreased with genistein in the three renal cancer cell lines (Figure 1D).
Figure 1.
Genistein effect on renal cancer cell lines (786-O, A-498 and Caki-2). Three cell lines were treated with DMSO or genistein (25 μℳ) for 4 days. (A) Cell viability assay. (B) Invasion assay. (C) Apoptosis assay-FACS based. (D) TCF/LEF reporter assays. Error bars represent ±s.d.
Relationship between miR-1260b expression level and clinical characteristics
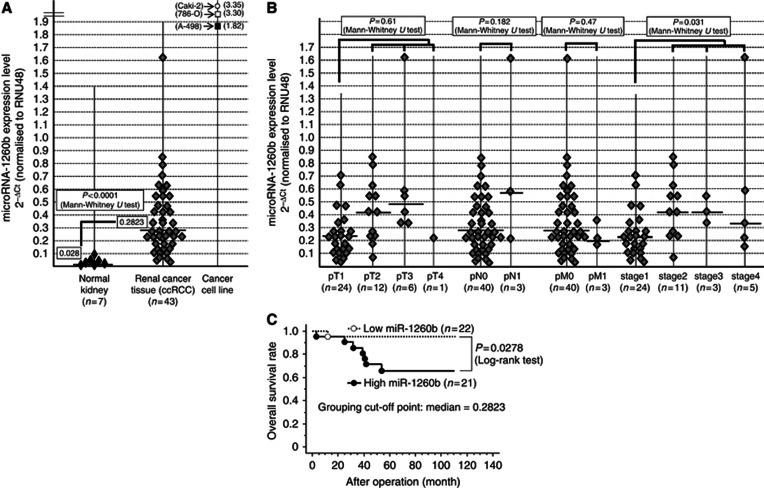
The miR-1260b expression levels were compared on the basis of real-time RT-PCR results. The miR-1260b expression level was significantly higher in renal cancer tissues (n=43, all cc-RCC patients) compared with normal kidney tissues (Figure 2A). MiR-1260b expression was compared with several clinical parameters, including grade, pathological tumor classification, pathological lymph node status, pathological metastasis status, stage and outcomes (overall survival). The expression of miR-1260b was significantly higher in stage 2+3+4 patients compared with stage 1 patients (Figure 2B). Overall survival based on miR-1260b expression was also compared in renal cancer tissues. Namely, we divided RCC patients with mean 2−ΔCt value (0.2823) of miR-1260b into two groups. Overall survival was significantly shorter in the high miR-1260b-expressing group (Figure 2C).
Figure 2.
miR-1260b expression and association with clinical parameters in renal cancer tissues. (A) miR-1260b expression in human clinical samples and renal cancer cell lines (786-O, A-498 and Caki-2). (B) Association of miR-1260b with clinic-pathological parameters. (C) Kaplan-Meier plots of overall survival.
Effect of miRNA-1260b on renal cancer cell viability, invasion and apoptosis
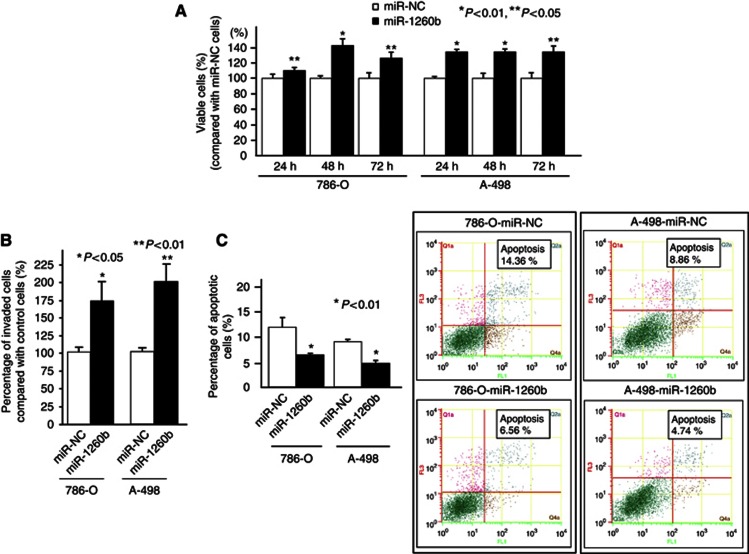
At 24 h after transfection of miR-NC or miR-1260b into renal cancer cells (786-O and A-498), increased miR-1260b expression level was verified by real-time RT-PCR (fold change; 5859 and 6240, respectively). Then, cell viability analysis (MTS assay) and cell invasion assays were performed using miRNA-1260b precursor or control transfectants of renal cancer cells (Figures 3A–C). We observed significantly increased cell viability (Figure 3A) and invasion (Figure 3B) in miR-1260b-transfected renal cancer cells compared with miR-NC-transfected cells. The percentages of apoptotic cells was significantly decreased in miR-1260b-transfected cells (Figure 3C).
Figure 3.
Effect of miR-1260b overexpression on renal cancer cell function (786-O and A-498).Two renal cancer cell lines (786-O and A-498) were transiently transfected with either miR-1260b precursor or control (miR-NC). (A) Cell viability assay. (B) Invasion assay. (C) Flow cytometric analysis of apoptosis in miR-NC or miR-1260b-transfected renal cancer cells.
Genistein, miR-1260b and TCF LEF reporter assay
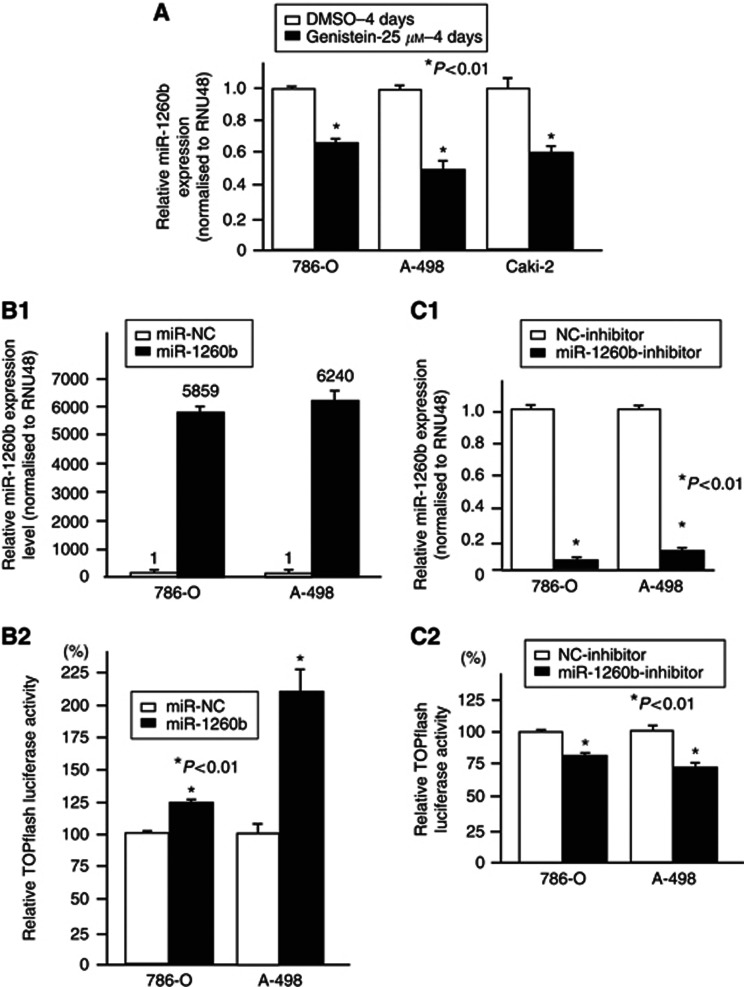
The expression of miR-1260b was significantly decreased by genistein treatment in three renal cancer cell lines (786-O, A-498 and Caki-2; Figure 4A). TOPflash reporter assay activity was significantly increased by overexpression of miR-1260b (Figure 4B). However, the TCF reporter activity was significantly decreased by knockdown of miR-1260b (Figure 4C).
Figure 4.
(A) Effect of genistein on miR-1260b expression in renal cancer cell lines, (B1) Overexpression of miR-1260b after miR-NC or miR-1260b precursor transfection. (B2) TCF/LEF reporter assays. (C1) Relative miR-1260b expression after NC-inhibitor or miR-1260b inhibitor transfection. (C2) TCF/LEF reporter assays.Genistein, miRNA-1260b and TCF reporter assay.
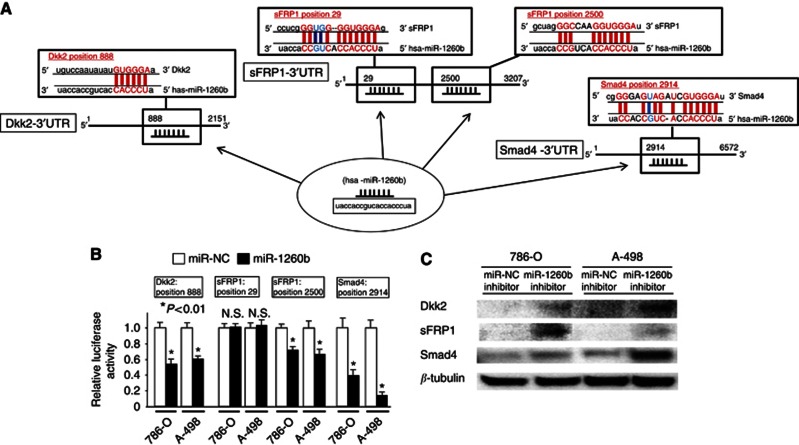
3′UTR luciferase assay and lower protein expression in miR-1260b transfectant
We identified two Wnt antagonist genes (Dkk2 and sFRP1) and one Wnt-related gene (Smad4) as potential target genes for miR-1260b, based on http://www.microRNA.org.
Dkk2 mRNA has one potential binding site for miR-1260b within its 3′UTR (Figure 5A).
Figure 5.
miR-1260b binds to the 3′UTR of Dkk2, sFRP1 and Smad4 mRNAs and downregulates expression. (A) Dkk2, sFRP1 and Smad4 3′UTR sequence and complementary miR-1260b binding site. (B) 3′UTR luciferase assay (miR-NC and miR-1260b precursor), error bars represent ±s.d.. (C) Protein expression of Dkk2, sFRP1, Smad4 and β-tubulin in miR-NC inhibitor or miR-1260b inhibitor-transfected renal cancer cells (786-O, A-498).
The relative luciferase activity was significantly decreased at position 888 in miR-1260b precursor-transfected cells (Figure 5B). The sFRP1 mRNA has two potential binding sites (position 29 and position 2500) for miR-1260b within its 3′UTR (Figure 5A). The relative luciferase activity is shown as a ratio of firefly and renilla luciferase activity for each sample. The relative luciferase activity was significantly decreased when the position 2500 construct was used in miR-1260b precursor-transfected cells (Figure 5B). Smad4 mRNA has one potential binding site for miR-1260b within its 3′UTR, and the relative luciferase activity was significantly decreased at position 2904 in miR-1260b precursor-transfected cells (Figure 5B). As target gene protein expression is low in 786-O and A-498, we performed western analysis to observe any changes in protein expression with miR-1260b inhibitor. As shown in Figure 5C, protein expression of target genes was significantly increased after miR-1260b inhibitor transfection (Figure 5C). This result suggests that the Dkk2, sFRP1 and Smad4 mRNAs are direct targets of miR-1260b.
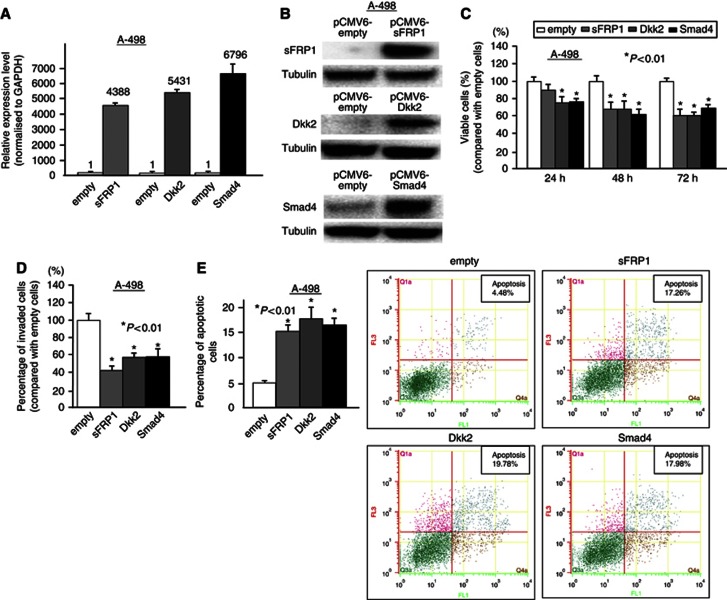
Effect of sFRP1, Dkk2 and Smad4 on renal cancer cell (A-498)
To assess whether miR-1260b exerts its function through sFRP1, Dkk2 and Smad4, we overexpressed these mRNAs. The overexpression effect was confirmed by measuring mRNA (Figure 6A) and protein levels (Figure 6B). As shown in Figures 6A and B, the relative sFRP1, Dkk2 and Smad4 mRNA and protein expression levels were significantly increased. Then we examined MTS, cell invasion and apoptosis assays in empty/sFRP1/Dkk2/Smad4-transfected A-498 cells. As shown in Figure 6, cell viability (Figure 6C) and invasion (Figure 6D) were significantly inhibited in sFRP1/Dkk2/Smad4-transfected A-498 cells. Also, percentages of apoptotic cells were significantly increased in sFRP1/Dkk2/Smad4-transfected A-498 cells compared with empty vector cells (Figure 6E).
Figure 6.
Effect of sFRP1, Dkk2 and Smad4 overexpression on renal cancer cell (A-498) function. At 24 h after transfection of either pCMV6-empty, pCMV6-sFRP1, pCMV6-Dkk2 or pCMV6-Smad4 into renal cancer cell (A-498), sFRP1, Dkk2 and Smad4 expression levels were verified by real-time RT-PCR (A) and western analysis (B). (C) Cell viability assay. (D) Invasion assay. (E) Apoptosis assay. Error bars represent ±s.d.
Discussion
Renal cancer is regarded as a chemotherapy-resistant and radiation-resistant cancer. Currently, multikinase inhibitors, mTOR inhibitors and a vascular endothelial growth factor (VEGF)-neutralising antibody have been approved for the treatment of advanced RCC (Pirrotta et al, 2011); however, more effective and less toxic treatments are needed for RCC patients. In this study, we focused on genistein because genistein has been used for cancer prevention and treatment in many cancers, and several studies have documented the effectiveness of genistein (Sarkar and Li, 2002). To understand the mechanism of genistein, some researchers have found that several cancer pathways such as Akt and NF-kappaB are repressed by genistein (Sarkar and Li, 2002). According to previous reports, renal cancer cell proliferation was decreased by genistein by inhibiting angiogenic factors such as VEGF and basic fibroblast growth factor in renal cancer cell lines (Sasamura et al, 2002). Our laboratory has reported that genistein decreased cell proliferation in A-498 and ACHN cells (Majid et al, 2009). In this study, we found that genistein inhibited cancer cell proliferation and invasion, and promoted apoptosis in three renal cancer cell lines. We also examined the relationship between genistein and the Wnt-signalling pathway in renal cancer cell lines as we have found that Wnt-signalling has an important role in renal cancer progression, invasion and metastasis. Wnt-signalling pathways include the β-catenin-dependent (canonical) and -independent pathways (non-canonical; (Katoh and Katoh 2007). The Wnt pathway is usually activated in cancer cells causing unphosphorylated β-catenin to accumulate in the cytoplasm, move to the nucleus and bind to TCF/LEF, thereby transcriptionally regulating Wnt target genes and promoting tumourigenesis (Ahmad et al, 2011). Thus, TCF reporter assays (TCF/LEF reporter assays) have been used to monitor the Wnt β-catenin-dependent pathway. We performed TCF/LEF reporter assays and found that TOPflash activity was significantly decreased in genistein-treated cells compared with controls. Although the effects of genistein on the β-catenin-dependent pathway have been shown in other cancers (Lee et al, 2012; Zhang and Chen 2011), our results appear to be the first for RCC cells.
Another aim of this study was to determine the effect of genistein on Wnt-signalling pathway in RCC. To investigate this, we focused on miRNA. The miRNA has emerged as a new regulator of gene expression in cancer pathways (Iorio and Croce, 2012). As numerous miRNAs have been identified, we used miRNA microarray to identify candidate miRNAs whose expression was altered by genistein in renal cancer cell lines. In the miRNA microarray platform used in this study, miR-1260b expression was the highest in two cancer cell lines (A-498 and Caki-2), and miR-1260b expression was significantly decreased by genistein. In addition, there are very few studies on concerning miR-1260b in the literature, so we focused on it in this study. As shown in Figure 2, miR-1260b expression was significantly higher in renal cancer compared with normal tissues, and its expression was significantly correlated with a shorter overall survival after radical nephrectomy in RCC patients based on Kaplan-Meier plots. Functional analysis also showed that miR-1260b overexpression increased renal cancer cell proliferation and invasion while apoptosis was inhibited. Therefore, this study indicates that miR-1260b functions as an oncogene in RCC. As onco-miR-1260b expression was significantly decreased by genistein, TOPflash activity was also altered by miR-1260b. Namely, when miR-1260b was overexpressed, TOPflash activity was increased, whereas in contrast TOPflash activity was decreased when miR-1260b was knocked down. These results show that miR-1260b regulates the β-catenin-dependent pathway in RCC cell lines.
As miRNAs exert their effects by regulating expression of target genes, we looked for target genes of miRNA that are involved in the Wnt-signalling pathway. On the basis of the http://www.microRNA.org computer algorithm, we identified three potential target genes of miRNA-1260b, two of which are Wnt antagonist genes (sFRP1 and Dkk2). The third, Smad4, has also been reported to inhibit the Wnt β-catenin signalling pathway (Freeman et al, 2012). 3′UTR luciferase assay and western analysis conclusively showed that miR-1260b binds to these three target genes and regulate their expression. Thus, our results indicate that sFRP1, Dkk2 and Smad4 are direct targets of miR-1260b. We investigated the function of these target genes by overexpressing them in a renal cancer cell line (A-498). As shown in Figure 6, sFRP1, Dkk2 and Smad4 decreased cancer cell proliferation and invasion, and the percentage of apoptotic cells was significantly increased by sFRP1, Dkk2 and Smad4 overexpression in A-498 cells.
Previous reports have shown that overexpression of sFRP1 and Dkk2 have an antitumor effect on renal cancer cells (Hirata et al, 2009; Cooper et al, 2012) and our results are similar to them. Loss of Smad4 has been reported to have an important role in initiating some cancers (Yang and Yang 2010), and our results indicate that Smad4 may have an important role in renal cancer.
Genes may be silenced or inactivated by loss of heterozygosity (LOH), gene mutation, DNA methylation, histone modification and miRNA post-transcriptionally. Dkk2 and sFRP1 genes have been reported to be silenced in renal cancer tissues by DNA methylation (Urakami et al 2006; Hirata et al 2009), and Smad4 gene silencing was reported to be associated with LOH in RCC (Hirata et al, 2005). Thus, the result of this study suggests that sFRP1, Dkk2 and Smad4 genes may be silenced by miRNA-1260b transcriptionally as well as DNA methylation and LOH.
In conclusion, this is the first report documenting that genistein decreased TCF reporter activity in renal cancer cell lines. In addition, miR-1260b expression was significantly higher in renal cancer tissues compared with normal kidney tissues, and its expression was significantly associated with shorter patient overall survival, suggesting that miR-1260b functions as an oncogene. As we have shown, miR-1260b expression was significantly decreased by genistein and TCF reporter activity paralleled that of miR-1260b expression in renal cancer cell lines. Our results also show that miR-1260b inhibits several tumour suppressor genes associated with Wnt-signalling, such as sFRP1, Dkk2 and Smad4. Our results indicate that genistein may be of therapeutic benefit to renal cancer patients.
Acknowledgments
We thank Dr Roger Erickson for his support and assistance with the preparation of the manuscript. This study was supported by National Center for Research Resources of the National Institutes of Health through Grant Number RO1CA138642, RO1CA130860, RO1CA160079, I01BX001123, VA Merit Review, VA Program Project (PI: R Dahiya) and Yamada Science Foundation.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Ahmad I, Morton JP, Singh LB, Radulescu SM, Ridgway RA, Patel S, Woodgett J, Winton DJ, Taketo MM, Wu XR, Leung HY, Sansom OJ. β-Catenin activation synergizes with PTEN loss to cause bladder cancer formation. Oncogene. 2011;30:178–189. doi: 10.1038/onc.2010.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Li Y, Wang Z, Sarkar FH. Multi-targeted therapy of cancer by genistein. Cancer Lett. 2008;269:226–242. doi: 10.1016/j.canlet.2008.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukowski RM. Natural history and therapy of metastatic renal cell carcinoma: the role of interleukin-2. Cancer. 1997;80:1198–1220. doi: 10.1002/(sici)1097-0142(19971001)80:7<1198::aid-cncr3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Cooper SJ, von Roemeling CA, Kang KH, Marlow LA, Grebe SK, Menefee ME, Tun HW, Colon-Otero G, Perez EA, Copland JA. Reexpression of tumor suppressor, sFRP1, leads to antitumor synergy of combined HDAC and methyltransferase inhibitors in chemoresistant cancers. Mol Cancer Ther. 2012;11:2105–2115. doi: 10.1158/1535-7163.MCT-11-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman TJ, Smith JJ, Chen X, Washington MK, Roland JT, Means AL, Eschrich SA, Yeatman TJ, Deane NG, Beauchamp RD. Smad4-mediated signaling inhibits intestinal neoplasia by inhibiting expression of β-catenin. Gastroenterology. 2012;142:562–571. doi: 10.1053/j.gastro.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gullett NP, Ruhul Amin AR, Bayraktar S, Pezzuto JM, Shin DM, Khuri FR, Aggarwal BB, Surh YJ, Kucuk O. Cancer prevention with natural compounds. Semin Oncol. 2010;37:258–281. doi: 10.1053/j.seminoncol.2010.06.014. [DOI] [PubMed] [Google Scholar]
- Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL, Stein GS, Lian JB. miR-218 directs a wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem. 2012;287:42084–42092. doi: 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata H, Hinoda Y, Nakajima K, Kawamoto K, Kikuno N, Kawakami K, Yamamura S, Ueno K, Majid S, Saini S, Ishii N, Dahiya R. Wnt antagonist gene DKK2 is epigenetically silenced and inhibits renal cancer progression through apoptotic and cell cycle pathways. Clin Cancer Res. 2009;15:5678–5687. doi: 10.1158/1078-0432.CCR-09-0558. [DOI] [PubMed] [Google Scholar]
- Hirata H, Matsuyama H, Matsumoto H, Korenaga Y, Ohmi C, Sakano S, Yoshihiro S, Naito K. Deletion mapping of 18q in conventional renal cell carcinoma. Cancer Genet Cytogenet. 2005;163:101–105. doi: 10.1016/j.cancergencyto.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, Dahiya R. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012;7:e51056. doi: 10.1371/journal.pone.0051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics. 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M. WNT signaling pathway and stem cell signaling network. Clin Cancer Res. 2007;13:4042–4045. doi: 10.1158/1078-0432.CCR-06-2316. [DOI] [PubMed] [Google Scholar]
- Lee J, Ju J, Park S, Hong SJ, Yoon S. Inhibition of IGF-1 signaling by genistein: modulation of E-cadherin expression and downregulation of β-catenin signaling in hormone refractory PC-3 prostate cancer cells. Nutr Cancer. 2012;64:153–162. doi: 10.1080/01635581.2012.630161. [DOI] [PubMed] [Google Scholar]
- Linehan WM, Bratslavsky G, Pinto PA, Schmidt LS, Neckers L, Bottaro DP, Srinivasan R. Molecular diagnosis and therapy of kidney cancer. Annu Rev Med. 2010;61:329–343. doi: 10.1146/annurev.med.042808.171650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, Saini S, Tanaka Y, Dahiya AV, Khatri G, Dahiya R. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009;30:662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta MT, Bernardeschi P, Fiorentini G. Targeted-therapy in advanced renal cell carcinoma. Curr Med Chem. 2011;18:1651–1657. doi: 10.2174/092986711795471293. [DOI] [PubMed] [Google Scholar]
- Sasamura H, Takahashi A, Miyao N, Yanase M, Masumori N, Kitamura H, Itoh N, Tsukamoto T. Inhibitory effect on expression of angiogenic factors by antiangiogenic agents in renal cell carcinoma. Br J Cancer. 2002;86:768–773. doi: 10.1038/sj.bjc.6600152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar FH, Li Y. Mechanisms of cancer chemoprevention by soy isoflavone genistein. Cancer Metastasis Rev. 2002;21:265–280. doi: 10.1023/a:1021210910821. [DOI] [PubMed] [Google Scholar]
- Urakami S, Shiina H, Enokida H, Hirata H, Kawamoto K, Kawakami T, Kikuno N, Tanaka Y, Majid S, Nakagawa M, Igawa M, Dahiya R. Wnt antagonist family genes as biomarkers for diagnosis, staging, and prognosis of renal cell carcinoma using tumor and serum DNA. Clin Cancer Res. 2006;12:6989–6997. doi: 10.1158/1078-0432.CCR-06-1194. [DOI] [PubMed] [Google Scholar]
- Walsh N, Larkin A, Kennedy S, Connolly L, Ballot J, Ooi W, Gullo G, Crown J, Clynes M, O'Driscoll L. Expression of multidrug resistance markers ABCB1 (MDR-1/P-gp) and ABCC1 (MRP-1) in renal cell carcinoma. BMC Urol. 2009;9:6. doi: 10.1186/1471-2490-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Q, Chen H. Genistein affects histone modifications on Dickkopf-related protein 1 (DKK1) gene in SW480 human colon cancer cell line. PLoS One. 2012;7:e40955. doi: 10.1371/journal.pone.0040955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Yang X. Smad4-mediated TGF-beta signaling in tumorigenesis. Int J Biol Sci. 2010;6:1–8. doi: 10.7150/ijbs.6.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman MS, Shahryari V, Deng G, Thamminana S, Saini S, Majid S, Chang I, Hirata H, Ueno K, Yamamura S, Singh K, Tanaka Y, Tabatabai ZL, Dahiya R. Up-regulation of microRNA-21 correlates with lower kidney cancer survival. PLoS One. 2012;7:e31060. doi: 10.1371/journal.pone.0031060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen H. Genistein attenuates WNT signaling by up-regulating sFRP2 in a human colon cancer cell line. Exp Biol Med. 2011;236:714–722. doi: 10.1258/ebm.2011.010347. [DOI] [PubMed] [Google Scholar]