Abstract
Background:
Autoantibodies have been detected in sera before diagnosis of cancer leading to interest in their potential as screening/early detection biomarkers. As we have found autoantibodies to MUC1 glycopeptides to be elevated in early-stage breast cancer patients, in this study we analysed these autoantibodies in large population cohorts of sera taken before cancer diagnosis.
Methods:
Serum samples from women who subsequently developed breast cancer, and aged-matched controls, were identified from UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) and Guernsey serum banks to formed discovery and validation sets. These were screened on a microarray platform of 60mer MUC1 glycopeptides and recombinant MUC1 containing 16 tandem repeats. Additional case–control sets comprised of women who subsequently developed ovarian, pancreatic and lung cancer were also screened on the arrays.
Results:
In the discovery (273 cases, 273 controls) and the two validation sets (UKCTOCS 426 cases, 426 controls; Guernsey 303 cases and 606 controls), no differences were found in autoantibody reactivity to MUC1 tandem repeat peptide or glycoforms between cases and controls. Furthermore, no differences were observed between ovarian, pancreatic and lung cancer cases and controls.
Conclusion:
This robust, validated study shows autoantibodies to MUC1 peptide or glycopeptides cannot be used for breast, ovarian, lung or pancreatic cancer screening. This has significant implications for research on the use of MUC1 in cancer detection.
Keywords: autoantibodies, MUC1 aberrant glycoforms, screening, breast cancer, early diagnosis
Early detection remains the most promising approach to improving survival of cancer patients. In breast cancer, mammographic screening significantly impacts on mortality (Kerlikowske et al, 1995), although controversy exists as to possible overdiagnosis (Gøtzsche and Nielsen, 2011). Use of serum biomarkers for the early detection of cancer, before development of clinical symptoms, is an attractive goal being minimally invasive and potentially highly cost-effective. Screening for autoantibodies rather than the antigens may improve sensitivity as substantial tumour mass may be required before the antigen can be detected in serum, whereas autoantibodies act as biological amplifiers increasing the detectable signal from the antigen. Indeed, specific autoantibodies have been reported to be present in sera of patients before clinical diagnosis of cancer (Lubin et al, 1995; Li et al, 2005; Zhong et al, 2006; Desmetz et al, 2011; Chapman et al, 2012; Lu et al, 2012; Pedersen et al, 2013) and are under trial for the detection of lung cancer (Chapman et al, 2012).
The antigen MUC1 is upregulated in breast and other cancers, and is also aberrantly glycosylated, adding another dimension to the cancer specificity. The mucin carries large numbers of O-linked glycans which in breast cancer are truncated, resulting in the appearance of cancer-specific glycopeptide epitopes, which are antigenically distinct (Sørensen et al, 2006; Tarp et al, 2007; Wandall et al, 2010). Using a novel O-glycopeptide/glycoprotein array-based assay detecting IgG antibodies, we have recently shown that autoantibodies reactive with the cancer-associated glycopeptide epitopes can be detected in sera from 30% of early breast cancer patients (Blixt et al, 2011). Moreover, high levels of autoantibodies were significantly associated with reduced risk of relapse and increased time to metastasis (Blixt et al, 2011). These encouraging results led us to explore whether autoantibodies to tumour-associated glycoforms of MUC1 could be used as a serum biomarker for detection of breast and other cancers before clinical diagnosis.
With a few exceptions that used prospective sera collections (Pinheiro et al, 2010; Chapman et al, 2012; Lu et al, 2012; Pedersen et al, 2013), most serum biomarker discovery studies for early detection of cancer have been carried out on sera collected from patients at diagnosis, (Chapman et al, 2007; Zhong et al, 2008; Boyle et al, 2011; Lacombe et al, 2013) or involved small cohorts with no independent validation (Lubin et al, 1995; Li et al, 2005; Robertson et al, 2005; Zhong et al, 2006; Pereira-Faca et al, 2007). Here, we report on MUC1 glycopeptide microarray analysis of serum samples from over 2000 women from the general population in nested breast cancer case–control studies involving two prospectively collected serum banks of initially healthy women: the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) with 202 638 women recruited between 2001–2005 (Menon et al, 2009) and the Guernsey island serum bank with 6500 women recruited between 1977–1991 (Fentiman et al, 2006). Complete follow-up for cancer and mortality was available for both cohorts. Moreover, it was possible to include an additional control group from the Guernsey cohort that consisted of women who had not developed any form of cancer up to 32 years (range 18–32) after serum donation. As MUC1 is expressed by most adenocarcinomas, we were also able to screen sera from apparently healthy women in the UKCTOCS bank who later developed lung, pancreatic or ovarian cancer and controls.
This robust, validated study reported here, which has been carried out in accordance with STARD guidelines, is important as considerable effort and resources are being focused on the analyses of autoantibodies for early cancer detection and MUC1 a commonly used antigen.
Materials and methods
Subjects
The cases and controls were identified from two cohorts (UKCTOCS and Guernsey) of women who were clinically healthy at recruitment. Serum samples from individuals participating in the multimodal arm of UKCTOCS trial (Menon et al, 2009) were included. In this trial, 50 640 women were randomised to the multimodal group between 2001 and 2005, and donated samples annually until 2011. All women were followed via electronic flagging for cancer registration and death through the Medical Research Information System in England and Wales and the Central Services Agency and Cancer Registry in Northern Ireland. The volunteers consented to use of their serum samples in further secondary studies and all exceptions to this were recorded and honoured. This current study was approved by the joint University College London (UCL)/University College London Hospital (UCLH) Committees on the Ethics of Human Research (Committee A; Ref 05/Q0505/57) on the 7th February 2008. Controls were women from the same trial centre who had no history of any cancer at last follow-up, and who had donated serial serum samples during the same period.
The Guernsey cohort consists of 6500 women aged 35 and over living on the island of Guernsey who were recruited between 1977–1991 (Fentiman et al, 2006). All women donated a serum sample at recruitment and underwent mammographic screening. Women were followed up by regular visits to Guernsey to access the hospital records and obtain copies of all female death certificates. Information was also received from the South West Cancer Registry for female Guernsey patients treated in Southampton (mainland UK). Thus, all cases of cancer have documentation by pathology report or death certificate, and occasionally radiology reports. Additionally, checks were made at the island registry (The Greffe) for changes of name through marriage or deed poll. Written informed consent was obtained from each volunteer. This consent covered use of the serum for the investigations of cancer biomarkers and access to the women's medical records. Ethical approval to allow the access to patients' medical records of the volunteers who donated sera to the Guernsey bank was obtained (Guernsey and Alderney Ethical Committee).
Samples
Breast cancer
Discovery sample set: Sera from the UKCTOCS women who went on to develop invasive breast cancer were identified. Women with previous cancer history at recruitment were excluded. The cases were matched to controls (healthy women with no notification of cancer when the case was identified) 1 : 1 on age at donation (±1 year) and length of frozen storage (±1 year).
Validation sample sets
UKCTOCS case–control set: Sera from women who developed invasive breast cancer after randomisation to UKCTOCS and sera donation (not included in the discovery set, no previous cancer history and had physician-confirmed breast cancer with data on histological subtype and either stage/grade or both) were included. These were matched to controls, women from UKCTOCS with no cancer history either at recruitment or when the case was identified, on age (±1 year), length of storage (±1 year) and trial centre.
Guernsey case–control set: Sera were identified from women who developed breast cancer up to 30 years post donation. Cases were matched to two sets of controls: (1) women who had no diagnosis of cancer at the time the case was diagnosed; (2) women who had not developed cancer during follow-up (range 18–32 years) on age (±1 year) and date of sample collection (±1 year).
Serum storage: UKCTOCS; all samples were stored in liquid nitrogen since collection. The aliquot used for this analysis had never been previously freeze thawed. Once the aliquot was thawed, it was divided into smaller aliquots and refrozen. Guernsey; all sample were stored aliquoted at −20 °C and the aliquot used for this analysis had never been freeze thawed. Once an aliquot was thawed, it was divided into smaller aliquots and refrozen.
Ovarian, lung and pancreatic cancer
Sera from UKCTOCS women who developed ovarian, lung and pancreatic cancers following randomisation to the trial were identified. Controls were healthy trial participants who did not have a notification of cancer at the time the case samples were identified. Cases were matched 1 : 1 to controls on the basis of age at donation (±1year) and time in freezer (±1year).
Microarray autoantibody assay
Glycopeptides and recombinant glycoprotein: Synthetic 60mer MUC1 peptides corresponding to 3 twenty amino-acid tandem repeats and MUC2 peptides were synthesised and glycosylated in preparative scale using recombinant enzymes produced in insect cells (Tarp et al, 2007; Wandall et al, 2010; Blixt et al, 2011), see Table 1 for a list of the glycopeptides, their glycan structure and sites of glycosylation. As with our previous study (Blixt et al, 2011) this study confirmed that the use of 20mers (one tandem repeat) or 60mers (three tandem repeats) gave comparable results (see Supplementary Figure 1). All structures were purified by preparative HPLC and analysed by MALDI-TOF as described (Tarp et al, 2007). Recombinant MUC1-based glycoproteins carrying ST and T were produced in CHOK1 cells as described by Bäckström et al (2003) and those without O-linked glycans or carrying the Tn glycan were produced in CHO ldlD cells.
Table 1. Structure of the MUC1 glycopeptides used on the arrays.
| Name | Peptide backbone | Glycan | Sites per repeat |
|---|---|---|---|
| MUC1 unglycosylated |
(VTSAPDTRPAPGSTAPPAHG)3 |
N/A |
N/A |
| MUC1 unglycosylatedRec |
16 tandem repeats plus the amino terminus expressed in CHO cells |
N/A |
N/A |
| MUC1core3 |
(VTSAPDTRPAPGSTAPPAHG)3 |
GlcNAcβ1,3GalNAcα- |
5 |
| MUC1STn |
(VTSAPDTRPAPGSTAPPAHG)3 |
Neu5Acα2,6GalNAcα- |
5 |
| MUC1T |
(VTSAPDTRPAPGSTAPPAHG)3 |
Galβ1,3GalNAcα- |
5 |
| MUC1TRec |
16 tandem repeats plus the amino terminus expressed in CHO cells |
Galβ1,3GalNAcα- |
Average of 4.3 |
| MUC1Tn |
(VTSAPDTRPAPGSTAPPAHG)3 |
GalNAcα- |
5 |
| MUC1TnRec |
16 tandem repeats plus the amino terminus expressed in CHO cells |
GalNAcα- |
Average of 3.8 |
| MUC1STRec |
16 tandem repeats plus the amino terminus expressed in CHO cells |
Neu5Acα2,3Galβ13GalNAcα- |
Average of 4.3 |
| MUC2Core3 |
|
GlcNAcβ1,3GalNAcα- |
11 |
| MUC2Tn | GalNAcα- | 11 |
Microarrays: Glycopeptides arrays were custom printed by ArrayIt (Sunnyvale, CA, USA) onto Schott Nexterion Slide H (Schott AG, Mainz, Germany) with 16 arrays per slide. Each peptide or glycopeptide was printed (0·5 nl) in triplicate and at three concentrations (50, 25 and 12·5 μℳ) and each recombinant protein at 250, 125 and 62·5 pg. The quality control of printed glycopeptides was visualised by staining with glycoform-specific lectins and antibodies as described previously (Blixt et al, 2011). Human IgG was also printed as a positive control for the second antibody and to orientate the arrays.
Sera were diluted 1 : 50 and the arrays screened as described by Blixt et al (2011). The slides were scanned in a PerkinElmer Scanarray and the images quantified with ProScanArray Express software programme (PerkinElmer, Cambridge, UK). Spots were identified using automated spot finding or manual adjustments for occasional irregularities.
All samples were screened in duplicate with blinding as to case or control. The same positive control serum from a breast cancer patient from the cohort used in our previous paper (Blixt et al, 2011) was used on every slide and where possible, cases and their controls were screened on the same slide. Sera were rescreened if the duplicates did not agree based on a similarity measure between them (see Supplementary Methods). If there was still inadequate agreement after the rescreen, the samples were removed from the analyses.
Statistical analysis
In order to quantitatively detect any difference between distributions, the data were split into quartiles. The quartile division was performed on the entire set of data with no information regarding the grouping of samples into cancer cases or controls. The null hypothesis was that the samples would be distributed randomly over quartiles. A χ2-test was performed to see if there was a significant difference between the numbers of samples in each quartile. This test was chosen to determine whether differences between the two groups exist.
In addition, using two s.d. values from the mean of the control sera for each antigen as cutoff, the fraction of autoantibody positive sera were compared between cases and controls in the two validation sets. Receiver operator characteristic (ROC) curves were constructed for each of the MUC1 antigens on the arrays and by giving equal weight to all features a generalised ROC curve was formed.
Results
Sample selection
From the UKCTOCS, 258 women who went on to develop invasive breast cancer up to 4 years following sample donation were identified for the discovery set. Eighteen women were ineligible because of a previous history of breast (12) or other cancer (6) at randomisation. From the remaining 240 cases, 273 serum samples were available meaning that 33 women donated two serum samples at different times prediagnosis. Analysis of these duplicate samples from the same woman showed no significant differences between the values (data not shown). There were 273 samples from 273 control women. There was no significant difference in baseline characteristics between cases and controls (Table 2A) although there was a trend for increased weight in the cases at randomisation, a known risk factor for breast cancer. All women were postmenopausal.
Table 2. Baseline characteristics of the cohorts used in the study.
| (A) Baseline characteristics of UKCTOCS cohorts used for the discovery and validation studies | ||||||
|---|---|---|---|---|---|---|
| |
Discovery set, median (25th–75th centiles) |
Validation set, median (25th–75th centiles) |
||||
| Controls (N=273) | Breast cancer cases (N=240) | Controls and BC (N=513) | Controls (N=431) | Breast cancer cases (N=431) | Controls and BC (N=862) | |
| Age (years) at randomisation |
60 (55–65) |
60 (56–66) |
60 (56–65) |
61 (55–66) |
61 (57–66) |
61 (57–66) |
| Years since last period at randomisation |
11 (5–19) |
10 (5–17) |
11 (5–18) |
12 (6–18) |
11 (5–18) |
11(6–18) |
| Duration of HRT use in those who were on HRT at randomisation (years) |
8 (5–12) |
8 (5–11) |
8 (5–12) |
8 (5–12) |
11 (6–13) |
10 (5–13) |
| Duration of OCP use (years) in those who had used it |
6 (3–10) |
6 (2–10) |
6 (3–10) |
5 (2–10) |
5 (2–10) |
5 (2–10) |
| Height (cms) |
163 (156–165) |
163 (158–168) |
163 (158–168) |
163 (158–165) |
163 (158–168) |
163 (158–168) |
| Weight (kg) | 67 (60–76) | 69 (63–76) | 68 (61–76) | 66 (60–76) | 68 (61–76) | 68 (60–76) |
| Ethnicity | Number (%) | Number (%) | ||||
|---|---|---|---|---|---|---|
| White | 262 (95·8%) | 234 (97·5%) | 496 (96.7%) | 417 (96·6%) | 426 (98·8%) | 843 (97·8%) |
| Other | 9 (3·3%) | 6 (2·5%) | 15 (2·9%) | 13 (3·0%) | 4 (0·9%) | 17 (2·0%) |
| Missing |
2(0·8%) |
0 (0%) |
2 (0·4%) |
1 (0·2%) |
1 (0·2%) |
2 (0·2%) |
| Hysterectomy |
51 (19·2%) |
46 (19·2%) |
97 (18.9%) |
75 (17·4%) |
83 (19·3%) |
158 (18·3%) |
| Ever use of OCP |
144 (53·3%) |
138 (57.5%) |
282 (55.0%) |
258 (59·8%) |
276 (64·0%) |
534 (62·0%) |
| Use of HRT at recruitment |
70 (26.3%) |
55 (23·0%) |
125 (24·6%) |
72 (16·7%) |
142 (33·0%) |
214 (24·9%) |
| Women having 1 or more pregnancies |
235 (85·4%) |
203 (84·6%) |
438 (85·4%) |
388 (90.0%) |
374 (86.8%) |
762 (88.4%) |
| Women having 1 or more miscarriages | 73 (25·8%) | 62 (25·8%) | 135 (26.3%) | 114 (26.55) | 131 (30.4%) | 245 (28.4%) |
| (B) Baseline characteristics of breast cancer cases and controls from the Guernsey cohort used in the Validation study | |||
|---|---|---|---|
| |
Median (25th–75th centiles) |
||
| Controls (N=664) | Breast cancer cases (N=332) | Controls and BC (N=996) | |
| Age (years) at serum donation |
50 (42–58) |
50 (43–57) |
50 (42–58) |
| Duration of HRT use in those who were on HRT at time of serum donation (years) |
7(3–20) |
11 (4–30) |
8 (3–23) |
| Duration of OCP use (years) in those who had used it |
3.8 (1–7.3) |
4(1–9.6) |
3.9 (1–9.6) |
| Height (cms) |
160 (155–164) |
161 (157–165) |
160 (156–165) |
| Weight (kg) | 64 (57–70) | 64 (60–72) | 64 (58–71) |
| Number (%) | |||
|---|---|---|---|
| Ethnicity: Missing |
664 (100%) |
332 (100%) |
996 (100%) |
|
Menopausal status | |||
| Pre | 259 (39·0%) | 114 (34·3%) | 373 (37·5%) |
| Peri | 54 (8·1%) | 22 (6·6%) | 76 (7·6%) |
| Post | 249 (37·5%) | 151 (45·6%) | 400 (40·2%) |
| Hysterectomy |
102 (15·4%) |
45 (13·6%) |
147 (14·6%) |
|
Use of oral contraceptive pill | |||
| Ever | 94 (14·1%) | 51 (15·4%) | 145 (14·6%) |
| Unknown |
439 (66·1%) |
221 (66·6%) |
660 (66·3%) |
| Use of HRT at recruitment |
36 (5·4%) |
20 (6·0%) |
56 (5·6%) |
| Women having 1 or more pregnancies |
560 (84·3%) |
288 (86·7%) |
848 (85·1%) |
| Women having 1 or more miscarriages | 144 (21·7%) | 86 (25·9%) | 230 (23·1%) |
Abbreviations: HRT=hormone replacement therapy; OCP=oral contraceptive pill; UKCTOCS=UK Collaborative Trial of Ovarian Cancer Screening.
The UKCTOCS validation set included a single serum sample from each of 431 cases and 431 controls. The Guernsey set included sera from 332 women who were later diagnosed with breast cancer, together with 664 age-matched controls (332 who did not have any type of cancer when their matched case was diagnosed and 332 who were alive and without cancer after up to 32 years follow-up (range 18–32 years). There was no difference in baseline characteristics between cases and controls for the 862 women in the UKCTOCS and 996 Guernsey cohorts in the validation set (see Table 2A and B respectively). The median age of the UKCTOCS cohort was 61 (IQR: 57–66) and all women were postmenopausal.
Time to diagnosis of breast cancer
In the UKCTOCS discovery set, the cases all donated sera up to 4 years before clinical diagnosis of breast cancer, 94% (257) preceding cancer diagnosis by 3 years or less. For the validation set, 95% (406 samples) of the breast cancer cases identified from the UKCTOCS cohort donated serum up to 4 years before clinical cancer diagnosis. In the Guernsey set, 25% of samples preceded cancer diagnosis by 6 months to 5 years with a further 27% collected 5–10 years before diagnosis. Supplementary Table 1 details the subtype, stage and grade of the tumours in women diagnosed with breast cancer.
Screening of discovery set
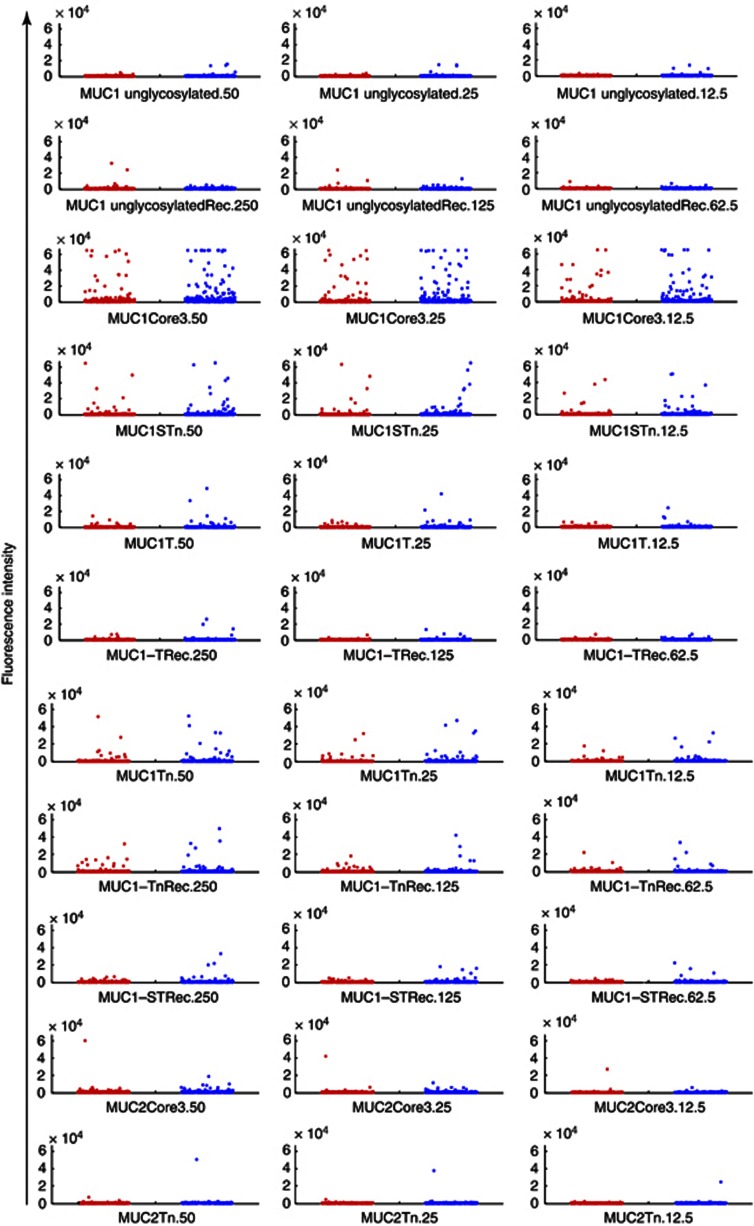
The detailed structures of the MUC1-based glycopeptides peptide and glycoproteins used in the microarray for screening the discovery set are listed in Table 1, and are based on the glycoforms used to detect reactive autoantibodies in sera from early-stage breast cancer patients (Blixt et al, 2011). The results are shown as a dot plot in Figure 1, and it can be seen that only two out of 273 samples from women from the breast cancer cases gave a positive reaction with unglycosylated recombinant MUC1 (16 tandem repeats).
Figure 1.
Autoantibodies to MUC1 in sera from women who subsequently developed breast cancer and matched controls. Dot blots showing the reactivity of autoantibodies present in the discovery sera from women who went on to develop breast cancer (red dots, n=273) and controls (blue dots, n=273), from the UKCTOCS discovery set. The peptide, glycopeptides and glycoproteins (Rec) present on the arrays are indicated beneath each dot plot. The numbers (50, 25, 12·5 and 250, 125, and 62·5) refer to the three concentrations spotted onto the arrays in μℳ for the peptide and glycopeptides, and in pg for the recombinant glycoproteins.
To statistically analyse the data, we investigated the distribution within quartiles (see Methods for description of quartiles). There was no significant difference in distribution of autoantibodies to MUC1 glycoforms between cases and controls over quartiles of reactivity (Table 3). There was, however, a trend for more cases than controls to be in highest quartile (Q4) for MUC1core3, MUC1STn and MUC1Tn (see Table 3A). While a number of sera in cases and control groups contained antibodies reactive to core3 or Tn when carried on MUC1, little reactivity was seen with MUC2 carrying these glycans indicating that the epitopes recognised consisted of the glycans and the MUC1 backbone (Figure 1).
Table 3. Reactivity of discovery set and validation set sera from women who subsequently developed breast cancer (cases) and controls.
| MUC1 peptide/glycopeptide antigen | Samples | Q1 | Q2 | Q3 | Q4 |
|---|---|---|---|---|---|
|
(A) Discovery set | |||||
| MUC1 peptide unglycosylated (60mer) | Cases | 66 | 70 | 61 | 76 |
| Controls | 70 | 67 | 75 | 61 | |
| |
P-value |
0·732 |
0·798 |
0·230 |
0·200 |
| MUC1core3 glycopeptide (60mer) | Cases | 63 | 63 | 66 | 81 |
| Controls | 73 | 73 | 71 | 56 | |
| |
P-value |
0·391 |
0·391 |
0·670 |
0·0327 |
| MUC1STn glycopeptide (60mer) | Cases | 67 | 68 | 60 | 78 |
| Controls | 69 | 68 | 77 | 59 | |
| |
P-value |
0·864 |
1 |
0·146 |
0·105 |
| MUC1Tn glycopeptides (60mer) | Cases | 62 | 60 | 69 | 82 |
| Controls | 74 | 77 | 67 | 55 | |
| |
P-value |
0·303 |
0·146 |
0·864 |
0·0210 |
| MUC1T glycopeptides (60mer) | Cases | 69 | 60 | 70 | 74 |
| Controls | 67 | 77 | 66 | 63 | |
| |
P-value |
0·864 |
0·146 |
0·732 |
0·347 |
| MUC1T CHO recombinant (16 tandem repeats) | Cases | 64 | 68 | 68 | 73 |
| Controls | 72 | 69 | 68 | 64 | |
| |
P-value |
0·492 |
0·932 |
1 |
0·442 |
| MUC1Tn CHO recombinant (16 tandem repeats) | Cases | 58 | 70 | 70 | 75 |
| Controls | 78 | 67 | 66 | 62 | |
| |
P-value |
0·086 |
0·798 |
0·732 |
0·267 |
| MUC1ST CHO recombinant (16 tandem repeats) | Cases | 62 | 61 | 73 | 77 |
| Controls | 74 | 75 | 64 | 60 | |
| |
P-value |
0·303 |
0·230 |
0·442 |
0·146 |
| MUC2core3 glycopeptide | Cases | 61 | 71 | 71 | 70 |
| Controls | 74 | 67 | 65 | 65 | |
| |
P-value |
0·263 |
0·733 |
0·607 |
0·798 |
|
(B) Validation set | |||||
|
UKCTOCS cohort |
|
|
|
|
|
| MUC1 peptide unglycosylated | Cases | 109 | 108 | 106 | 103 |
| (60mer) | Controls | 104 | 105 | 107 | 110 |
| |
P-value |
0·732 |
0·837 |
0·945 |
0·631 |
| MUC1core3 glycopeptides (60mer) | Cases | 108 | 97 | 114 | 107 |
| Controls | 105 | 116 | 99 | 106 | |
| |
P-value |
0·837 |
0·193 |
0·304 |
0·945 |
| MUC1STn glycopeptides (60mer) | Cases | 103 | 108 | 110 | 105 |
| Controls | 110 | 105 | 103 | 108 | |
| |
P-value |
0·631 |
0·837 |
0·631 |
0·837 |
| MUC1Tn glycopeptides (60mer) | Cases | 115 | 93 | 105 | 113 |
| Controls | 98 | 120 | 108 | 100 | |
| |
P-value |
0·244 |
0·064 |
0·837 |
0·373 |
| MUC1T CHO recombinant | Cases | 105 | 113 | 96 | 112 |
| (16 tandem repeats) | Controls | 108 | 100 | 117 | 101 |
| |
P-value |
0·837 |
0·373 |
0·150 |
0·451 |
| MUC1Tn CHO recombinant | Cases | 103 | 103 | 107 | 113 |
| (16 tandem repeats) | Controls | 110 | 110 | 106 | 100 |
| |
P-value |
0·631 |
0·631 |
0·945 |
0·373 |
| MUC1ST CHO recombinant | Cases | 115 | 106 | 98 | 107 |
| (16 tandem repeats) | Controls | 98 | 107 | 115 | 106 |
| P-value | 0·244 | 0·945 | 0·244 | 0·945 | |
|
Guernsey cohort | |||||
| MUC1 peptide unglycosylated | Cases | 74 | 66 | 78 | 85 |
| (60mer) | Control 1 | 77 | 69 | 86 | 71 |
| Control 2 | 76 | 92 | 64 | 71 | |
| |
P-value |
0·806 |
0·796 |
0·532 |
0·262 |
| MUC1core3 glycopeptides (60mer) | Cases | 77 | 68 | 84 | 74 |
| Control 1 | 78 | 79 | 76 | 70 | |
| Control 2 | 72 | 80 | 68 | 83 | |
| |
P-value |
0·936 |
0·364 |
0·527 |
0·739 |
| MUC1STn glycopeptides (60mer) | Cases | 79 | 78 | 72 | 74 |
| Control 1 | 72 | 78 | 77 | 76 | |
| Control 2 | 76 | 71 | 79 | 77 | |
| |
P-value |
0·569 |
1 |
0·682 |
0·870 |
| MUC1Tn glycopeptides (60mer) | Cases | 73 | 72 | 76 | 82 |
| Control 1 | 81 | 79 | 79 | 64 | |
| Control 2 | 73 | 76 | 73 | 81 | |
| |
P-value |
0·519 |
0·569 |
0·809 |
0·136 |
| MUC1T CHO recombinant (16 tandem repeats) | Cases | 83 | 74 | 71 | 75 |
| Control 1 | 70 | 80 | 74 | 79 | |
| Control 2 | 74 | 73 | 83 | 73 | |
| |
P-value |
0·293 |
0·629 |
0·803 |
0·240 |
| MUC1Tn CHO recombinant (16 tandem repeats) | Cases | 84 | 75 | 66 | 78 |
| Control 1 | 75 | 82 | 82 | 64 | |
| Control 2 | 68 | 70 | 80 | 85 | |
| |
P-value |
0·475 |
0·576 |
0·188 |
0·240 |
| MUC1ST CHO recombinant (16 tandem repeats) | Cases | 78 | 81 | 74 | 70 |
| Control 1 | 75 | 70 | 71 | 87 | |
| Control 2 | 74 | 76 | 83 | 70 | |
| P-value | 0·808 | 0·370 | 0·803 | 0·175 | |
Abbreviation: UKCTOCS=UK Collaborative Trial of Ovarian Cancer Screening.
Cases and controls were divided into quartiles dependent on the reactivity of their sera with the indicated antigens.
As we are hypothesising that the presence of autoantibodies to aberrant glycoforms of MUC1 is an antigen driven immune response arising from a clinically undetectable tumour, and as autoantibodies to other antigens such as p53 in colon cancer (Pedersen et al, 2013) and in lung cancer (Lubin et al, 1995; Li et al, 2005), and alpha-fetoprotein in hepatic cancer (Zhang and Tan, 2010) can only be detected within 3 years of cancer diagnosis, we investigated if the presence of antoantibodies to MUC1 glycoforms is associated with breast cancer development in cases who developed breast cancer within 3 years of donating sera. The cases were stratified into cohorts who developed breast cancer within 1 year, 1–2 years and 2–3 years of sera donation. Table 4A shows that even when sera were taken 1 year or less before breast cancer was diagnosed, there was no significant difference between the presence of autoantibodies to MUC1 VNTR peptide or MUC1 glycopeptides in the cases compared with the age-matched controls.
Table 4. Comparison of autoantibodies to MUC1 glycoforms in cases of breast cancer taken up to 3 years before diagnosis vs controls.
| (A) Discovery set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
|
MUC1 glycoforms
P-value breast cancer cases
vs
controls |
||||||||
| Time to diagnosis in years | No. of sera | MUC1 ungly | MUC1 unglyRec | MUC1core3 | MUC1STn | MUC1T | MUC1TRec | MUC1Tn | MUC1TnRec | MUC1STRec |
| 0–1 |
93 |
0.245 |
0.598 |
0.124 |
0.374 |
0.362 |
0.560 |
0.171 |
0.418 |
0.299 |
| 1–2 |
90 |
0.121 |
0.409 |
0.024 |
0.311 |
0.373 |
0.359 |
0.320 |
0.453 |
0.204 |
| 2–3 | 74 | 0.347 | 0.110 | 0.185 | 0.583 | 0.251 | 0.322 | 0.763 | 0.961 | 0.331 |
|
(B) Validation set | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| UKCTOCS cohort | |||||||||
| |
|
MUC1 glycoforms
P-value breast cancer cases
vs
controls |
|||||||
| Time to diagnosis in years | No. of sera | MUC1 ungly | MUC1 ungly Rec | MUC1core3 | MUC1STn | MUC1TRec | MUC1Tn | MUC1TnRec | MUC1STRec |
| 0–1 |
133 |
0.793 |
0.340 |
0.386 |
0.842 |
0.721 |
0.789 |
0.762 |
0.799 |
| 1–2 |
87 |
0.391 |
0.661 |
0.364 |
0.458 |
0.639 |
0.887 |
0.942 |
0.591 |
| 2–3 | 94 | 0.155 | 0.763 | 0.484 | 0.472 | 0.334 | 0.435 | 0.559 | 0.477 |
| Guernsey cohort | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |
|
MUC1 glycoforms
P-value breast cancer cases
vs
all controls |
|||||||
| Time to diagnosis in years | No. of sera | MUC1 ungly | MUC1 ungly Rec | MUC1core3 | MUC1STn | MUC1TRec | MUC1Tn | MUC1TnRec | MUC1STRec |
| 0–3 | 42 | 0.594 | 0.520 | 0.957 | 0.311 | 0.834 | 0.453 | 0.527 | 0.435 |
Abbreviation: UKCTOCS=UK Collaborative Trial of Ovarian Cancer Screening.
Screening of validation set
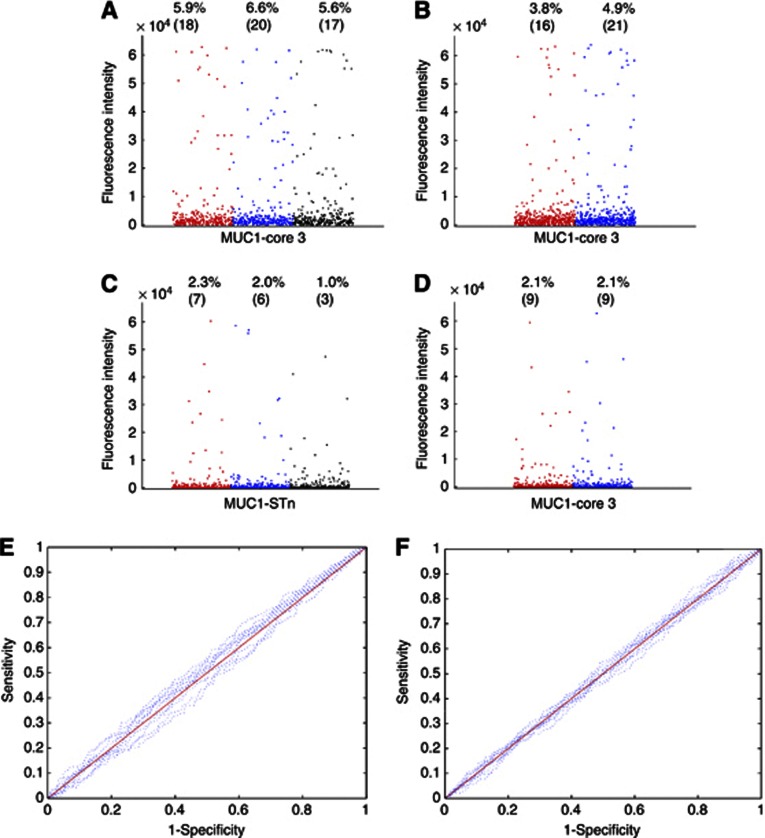
The coded sera were screened on the microarrays. Five samples from the UKCTOCS cases and 29 samples from the Guernsey cases had to be removed from the analysis because the duplicates did not agree based on a similarity measure (described in Supplementary Methods), and rescreening the sera still showed disagreement. The relevant controls were also removed from the analysis. The final analysis, therefore, included 426 UKCTOCS and 303 Guernsey cases samples, with their matched controls. Figure 2 shows a dot blot of the results obtained from both sera sets for MUC1core3 and MUC1STn, the two glycopeptides that gave the highest levels of antibodies in the discovery set. There was no difference in the percentage of sera showing MUC1core3 or MUC1STn autoantibodies between the cases and the controls from either serum bank (Figure 2) or between the Guernsey breast cancer cases and the controls who did not develop cancer within the extended follow-up period (18–32 years).
Figure 2.
Autoantibodies to MUC1 glycopeptides do not distinguish breast cancer cases from controls. (A, B, C ,D) dot blots showing the reactivity of autoantibodies present in the validation sera from women who went on to develop breast cancer and controls. (A, B) Reactivity on 50 μM of MUC1 core3 glycopeptide; (C, D) Reactivity on 50 μℳ of MUC1STn glycopeptide. (A, C) Sera were identified from the Guernsey serum bank who subsequently developed breast cancer (red dots, n=303), matched controls who were not diagnosed with cancer at the time of diagnosis of the cases (blue dots, n=303) and a second cohort of matched controls consisting of sera from 303 women who had not developed cancer up to 32 years after donation of blood (black dots). (B, D) A second cohort of sera identified from the UKCTOCS bank from women who subsequently developed breast cancer (red dots, n=426) and matched controls (blue dots, n=426). Percentages refer to the percentage of samples giving values higher than two s.d. values above the mean of the controls, and (n) refers to the number of women. (E, F) Receiver operating characteristics of individual and combined features for E, samples from the Guernsey bank and F, samples from UKCTOCS. Solid red lines represent the combination of all MUC1 antigens (see Table 1 for list of antigens) and dotted blue lines represent the individual antigens.
The distribution of levels of autoantibodies in cases and controls over quartiles of reactivity also showed no significant differences between cases and controls in the two independent banks when analysed for autoantibodies to all glycopeptides, glycoproteins or unglycosylated MUC1 peptides. Also, the trend observed in the discovery set of more cases in the highest (Q4) quartile of MUC1core3, MUC1Tn and MUC1STn was not observed (see Table 3B). Furthermore, a heat map analysis suggested no correlation was seen between the presence of autoantibodies and time to diagnosis (see Supplementary Figure 1). However, to analyse this in greater details we again stratified the samples from the UKCTOCS bank into those donated 0–1 years, 1–2 years and 2–3 years before breast cancer diagnosis. As there were fewer samples from the Guernsey cohort with shorter times to diagnosis, we analysed as a single stratification samples taken 0–3 years before diagnosis. As can be seen from Table 4B there was no significant differences between the cases and controls in autoantibodies even at 0–1 year preclinical diagnoses, in agreement with the data obtained with the discovery set. The Guernsey serum samples taken 1–3 years before diagnosis were compared with both sets of controls and again, no significant difference was obtained. For clarity, the results presented in Table 4B show the cases compared with the two sets of controls combined.
In addition, ROC curves for each of the MUC1 glycopeptides on the arrays fit the perfect diagonal and the areas under the curve did not significantly differ from 0.5 indicating that no distinction between the real data and data generated randomly could be made (see Figures 2E and F). Thus, autoantibodies to the MUC1 glycopeptides cannot be used to distinguish cases from the controls.
MUC1 autoantibodies in ovarian, lung and pancreatic cancer
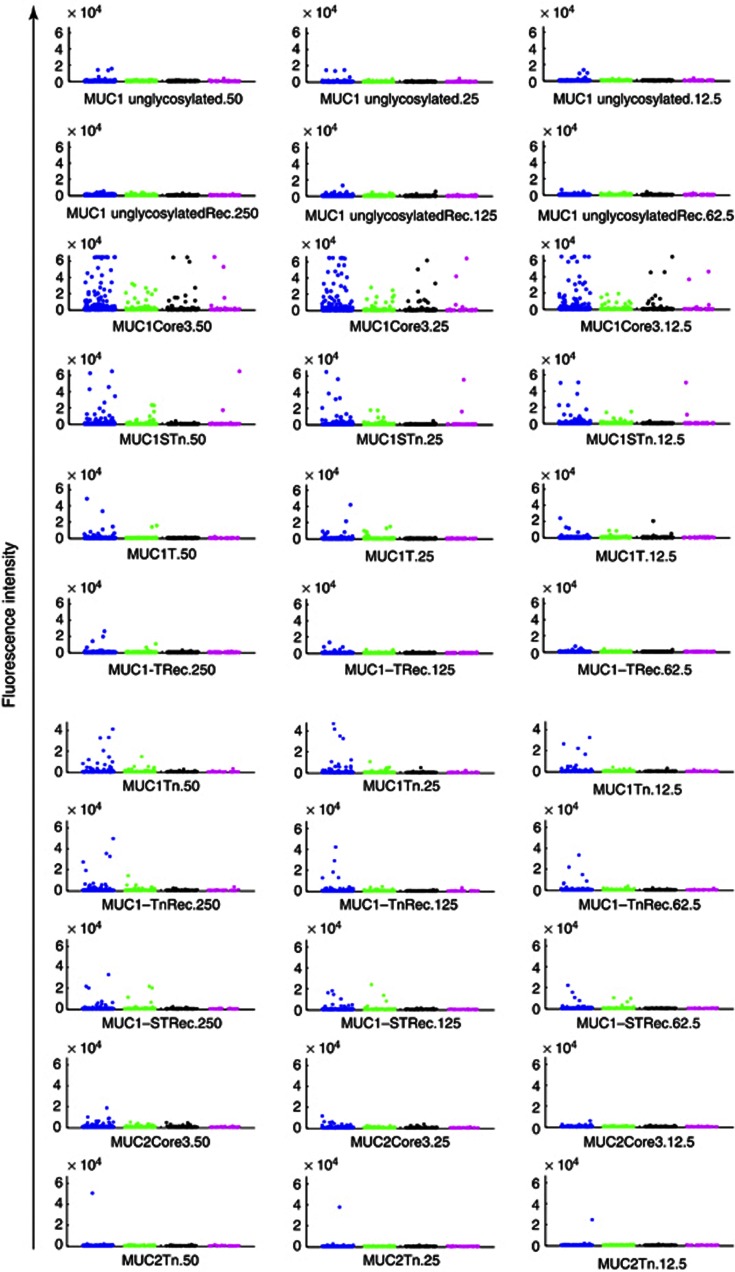
Eighty-nine serum samples taken from 86 women with ovarian cancer, preceding diagnosis by a mean of 1 year (IQR: 0·4–1·5), 123 sera taken from 123 women preceding lung cancer diagnosis by a mean of 1·6 years (IQR: 1·0–2·2) and 35 samples taken from 35 women preceding pancreatic cancer by a mean of 1 year (IQR: 0·8–2·0) and matched controls (247) were identified from the UKCTOCS serum bank. Baseline characteristics are presented in Supplementary Table 2, and tumour characteristics in Supplementary Table 3. The samples were screened on the glycopeptides arrays and there was no difference in autoantibodies to MUC1core3 and MUC1STn between cases and controls (Figure 3). Although there appear to be more sera, which are positive for antibodies to MUC1STn and MUC1core3 in the control sera, this is because there are more control samples (247, see Supplementary Table 2), and there were only minor differences in rates of positivity between controls (see legend to Figure 3). Moreover, stratifying the samples into cohorts of 0–1, 1–2 and 2–3 years before cancer diagnosis did not shown any difference between cases and controls (data not shown).
Figure 3.
Elevated levels or increased frequency of autoantibodies to MUC1 are not found in sera from ovarian, pancreatic or lung cancer patients before clinical diagnosis. Dot blots showing the reactivity of autoantibodies present in the sera of women who went on to develop lung cancer (green dots, n=123), ovarian cancer (black dots, n=89), pancreatic cancer (magenta dots, n=35) or matched controls (blue dots n=247). The peptide, glycopeptides and glycoproteins (Rec) present on the arrays are indicated beneath each dot blot. The numbers (50, 25, 12·5 and 250, 125, and 62·5) refer to the three concentrations spotted onto the arrays in μℳ for the peptide and glycopeptides, and in pg for the recombinant glycoproteins. Positive samples were defined as samples giving values higher than two s.d. values above the mean of the controls and for MUC1core3 and MUC1STn were as follows: MUC1core3; controls 5.9% positive, pancreatic cancer 5.7% positive, ovarian 3.4% positive, lung 0% positive. MUC1STn; controls 2.3% positive, pancreatic 2.9% positive, ovarian cancer 0%.
Discussion
This is the largest case–control study that we are aware of exploring MUC1 autoantibody profile before diagnosis of breast and other adenocarcinomas. No differences were observed in autoantibodies recognising MUC1 tumour-associated glycopeptides in the nested case–control study involving over 1000 serum samples from women who later developed breast cancer and over 1300 matched controls in two independent cohorts (UKCTOCS and Guernsey). This was irrespective of the time between serum donation and diagnosis of cancer with 273 of the samples analysed being from women who were diagnosed with breast cancer within 1 year of serum donation. This result was totally unexpected as we have previously shown that autoantibodies to MUC1 glycoforms can be detected in sera from early-stage breast cancer patients when the sera were taken at or just after the time of diagnosis (Blixt et al, 2011). Unfortunately, sera were not available at the time of diagnosis from the cases studied in the present paper. It should be noted that we did not assay autoantibodies on MUC1 purified from tumours. However, such material is limited in quantities and it is very difficult to obtain homogeneous material that is standardised from one preparation to the next or from different individuals.
Similar results were obtained for ovarian, lung and pancreatic cancer. Our findings suggest that detection of autoantibodies to MUC1 VNTR peptides, or to glycopeptides and full-length glycoforms carrying cancer-associated glycans, cannot be used as a screening tool for early detection of these cancers in the general population. The results of this robust, validated large-scale prospective-specimen collection, retrospective-blinded evaluation study have significant implications, as MUC1 has been the focus of several studies aiming for early detection of breast cancer (Chapman et al, 2007; Pinheiro et al, 2010; Wandall et al, 2010; Zhang and Tan, 2010; Blixt et al, 2011; Lacombe et al, 2013).
The robustness of the study design, and the large number of sera screened gives us confidence of the validity of the results. The strengths of the study include (1) a microarray approach, which allowed simultaneous screening for autoantibodies to unglycosylated MUC1 (consisting of three tandem repeats of MUC1), to MUC1 60mer glycopeptides and to recombinant MUC1 produced in CHO cells and carrying no or defined O-linked glycans, (2) use of a prospective-specimen collection, with retrospective-blinded evaluation study design (Pepe et al, 2008), (3) validation of results on separate case–control sets including one from an independent serum bank, (4) additional controls from the Guernsey cohort with up to 32 years follow-up, (5) further evaluation of sera from individuals who later developed other cancers known to express MUC1, namely ovarian, pancreatic and lung, (6) matching for age and storage time of samples and (7) well balanced baseline characteristics between cases and controls. Limitations include the fact that sera were not available at the time of diagnosis from the cases studied in the present paper and the sera had been stored for a number of years before autoantibody determination. However, it is unlikely that this resulted in the lost of autoantibody activity as antibodies to p53 have been shown to be present (Pedersen et al, 2013) and in our previous study autoantibodies to MUC1 glycopeptides were found in the sera from breast cancer patients after a storage time of 30 years (Blixt et al, 2011). In addition, a significant proportion of the breast cancer cases would have been screen detected as a result of the national mammography screening programme and some of the ovarian cancer cases could also have been screen detected as UKCTOCS is an ovarian cancer screening trial. On the other hand, similar results were obtained when we used preclinical samples from other cancers especially lung and pancreas for which no screening was available in the UK.
When determining the use of anti-MUC1 antibodies for early detection or cancer risk, most previous studies have looked for antibodies in sera from cancer patients (Chapman et al, 2007; Desmetz et al, 2011; Pedersen et al, 2011) and extrapolated the results to suggest the assay's usefulness in early detection. We too have previously shown that more sera from stage I and II breast cancer patients contain autoantibodies compared with aged-matched controls and hypothesised that this might aid early detection (Blixt et al, 2011). This is in keeping with most biomarker discovery studies for early detection of cancer, which are usually carried out on sera collected from patients with clinical disease (Chapman et al, 2007; Zhong et al, 2008; Boyle et al, 2011; Lacombe et al, 2013), or small cohorts with lack of independent validation of the findings (Lubin et al, 1995; Li et al, 2005; Robertson et al, 2005; Zhong et al, 2006; Pereira-Faca et al, 2007). There are only a few studies that have used a prospective sera collection (Pinheiro et al, 2010; Chapman et al, 2012; Pedersen et al, 2013). The other study where preclinical cancer samples were screened for the presence of autoantibodies to the unglycosylated MUC1 VNTR was the case–control study from the Nurses Health cohort involving sera from women who went on to develop ovarian cancer and healthy controls (Pinheiro et al, 2010). Autoantibodies to a MUC1 tandem repeat peptide (consisting of five tandem repeats) were found to be associated with a lower risk of developing ovarian cancer in those under 64 years of age and higher risk in women more than 64-years-old. However, the study only included 117 cases with only 27 over 64 years of age, making data interpretation difficult.
Our findings show the importance of validating initial findings in a larger sample set, as the trend towards more cases being in the highest quartile compared with controls observed in our discovery set was subsequently not validated in two independent sets.
Our results are in contrast to results with p53 as autoantibodies to p53 were detected in sera from UKCTOCS women who went on to develop colon cancer (Pedersen et al, 2013), providing support for the fitness of the UKCTOCS serum bank samples for study of autoantibodies. There is considerable effort directed to developing a screen for antibodies to cancer antigens for individuals at high risk for lung cancer. In this context, antigen panels which can include, p53, 14-3-3, Annexin 1 or NY-ESO-1 show promise and are being evaluated in larger cohorts (Lubin et al, 1995; Li et al, 2005; Pereira-Faca et al, 2007; Qui et al 2008; Boyle et al, 2011; Chapman et al, 2012).
p53 is a nuclear protein, as are some of the other antigens showing promise as inducing autoantibodies before clinical diagnosis of cancer (Desmetz et al, 2011), while MUC1 is a membrane antigen. It is not clear whether a difference in localisation could relate to the early induction of autoantibodies in cancer patients, unless there is a more stringent tolerance of the adaptive response to the surface molecules, requiring higher levels of membrane antigen. Certainly as long as the normal polarity of the epithelial cells is intact the MUC1 glycoprotein will be on the luminal surface and less accessible to circulating immune cells. Moreover, while the change in glycosylation of MUC1 is seen in early-stage cancers, (clinically diagnosed), the timing of this change in the initiation and progression to malignancy before clinical diagnosis is not known, and may correlate with a certain level of loss of ordered tissue architecture. Nonetheless, autoantibodies to MUC1 do appear in the sera of a proportion of early-stage breast cancer patients at the time of diagnosis, whereas patients with benign breast disease have similar levels to controls (Blixt et al, 2011). However, although the data from this study show that autoantibodies to MUC1 may be useful for determining prognosis in women with early breast cancer, the results presented here show that an autoantibody profile to MUC1 is unlikely to be useful as a screening test for cancer within the general population. A considerable amount of time and resources are devoted to developing MUC1-based autoantibody assays and our results suggest that these should be focused on other tumour-associated antigens, possibly nuclear antigens, for early cancer detection and risk stratification.
Acknowledgments
We are particularly grateful to women who contributed their samples to this study and to the medical, nursing and administrative staff who worked on the UKCTOCS/ Guernsey study. UKCTOCS was core funded by the Medical Research Council, Cancer Research UK and the NIHR with additional support from the Eve Appeal, Special Trustees of Bart's and the London and Special Trustees of UCLH. This study was supported by an EU FP7 collaborative project grant, O-PTM Biomarkers, grant agreement number 201381 and NIH grant U01CA128437-03. We also acknowledge financial support from the Experimental Cancer Medicine Centre at King's College London and the National Institute for Health Research via CBRC funding (women's health theme) to University College London Hospitals (UCLH) in partnership with University College London, and the Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King's College London. We also acknowledge financial support from the Excellence programme at University of Copenhagen, the Danish Research Council and National Research Foundation. The researchers are independent from the funders and the views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. UKCTOCS was core funded by the Medical Research Council, Cancer Research UK, and the NIHR with additional support from the Eve Appeal, Special Trustees of Bart's and the London, and Special Trustees of UCLH.
UM and IJJ have a financial interest through UCL Business and Abcodia Ltd in the third party exploitation of clinical trials biobanks, which have been developed through the research at UCL. OB, HHW, and HC have financial interests in GlycoZym Inc. UM and HC have a patent application claiming use of MUC1 glycoforms in therapeutic and preventive applications. JT-P and JB have patents for the use of MUC1 in therapeutic applications. The remaining authors have no conflicts of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Blixt O, Bueti D, Burford B, Allen D, Julien S, Hollingsworth M, Gammerman A, Fentiman I, Taylor-Papadimitriou J, Burchell J. Autoantibodies to aberrantly glycosylated MUC1 in early stage breast cancer are associated with a better prognosis. Breast Cancer Res. 2011;8 (13:R25. doi: 10.1186/bcr2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Chapman CJ, Holdenrieder S, Murray A, Robertson C, Wood WC, Maddison P, Healey G, Fairley GH, Barnes AC, Robertson JF. Clinical validation of an autoantibody test for lung cancer. Ann Oncol. 2011;22:383–389. doi: 10.1093/annonc/mdq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckström M, Link T, Olson FJ, Karlsson H, Graham R, Picco G, Burchell J, Taylor-Papadimitriou J, Noll T, Hansson GC. Recombinant MUC1 mucin with a breast cancer-like O-glycosylation produced in large amounts in Chinese-hamster ovary cells. Biochem J. 2003;376:677–686. doi: 10.1042/BJ20031130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman C, Murray A, Chakrabarti J, Thorpe A, Woolston C, Sahin U, Barnes A, Robertson J. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol. 2007;18:868–873. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- Chapman CJ, Healey GF, Murray A, Boyle P, Robertson C, Peek LJ, Allen J, Thorpe AJ, Hamilton-Fairley G, Parsy-Kowalska CB, Macdonald IK, Jewell W, Maddison P, Robertson JF. EarlyCDT-Lung test: improved clinical utility through additional autoantibody assays. Tumour Biol. 2012;33:1319–1226. doi: 10.1007/s13277-012-0379-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmetz C, Mange A, Maudelonde T, Solassol J. Autoantibody signatures: progress and perspectives for early cancer detection. J Cell Mol Med. 2011;15:2013–2024. doi: 10.1111/j.1582-4934.2011.01355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fentiman IS, Hanby A, Allen DS, Key T, Meilahn EN. Hormone dependency of breast tumours developing in the Guernsey Cohort study. Breast Cancer Res Treat. 2006;97:205–208. doi: 10.1007/s10549-005-9113-8. [DOI] [PubMed] [Google Scholar]
- Gøtzsche PC, Nielsen M. Screening for breast cancer with mammography. Cochrane Database. Syst Rev. 2011;1:CD001877. doi: 10.1002/14651858.CD001877.pub4. [DOI] [PubMed] [Google Scholar]
- JFR Robertson, Chapman C, Cheung K-L, Murray A, Pinder SE, Price MR, Graves RL. Autoantibodies in early breast cancer. J Clin Oncol. 2005;23:549. [Google Scholar]
- Kerlikowske K, Grady D, Rubin SM, Sandrock C, Ernster VL. Efficacy of screening mammography. A meta-analysis. JAMA. 1995;273:149–154. [PubMed] [Google Scholar]
- Lacombe J, Mangé A, Jarlier M, Bascoul-Mollevi C, Rouanet P, Lamy PJ, Maudelonde T, Solassol J. Identification and validation of new autoantibodies for the diagnosis of DCIS and node negative early stage-stage breast cancers. Int J Cancer. 2013;132:1105–1113. doi: 10.1002/ijc.27766. [DOI] [PubMed] [Google Scholar]
- Li Y, Karjalainen A, Koskinen H, Hemminki K, Vainio H, Shnaidman M, Ying Z, Pukkala E, Brandt-Rauf PW. p53 autoantibodies predict subsequent development of cancer. Int J Cancer. 2005;114:157–160. doi: 10.1002/ijc.20715. [DOI] [PubMed] [Google Scholar]
- Lu H, Ladd J, Feng Z, Wu M, Goodell V, Pitteri SJ, Li CI, Prentice R, Hanash SM, Disis ML. Evaluation of known oncoantibodies, HER2, p53, and cyclin B1, in pre-diagnostic breast cancer sera. Cancer Prev Res (Phila) 2012;5:1036–1043. doi: 10.1158/1940-6207.CAPR-11-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin R, Zalcman G, Bouchet L, Trédanel J, Legros Y, Cazals D, Hirsch A, Soussi T. Serum p53 antibodies as early markers of lung cancer. Nat Med. 1995;1:701–702. doi: 10.1038/nm0795-701. [DOI] [PubMed] [Google Scholar]
- Menon U, Gentry-Maharaj A, Hallett R, Ryan A, Burnell M, Sharma A, Lewis S, Davies S, Philpott S, Lopes A, Godfrey K, Oram D, Herod J, Williamson K, Seif MW, Scott I, Mould T, Woolas R, Murdoch J, Dobbs S, Amso NN, Leeson S, Cruickshank D, McGuire A, Campbell S, Fallowfield L, Singh N, Dawnay A, Skates SJ, Parmar M, Jacobs I. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncol. 2009;10:327–340. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- Pedersen JW, Blixt O, Bennett EP, Tarp MA, Dar I, Mandel U, Poulsen SS, Pedersen AE, Rasmussen S, Jess P, Clausen H, Wandall HH. Seromic profiling of colorectal cancer patients with novel glycopeptides microarrays. Int J Cancer. 2011;128:1860–1871. doi: 10.1002/ijc.25778. [DOI] [PubMed] [Google Scholar]
- Pedersen JW, Gentry-Maharaj A, Fourkala E-O, Dawnay A, Burnell M, Zaikin A, Pedersen AE, Jacobs I, Menon U, Wandall HH. Early detection of cancer in the general population - a blinded case control study of p53 auto-antibodies in colorectal cancer. Br J Cancer. 2013;108:107–114. doi: 10.1038/bjc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe MS, Feng Z, Janes H, Bossuyt PM, Potter JD. Pivotal evaluation of the accuracy of a biomarker used for classification or prediction: standards for study design. J Natl Cancer Inst. 2008;100:1432–1438. doi: 10.1093/jnci/djn326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Faca SR, Kuick R, Puravs E, Zhang Q, Krasnoselsky AL, Phanstiel D, Qiu J, Misek DE, Hinderer R, Tammemagi M, Landi MT, Caporaso N, Pfeiffer R, Edelstein C, Goodman G, Barnett M, Thornquist M, Brenner D, Hanash SM. Identification of 14-3-3 theta as an antigen that induces a humoral response in lung cancer. Cancer Res. 2007;67:12000–12006. doi: 10.1158/0008-5472.CAN-07-2913. [DOI] [PubMed] [Google Scholar]
- Pinheiro SP, Hankinson SE, Tworoger SS, Rosner BA, McKolanis JR, Finn OJ, McKolanis JR, Finn OJ, Cramer DW. Anti-MUC1 antibodies and ovarian cancer risk: prospective data from the Nurses' Health Studies. Cancer Epidemiol Biomarkers Prev. 2010;19:1595–1601. doi: 10.1158/1055-9965.EPI-10-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Choi G, Li L, Wang H, Pitteri SJ, Pereira-Faca SR, Krasnoselsky AL, Randolph TW, Omenn GS, Edelstein C, Barnett MJ, Thornquist MD, Goodman GE, Brenner DE, Feng Z, Hanash SM. Occurrence of autoantibodies to annexin I, 14-3-3 theta and LAMR1 in prediagnostic lung cancer sera. J Clin Oncol. 2008;26:5060–5066. doi: 10.1200/JCO.2008.16.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen AL, Reis CA, Tarp MA, Mandel U, Ramachandran K, Sankaranarayanan V, Schwientek T, Graham R, Taylor-Papadimitriou J, Hollingsworth MA, Burchell J, Clausen H. Chemoenzymatically synthesized multimeric Tn/STn MUC1 glycopeptides elicit cancer-specific anti-MUC1 antibody responses and override tolerance. Glycobiology. 2006;16:96–107. doi: 10.1093/glycob/cwj044. [DOI] [PubMed] [Google Scholar]
- Tarp MA, Sørensen AL, Mandel U, Paulsen H, Burchell J, Taylor-Papadimitriou J, Clausen H. Identification of a novel cancer-specific immunodominant glycopeptide epitope in the MUC1 tandem repeat. Glycobiology. 2007;17:197–209. doi: 10.1093/glycob/cwl061. [DOI] [PubMed] [Google Scholar]
- Wandall HH, Blixt O, Tarp MA, Pedersen JW, Bennett EP, Mandel U, Ragupathi G, Livingston PO, Hollingsworth MA, Taylor-Papadimitriou J, Burchell J, Clausen H. Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 2010;70:1306–1313. doi: 10.1158/0008-5472.CAN-09-2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J-Y, Tan EN. Autoantibodies to tumour-associated antigens as diagnostic biomarkers in hepatocellular carcinoma and other solid tumours. Expert Rev Mol Diagn. 2010;10:321–328. doi: 10.1586/erm.10.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, Coe SP, Stromberg AJ, Khattar NH, Jett JR, Hirschowitz EA. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J Thorac Oncol. 2006;1:513–519. [PubMed] [Google Scholar]
- Zhong L, Ge K, Zu JC, Zhao LH, Shen WK, Wang JF, Zhang XG, Gao X, Hu W, Yen Y, Kernstine KH. Autoantibodies as potential biomarkers for breast cancer. Breast Cancer Res. 2008;10:R40. doi: 10.1186/bcr2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.