Abstract
Background:
This study evaluates the geographic expression pattern of Raf-1 Kinase Inhibitor Protein (RKIP) in colorectal cancer (CRC) in correlation with clinicopathological and molecular features, markers of epithelial-mesenchymal transition (EMT) and survival outcome.
Methods:
Whole-tissue sections of 220 well-characterised CRCs were immunostained for RKIP. NF-κB and E-Cadherin expression was assessed using a matched multi-punch tissue microarray. Analysis of mismatch repair (MMR) protein expression, B-Raf and KRAS mutations was performed. RKIP expression in normal mucosa, tumour centre, invasion front and tumour buds was each assessed for clinical relevance.
Results:
RKIP was diffusely expressed in normal mucosa and progressively lost towards tumour centre and front (P<0.0001). Only 0.9% of tumour buds were RKIP-positive. In the tumour centre, RKIP deficiency predicted metastatic disease (P=0.0307), vascular invasion (P=0.0506), tumour budding (P=0.0112) and an invasive border configuration (P=0.0084). Loss of RKIP correlated with NF-κB activation (P=0.0002) and loss of E-Cadherin (P<0.0001). Absence of RKIP was more common in MMR-deficient cancers (P=0.0191), while no impact of KRAS and B-Raf mutation was observed. RKIP in the tumour centre was identified as a strong prognostic indicator (HR (95% CI): 2.13 (1.27–3.56); P=0.0042) independently of TNM classification and therapy (P=0.0474).
Conclusion:
The clinical relevance of RKIP expression as an independent prognostic factor is restricted to the tumour centre. Loss of RKIP predicts features of EMT and correlates with frequent distant metastasis.
Keywords: colorectal cancer, Raf kinase inhibitor protein, RKIP, prognostic factor, therapy response
RKIP is a ubiquitously expressed phospholipid-binding protein that functions as an endogenous inhibitor of the Ras/Raf/MEK/ERK signalling cascade (Yeung et al, 1999). RKIP inhibits this pathway at the level of Raf. It binds both Raf and MEK, inhibiting their association and the phosphorylation of MEK, thereby turning off the signalling pathway (Supplementary Figure 1A). Subsequently, it was shown that RKIP suppresses the activation of the NF-κB/SNAIL circuit (Supplementary Figure 1B) (Wu and Bonavida, 2009). This pathway has an important role for the induction of epithelial-mesenchymal transition (EMT) of cancer cells as one of the initial steps for the induction of metastasis (Wu and Bonavida, 2009). The histopathological hallmark of EMT is thought to be the presence of single tumour cells or small clusters (<5) of dedifferentiated cells at the invasive front of CRC, termed tumour budding (Jass et al, 1986).
A reduced expression of RKIP has been identified as a common feature in malignant tumours with a further decrease in metastatic disease (Fu et al, 2003; Hagan et al, 2005). Using animal models, conclusive evidence has been provided that RKIP impairs invasiveness and the ability to form metastases in cancer cells (Fu et al, 2003). Loss of RKIP protein has been identified as a significant prognostic factor in prostate cancer (Fu et al, 2006), CRC (Al-Mulla et al, 2006; Minoo et al, 2007; Zlobec et al, 2008; Doyle et al, 2013), GIST (Martinho et al, 2009), pancreas (Kim et al, 2010), lung (Huerta-Yepez et al, 2011), and very recently in oesophagus (Kim et al, 2012), stomach (Fujimori et al, 2012) and malignant glioma (Martinho et al, 2012). The fact that the protein expression of RKIP shows a strong correlation with survival outcome in a variety of malignancies of epithelial, mesenchymal and glial origin highlights the potential value of this biomarker for prospective clinical trials.
In colorectal cancer (CRC), several studies based on single-punch tissue microarrays (TMA) demonstrated a significant correlation between RKIP expression and overall survival in CRC: Al-Mulla et al (2006) provided first evidence that RKIP protein expression in primary CRC strongly correlates with the overall survival independent of stage and may be used efficiently for risk stratification. Consecutively, Minoo et al (2007) demonstrated that the assessment of RKIP protein expression by immunohistochemistry is highly reproducible (ICC=0.75) and confirmed the predictive value of RKIP expression for overall survival and metastases in both MMR-proficient and deficient tumours in a large cohort of over 1000 CRC patients. Further, it was demonstrated by our group that RKIP status can be combined with N-stage and vascular invasion to provide independent predictive information on metastatic disease (Zlobec et al, 2008). In TMA-based profiling of multi-marker phenotypes of CRC, we identified RKIP as a predictor of high-grade tumour budding in univariate, but not in multivariate analysis (Karamitopoulou et al, 2010) that is differentially expressed between tumour centre and tumour front (Karamitopoulou et al, 2011).
Taken together, these studies suggest that RKIP may be able to serve as a valuable biomarker to identify aggressive CRC that should be considered for adjuvant therapy. However, recent studies provide conclusive evidence that malignant tumours demonstrate extensive genetic heterogeneity (Gerlinger et al, 2012). It is therefore important to evaluate the intratumoural heterogeneity of immunohistochemical marker expression on large bore TMAs or whole-tissue sections to confirm the prognostic and predictive value of promising biomarkers in the context of molecular characteristics. Consequently, the aim of this study was the geographic analysis of RKIP expression in all histological areas of CRC and to determine the correlation of RKIP expression pattern with clinicopathological characteristics, molecular features and survival outcome.
Materials and methods
Patient cohort
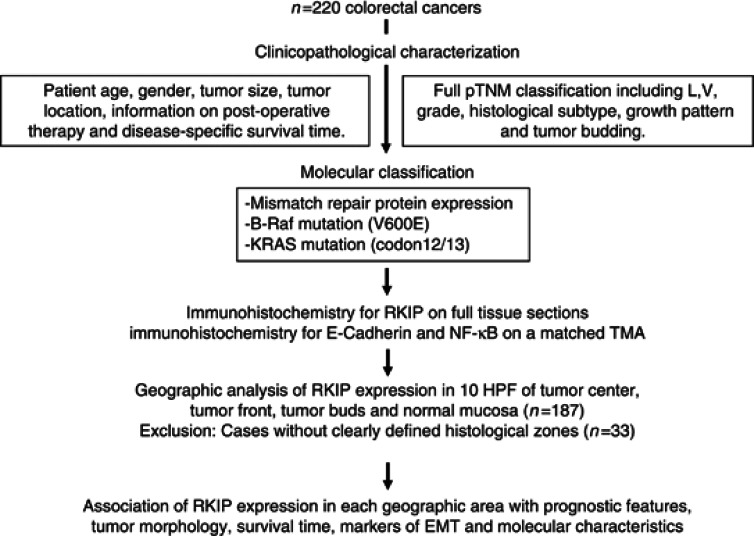
Two hundred and twenty unselected, non-consecutive CRC patients treated at the Aretaieion University Hospital, University of Athens, Greece between the years 2004 and 2007 were included in this study (Figure 1). Clinical data including patient gender, age at diagnosis, tumour diameter, tumour location, post-operative therapy information and disease-specific survival time were retrieved from patient records. The histomorphological data were reviewed by an experienced gastrointestinal pathologist (EK) and includes histological subtype, tumour grade, pT classification, lymphatic and venous invasion, nodal status, tumour growth pattern and tumour budding. Tumour budding was quantified in 10HPF along the invasive front of each case (Karamitopoulou et al, 2013). The average survival time was 47 months (35–58 months). Patient characteristics are summarised in Table 1; where indicated by a lower case number, information for single patients on one of the clinicopathological features was not available. Information on the number of patients receiving chemotherapeutic treatment within each subset is indicated in Tables 2 and 3 and Supplementary Table S3; further information on therapy and performance status is provided in Supplementary Tables S1 and S2. The use of patient data has been approved by the local Ethics Committee of the University of Athens, Greece.
Figure 1.
Study design: 220 CRC patients were entered into the study. Cases were reviewed for clinicopathological features and characterised for B-Raf, KRAS mutations and MMR protein expression. Immunohistochemistry for RKIP was performed on full-tissue sections. Geographic analysis of RKIP expression patterns was performed in 10HPF each of tumour centre, tumour front and tumour buds as well as normal mucosa (5 HPF). Expression of E-Cadherin and NF-κB was assessed on a matched multi-punch TMA. The association of RKIP protein expression with prognostic features, tumour morphology, survival time, markers of EMT and molecular characteristics was carried out for each histological zone.
Table 1. Patient characteristics (n=187).
| Feature | Frequency (n, %) |
|---|---|
|
Gender (n=186) | |
| Male | 92 (49.5) |
| Female |
94 (50.5) |
|
Age (years) | |
| Mean (min, max) |
68.1 (35–93) |
|
Tumor size (mm) | |
| Mean (min, max) |
4.5 (1–12) |
|
Histological subtype (n=187) | |
| Non-mucinous | 163 (87.2) |
| Mucinous |
24 (12.8) |
|
Tumor location (n=186) | |
| Left-side | 115 (61.8) |
| Right-side | 45 (24.2) |
| Rectum |
26 (14.0) |
|
Tumor grade (n=187) | |
| G1–2 | 118 (63.1) |
| G3 |
69 (36.9) |
|
pT (n=187) | |
| pT1–2 | 49 (26.2) |
| pT3–4 |
138 (73.8) |
|
pN (n=187) | |
| pN0 | 95 (50.8) |
| pN1–2 |
92 (49.2) |
|
pM (n=186) | |
| pM0 | 169 (90.9) |
| pM1 |
17 (9.1) |
|
V (n=187) | |
| V+ | 29 (15.5) |
| V− |
158 (84.5) |
|
L (n=187) | |
| L+ | 75 (40.1) |
| L− |
112 (59.9) |
|
Tumor budding (n=187) | |
| Low-grade | 105 (56.2) |
| High-grade |
82 (43.9) |
|
Tumor growth pattern (n=187) | |
| Infiltrating | 134 (71.7) |
| Pushing |
53 (28.3) |
|
Post-operative therapy (n=187) | |
| None | 69 (36.9) |
| Chemo | 75 (40.1) |
| Chemo+radio | 42 (22.5) |
| Radio |
1 (0.5) |
|
Mismatch repair status (n=186) | |
| Proficient | 173 (93.0) |
| Deficient |
13 (7.0) |
|
KRAS mutation (codon 12/13), (n=171) | |
| Wild type | 113 (66.1) |
| Mutated |
58 (33.9) |
|
Survival time (n=186) | |
| Months | 47 (35–58) |
Table 2. Expression of RKIP in the tumor centre and association with clinicopathological features (n=187).
| |
RKIP (n, %) |
|
|
|---|---|---|---|
| Feature | Negative (<10%) | Positive (⩾10%) | P-value |
|
Gender (n=186) | |||
| Male | 43 (50.6) | 49 (48.5) | 0.7781 |
| Female |
42 (49.4) |
52 (41.5) |
— |
|
Age (years) | |||
| Mean (min, max) |
67.4 (35, 89) |
68.7 (36, 93) |
0.4496 |
|
Tumor size (mm) | |||
| Mean (min, max) |
4.7 (1, 12) |
4.3 (1.2, 9.0) |
0.2931 |
|
Histological subtype (n=187) | |||
| Non-mucinous | 73 (85.9) | 90 (88.2) | 0.6319 |
| Mucinous |
12 (14.1) |
12 (11.8) |
— |
|
Tumor location (n=186) | |||
| Left-side | 53 (63.1) | 62 (60.8) | 0.9016 |
| Right-side | 12 (14.3) | 14 (13.7) | — |
| Rectum |
19 (22.6) |
26 (25.5) |
— |
|
Tumor grade (n=187) | |||
| G1–2 | 48 (56.5) | 70 (68.6) | 0.0863 |
| G3 |
37 (43.5) |
32 (31.4) |
— |
|
pT (n=187) | |||
| pT1–2 | 23 (27.1) | 26 (25.5) | 0.8081 |
| pT3–4 |
62 (72.9) |
76 (74.5) |
— |
|
pN (n=187) | |||
| pN0 | 41 (48.2) | 54 (52.9) | 0.5216 |
| pN1–2 |
44 (51.8) |
48 (47.1) |
— |
|
pM (n=186) | |||
| pM0 | 73 (85.9) | 96 (95.1) | 0.0307 |
| pM1 |
12 (14.1) |
5 (5.0) |
— |
|
V (n=187) | |||
| V+ | 18 (21.2) | 11 (10.8) | 0.0506 |
| V− |
67 (78.8) |
91 (89.2) |
— |
|
L (n=187) | |||
| L+ | 39 (45.9) | 36 (35.3) | 0.1413 |
| L− |
46 (54.1) |
66 (64.7) |
— |
|
Tumor budding (n=148) | |||
| Low-grade | 47 (72.3) | 43 (51.8) | 0.0112 |
| High-grade |
18 (27.7) |
40 (48.2) |
— |
|
Tumor growth pattern (n=187) | |||
| Infiltrating | 69 (81.2) | 65 (63.7) | 0.0084 |
| Pushing |
16 (18.8) |
37 (36.3) |
— |
|
Mismatch repair status (n=186) | |||
| Proficient | 75 (88.2) | 98 (97.0) | 0.0191 |
| Deficient |
10 (11.8) |
3 (3.0) |
— |
|
B-RAF mutation (V600E) (n=180) | |||
| Wild type | 70 (87.5) | 93 (93.0) | 0.2099 |
| Mutated |
10 (12.5) |
7 (7.0) |
— |
|
KRAS mutation (codon 12/13), (n=171) | |||
| Wild type | 51 (67.1) | 62 (65.3) | 0.8004 |
| Mutated |
25 (32.9) |
33 (34.7) |
— |
|
Hazard ratio for death with loss of RKIP (n=186) | |||
| HR (95%CI) |
2.13 (1.27–3.56) |
0.0042 |
|
|
Adjuvant therapy (n=187) | |||
| None | 30 (43.5) | 55 (46.6) | 0.6781 |
| Treated | 39 (56.5) | 63 (53.4) | — |
Table 3. Expression of RKIP in the tumor front and association with clinicopathological features (n=187).
| |
RKIP (n, %) |
|
|
|---|---|---|---|
| Feature | Negative (<10%) | Positive (⩾10%) | P-value |
|
Gender (n=186) | |||
| Male | 43 (50.0) | 49 (49.0) | 0.8918 |
| Female |
43 (50.0) |
51 (51.0) |
— |
|
Age (years) | |||
| Mean (min, max) |
67.6 (35, 89) |
68.6 (36, 93) |
0.5366 |
|
Tumor size (mm) | |||
| Mean (min, max) |
4.9 (1, 12) |
4.2 (1.2, 3.0) |
0.0229 |
|
Histological subtype (n=187) | |||
| Non-mucinous | 73 (84.9) | 90 (89.1) | 0.3893 |
| Mucinous |
13 (15.1) |
11 (10.9) |
— |
|
Tumor location (n=186) | |||
| Left-side | 54 (63.5) | 61 (60.4) | 0.6518 |
| Right-side | 13 (15.3) | 13 (12.9) | — |
| Rectum |
18 (21.2) |
27 (26.7) |
— |
|
Tumor grade (n=187) | |||
| G1–2 | 48 (55.8) | 70 (69.3) | 0.0567 |
| G3 |
38 (44.2) |
31 (30.7) |
— |
|
pT (n=187) | |||
| pT1–2 | 21 (24.4) | 28 (27.7) | 0.6086 |
| pT3–4 |
65 (75.6) |
73 (72.3) |
— |
|
pN (n=187) | |||
| pN0 | 39 (45.4) | 56 (55.5) | 0.1687 |
| pN1–2 |
47 (54.7) |
45 (44.6) |
— |
|
pM (n=186) | |||
| pM0 | 75 (87.2) | 94 (94.0) | 0.1091 |
| pM1 |
11 (12.8) |
6 (6.0) |
— |
|
V (n=187) | |||
| V+ | 18 (20.9) | 11 (10.9) | 0.0587 |
| V− |
68 (79.1) |
90 (89.1) |
— |
|
L (n=187) | |||
| L+ | 41 (47.7) | 34 (33.7) | 0.0514 |
| L− |
45 (52.3) |
67 (66.3) |
— |
|
Tumor budding (n=148) | |||
| Low-grade | 50 (74.6) | 40 (49.4) | 0.0017 |
| High-grade |
17 (25.4) |
41 (50.6) |
— |
|
Tumor growth pattern (n=187) | |||
| Infiltrating | 68 (79.1) | 66 (65.4) | 0.038 |
| Pushing |
18 (20.9) |
35 (34.7) |
— |
|
Mismatch repair status (n=186) | |||
| Proficient | 77 (89.5) | 96 (96.0) | 0.0847 |
| Deficient |
9 (10.5) |
4 (4.0) |
— |
|
B-RAF mutation (V600E) (n=180) | |||
| Wild type | 71 (86.6) | 92 (93.9) | 0.0957 |
| Mutated |
11 (13.4) |
6 (6.1) |
— |
|
KRAS mutation (codon 12/13), (n=171) | |||
| Wild type | 54 (71.1) | 59 (62.1) | 0.2194 |
| Mutated |
22 (29.0) |
36 (37.9) |
— |
|
Hazard ratio for death with loss of RKIP (n=186) | |||
| HR (95%CI) |
1.37 (0.84–2.22) |
0.2082 |
|
|
Adjuvant therapy (n=187) | |||
| None | 30 (43.5) | 56 (47.5) | 0.5983 |
| Treated | 39 (56.5) | 62 (52.5) | — |
Analysis
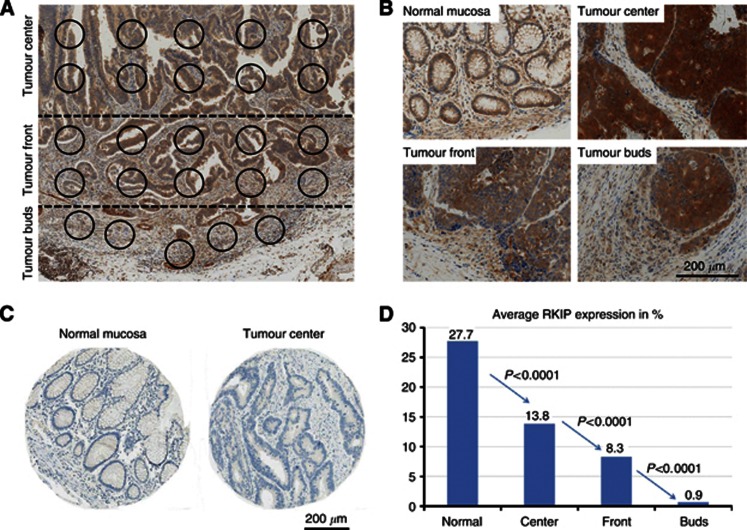
Antibodies for RKIP (Merck Millipore, Billerica, MA, USA; Cat no. 07–137; rabbit anti-human RKIP, polyclonal) were used at a dilution of 1 : 1000. RKIP staining was performed using a Bond Max Autostainer from Leica Microsystems (Wetzlar, Germany) with antigen retrieval performed in citrate buffer at 100 °C for 20 min. For every case, the percentage of RKIP-positive cells per HPF was evaluated in a total of 35 randomly selected high-power fields (HPFs) in the tumour centre, invasion front, tumour buds (10HPF each) and normal mucosa (5 HPF) independent of staining intensity (Figure 2A and B). Normal colonic mucosa and nerve tissue served as an internal positive control; negative controls were performed without primary antibody. Isotype controls were established using a matched multi-punch test TMA of CRC and normal mucosa (Figure 2C). Based on receiver operating characteristic (ROC)-curve analysis for survival, tumour regions with >10% of RKIP-positive cells were classified as positive. Cases that did not allow a clear definition of the different histological zones were excluded from the analysis (33 cases). This exclusion criterion occurred randomly. The average expression in tumour centre, front and tumour buds was assessed for its correlation with clinicopathological features and molecular characteristics of each case. Analysis of mismatch-protein expression was performed using a four-marker panel (MLH1, MSH2, MSH6 and PMS2) as previously described (Koelzer et al, 2012). For markers of EMT, staining for NF-κBp65 and E-Cadherin was performed using a matched multi-punch TMA from the same cohort containing two spots each from the tumour centre and tumour front. NF-κBp65 immunostaining (Merck Millipore; Cat. no. D14E12; 1 : 1000) was performed using a Bond Max Autostainer from Leica Microsystems (Wetzlar, Germany) with antigen retrieval in citrate buffer at 100 °C for 30 min; staining for E-cadherin (Dako, Baar, Switzerland; Cat. no. NCH-38; 1 : 100) was performed as previously described (Karamitopoulou et al, 2010). KRAS mutational status (codon 12/13) and B-Raf mutational status was established using pyrosequencing.
Figure 2.
(A) RKIP-stained full-tissue section with exemplary identification of the geographic areas tumour centre, tumour front and tumour buds and high-power fields selected for evaluation (B) Immunohistochemistry for RKIP showing normal mucosa, tumour centre, tumour front and tumour buds (all 100 × ). (C) Isotype controls for RKIP staining on normal mucosa and tumour tissue. (D) Average expression of RKIP (% of positive cells in 10HPF) in the different histological zones of CRC. Expression of RKIP is progressively lost from normal mucosa to tumour centre, tumour front and tumour buds (P<0.0001).
Statistical analysis
Differences in RKIP expression by tissue type were evaluated using the Kruskal–Wallis test. Cutoff for RKIP expression was derived using ROC curve analysis and the end point survival (Zlobec et al, 2007), based on the current cohort. The association of RKIP loss with clinicopathological features was assessed using the χ2, or Fisher's exact test where appropriate. Differences in survival time were displayed using Kaplan–Meier curves and tested using the log-rank test in univariate analysis. The independent prognostic effect of RKIP after adjusting for other potential confounding factors was analysed using a Cox regression model, after verification of the proportional hazards assumption. Adjustment for multiple hypothesis testing was not undertaken (Perneger, 1998), rather P-values <0.05 were considered statistically significant. Analyses were carried out using SAS (V9.2, SAS Institute, Cary, NC, USA). This study complied with the REporting recommendations for tumour MARKer prognostic studies (REMARK) criteria (McShane et al, 2005).
Results
Geographic analysis of RKIP protein expression
The highest average expression of RKIP was consistently observed in normal mucosa (27% RKIP+ cells), with a significant reduction in cancer tissue (11% RKIP+ cells across all areas). Interestingly, within CRCs, RKIP expression demonstrated considerable heterogeneity: RKIP was lost progressively from the tumour centre towards the invasive front and tumour buds (13.8%, 8.3% and 0.9% RKIP-positivity, respectively; P<0.0001) (Figure 2D). Tumour buds consistently showed the lowest frequency of RKIP expression, in comparison to tumour centre and front, with 60.8% of cases showing complete loss of RKIP expression in this area.
Association of RKIP expression with clinicopathological features
RKIP loss in the tumour centre independently predicted more frequent distant metastatic disease (P=0.0307). Of 17 patients with distant metastasis, 12 showed loss of RKIP expression in the primary tumour (Table 2). Further, a strong trend towards vascular invasion (P=0.0506) was observed. In patients with loss of RKIP expression, vascular invasion was observed in 21.2% of cases, while only 10.8% of patients with maintained RKIP expression in their primary tumours presented with this feature. Further, RKIP-negative tumours showed an aggressive phenotype with invasive tumour border configuration in up to 81.2% of cases, independent of the geographic distribution of RKIP loss (infiltrative tumour growth, centre: P=0.0084; front P=0.038; (Tables 2 and 3). At the same time, intense tumour budding (>10 tumour buds per HPF) was observed in tumours with RKIP loss in tumour centre or front (centre: P=0.0112; front P=0.0017). RKIP loss at the invasive front correlated inversely with tumour size (P=0.0229), and identified a trend towards vascular and lymphatic invasion (P=0.0587 and P=0.0514 respectively), while no association with nodal or distant metastatic disease was observed (Table 3). Loss of RKIP was frequent in tumour buds, with 60.8% of cases demonstrating complete loss in this geographic area of CRC. Within tumour buds themselves, no association of RKIP expression with clinicopathological features was observed (Supplementary Table S3).
Association of RKIP expression with survival
Loss of RKIP expression in the tumour centre predicted a significantly increased hazard ratio for death (HR (95%CI): 2.13 (1.27–3.56); P=0.0042), independently of TNM classification and therapy (P=0.0474) (Figure 3A, Tables 2 and 4). In contrast, RKIP expression at the front and in tumour buds did not show utility for the prediction of survival (HR (95% CI): 1.37 (0.84–2.22); P=0.2082 and HR (95% CI): 0.98 (0.58–1.68); P=0.9467, respectively) (Figure 3B and C). We further analysed the predictive value of RKIP expression for survival in the tumour centre stratified by stage: In stage III CRC, a significant correlation between maintained RKIP expression and prolonged survival was observed (P=0.01), while early-stage patients did not derive prognostic benefit (P=0.9585) (Figure 4A and B). Further subgroup analysis demonstrated that the prognostic value of RKIP expression in the tumour centre was maintained in stage III CRC patients receiving chemotherapy (P=0.002), (Figure 4C).
Figure 3.
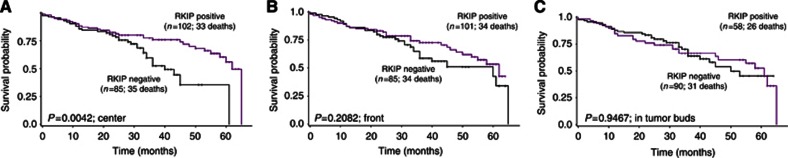
Prognostic effects of RKIP expression in tumour centre, tumour front and tumour buds. (A) Loss of RKIP expression in the tumour centre predicted a significantly increased hazard ratio for death (HR (95% CI): 2.13 (1.27–3.56); P=0.0042). (B, C) In contrast, no impact of RKIP expression on outcome was observed at the tumour front and in tumour buds (HR (95% CI): 1.37 (0.84–2.22); P=0.2082 and HR (95% CI): 0.98 (0.58–1.68); P=0.9467, respectively).
Table 4. Multivariate Cox regression analysis of RKIP expression in the tumor center and other established prognostic variables.
| HR (95% CI) | P-value | |
|---|---|---|
|
RKIP expression | ||
| Negative (<10%) | 1.78 (1.01–3.07) | 0.0474 |
| Positive (>10%) |
1.0 |
— |
|
T-Stage | ||
| pT1–2 | 1.0 | 0.5021 |
| pT3–4 |
1.34 (0.57–3.17) |
— |
|
N-Stage | ||
| pN0 | 1.0 | 0.0001 |
| pN1–2 |
4.15 (2.0–8.6) |
— |
|
M-Stage | ||
| pM0 | 1.0 | 0.0002 |
| pM1 |
3.44 (1.79–6.6) |
|
|
Postop therapy | ||
| No | 1.0 | 0.1127 |
| Yes | 0.61 (0.33–1.12) | — |
Figure 4.
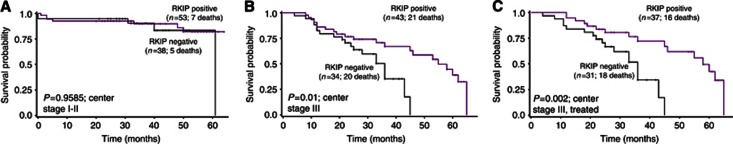
Prognostic effects of RKIP expression in tumour centre stratified by stage and therapy. (A) No impact on prognosis was observed in early-stage patients (P=0.9585). (B) Stage III patients with maintained RKIP expression derived a significant survival benefit (P=0.01). (C) The prognostic value of RKIP in the tumor centre was maintained in stage III CRC patients receiving adjuvant chemotherapy (P=0.002).
Correlative analysis of RKIP Expression with E-Cadherin and NF- κ B
Correlative analysis of average RKIP expression in the tumour centre with markers of EMT demonstrated a significant positive correlation with staining for E-Cadherin in this geographic area of CRC (P<0.0001; r=0.4). Cases with loss of RKIP infrequently demonstrated maintained E-Cadherin expression, indicating loss of cell adhesion. Further, RKIP expression in the tumour centre correlated inversely with an increased frequency of nuclear translocation of NF-κB (P=0.0002; r=−0.156), indicating that loss of RKIP correlates with more nuclear positivity for the marker and increased activation of the NF-κB signalling pathway.
Analysis of RKIP expression in a molecular pathology context
As RKIP serves a central role in the regulation of the mitogen-activated kinase (MAPK) signalling cascade, which is initialised by Raf-1, RKIP protein expression was analysed in the context of KRAS and activating B-Raf (V600) mutation frequency. Interestingly, both KRAS and activating B-Raf mutations did not show an impact on RKIP protein expression in any of the areas analysed (Tables 2 and 3 and Supplementary Table S3). RKIP maintained its prognostic significance when adjusting for B-Raf status (P=0.0051; HR (95% CI): 2.15 (1.26–3.67)) as well as KRAS-mutation (P=0.0029; HR (95% CI): 2.28 (1.3–3.9)) in multivariate analysis. B-Raf and KRAS mutations were not prognostic in our series (P=0.958 and P=0.1986 respectively). Loss of RKIP expression in the tumour centre and in tumour buds was significantly more common in MMR-deficient cancers (P=0.0191 and P=0.05, respectively), while no association was observed at the tumour front.
Discussion
This study presents a novel geographic analysis of RKIP expression pattern in primary CRC in accordance with the REMARK guidelines for biomarker prognostic studies (McShane et al, 2005). By these means, we identify considerable intratumoural heterogeneity of RKIP expression in primary CRC. Interestingly, the prognostic value of RKIP expression in CRC is shown to be limited to the tumour centre. In this geographic area, loss of RKIP is a valuable marker for the prediction of aggressive histopathological features, metastatic relapse and poor outcome in CRC patients and correlates with histomorphological features of EMT, loss of E-Cadherin and NF-κB activation.
The present study contributes to the understanding of the role of RKIP in CRC in several ways. In a first step, we analyse RKIP expression in a well-characterised set of CRC-patients using a novel geographic approach towards protein expression analysis by immunohistochemistry. Our results illustrate a considerable biological variation of RKIP protein expression in primary CRC with progressive loss of RKIP from the tumour centre towards the tumour front and almost complete absence in tumour buds. Previously conducted studies have been based on tissue microarray analysis (Al-Mulla et al, 2006; Minoo et al, 2007; Zlobec et al, 2008; Doyle et al, 2013), which may cause considerable underestimation of the true distribution of RKIP expression based on the well-described genetic heterogeneity of CRC (Gerlinger et al, 2012). A first indication of intratumoural heterogeneity of RKIP expression in CRC was provided by systematic analysis of proteins from different signalling pathways in the tumour centre and invasive front in a previously conducted study by our group (Karamitopoulou et al, 2011). However, a systematic protein expression analysis in all histological areas of CRC, the examination of RKIP expression in tumour buds and the systematic investigation of the molecular pathology context of RKIP expression was not performed.
Second, we show that loss of RKIP expression in the tumour centre is strongly associated with aggressive histopathological features, metastatic relapse and poor clinical outcome independent of TNM stage and postoperative therapy. This data confirms previous studies that identify RKIP as a predictive marker of metastasis and decreased survival outcome in CRC (Al-Mulla et al, 2006; Minoo et al, 2007; Zlobec et al, 2008; Doyle et al, 2013). In these studies, both single-punch (Al-Mulla et al, 2006; Minoo et al, 2007; Zlobec et al, 2008) and multi-punch TMAs (Doyle et al, 2013) from the tumour centre were used in non-geographic analysis. Interestingly, the prognostic value of RKIP in geographic analysis is restricted to the tumour centre, confirming these previous studies that were limited to tumor samples of this area. In correlative subgroup analysis, stage III patients of our series with maintained RKIP expression in the tumor centre derived significant prognostic benefit, while the prognosis of stage II patients was not influenced by RKIP expression. These results based on geographic analysis of RKIP protein expression contrast with previously published studies that identify a significant prognostic impact of RKIP expression in early-stage CRC patients (Al-Mulla et al, 2006; Zlobec et al, 2008; Doyle et al, 2013). Differences in study design, analytical methods (antibodies, staining protocols), the material used (TMA vs full tissue sections), therapy and patient selection may contribute to the observed discrepancies in the survival analysis of the early-stage subset. Clearly, the prognostic value of RKIP expression of the stage II subset needs to be further studied, ideally in randomized prospective clinical trials.
Third, we demonstrate a maintained correlation between RKIP expression in the tumour centre and improved survival outcome in stage III patients receiving chemotherapy. A direct comparison between treated and untreated stage III patients was not possible with adequate statistical power in our study cohort. This is due to the low frequency of stage III patients who did not receive chemotherapeutic agents in our series, as adjuvant treatment corresponds to the standard of care for CRC patients presenting with nodal metastasis. Consequently, studies analysing impact of RKIP on therapy response in CRC are few: Minoo et al (2007) first demonstrated an increased response rate in a retrospective analysis of CRC patients receiving adjuvant therapy. An enhanced response to chemotherapeutic agents dependent on RKIP expression has also previously been demonstrated in several other epithelial malignancies, including breast and prostate cancer (Chatterjee et al, 2004). On a mechanistic level, RKIP has been shown to regulate tumour-cell resistance to apoptotic stimuli through the induction of TRAIL (Baritaki et al, 2007). Al-Mulla et al (2012) have provided further insights into how RKIP expression effectively sensitises CRC cell lines to chemotherapeutic and immunotherapeutic drugs: RKIP enhances the stability of Kelch-like ECH-associated protein 1 (KEAP 1) inhibiting the activation of NF-E2-related nuclear factor 2 (NRF2)-responsive genes, including intracellular antioxidants and redox-regulating genes. Based on these mechanistic insights and the available retrospective clinical data, RKIP expression in the tumour centre of CRC may be a promising future predictive marker for evaluation in randomized multicenter trials and prospective studies.
Fourth, the current study demonstrates a strong correlation of the loss of RKIP with morphological hallmarks of EMT such as an invasive border configuration and strong tumour budding activity both in the tumour centre and tumour front. On a signalling level, we identify strong correlations of RKIP-loss in the tumour centre with nuclear translocation of NF-κB and loss of E-cadherin expression. This confirms previous data by our group that identified loss of RKIP as a potential feature of a protein phenotype for the prediction of tumour budding in CRC (Karamitopoulou et al, 2010). We now further advance this analysis by correlation of geographic expression patterns of RKIP with features of EMT both on a morphological and signalling level and RKIP expression profiling of tumour buds using full-tissue sections. These data provide further evidence of the association of RKIP loss with features of EMT as histological hallmarks of biological – and clinical – aggressiveness in CRC. RKIP has been suggested to have a major and pleiotropic role for negative regulation of EMT through inhibition of both Raf-1/MEK/ERK and NF-κB mediated signalling (Wu and Bonavida, 2009). RKIP also negatively influences the expression of β-catenin, SNAIL and SLUG by enhancing glycogen synthase kinase-3β signalling, and downregulates central mediators of cell cycle progression and cell motility (Al-Mulla et al 2011a). Loss of RKIP disinhibits NF-κB, which contributes to the induction of metastasis through the activation of its downstream transcriptional target SNAIL in cancer cells (Wu et al, 2009); SNAIL contributes to the induction of EMT through positive-feedback regulation of WNT-signalling resulting in downregulation of E-Cadherin and nuclear translocation of β-catenin (Wu and Bonavida, 2009). The current study provides further evidence of a link between loss of RKIP and activation of EMT on a histopathologic level based on the presence of tumour budding and an invasive tumour border configuration in cases with loss of RKIP at the tumour centre or front. Further, we demonstrate frequent RKIP loss in tumour buds themselves, implicating that loss of RKIP may be an early event during EMT. In correlative analysis, we identify a significant correlation between loss of RKIP, E-Cadherin negativity and nuclear translocation of NF-κB, in agreement with the abovementioned mechanistic studies.
Next, we performed a novel correlative analysis of RKIP protein expression in the context of activating B-Raf (V600) and KRAS mutations in each histological zone. Interestingly, no impact of B-Raf and KRAS mutational status on RKIP protein expression or survival was observed. While the inhibitory function of RKIP on Raf-1 signalling is well described, previous studies have provided conflicting results on the ability of RKIP to inhibit wild-type B-Raf kinase (Park et al, 2005; Trakul et al, 2005). For KRAS, no direct regulatory effect of RKIP has been identified, yet RKIP interferes with the association of the central RAS-target Raf-1 with MEK (Yeung et al, 1999). It has not been investigated whether the oncogenic effects of a constitutive activation of the B-Raf or KRAS-pathway may be limited by maintained RKIP expression. Here, we demonstrate first evidence that both B-Raf and KRAS mutation do not correlate with a change of RKIP protein expression in CRC in a well-characterised cohort.
In the context of MMR deficiency, an increased frequency of RKIP-loss was observed in the tumour centre and in tumour buds, yet no association was observed at the tumour front. Previous studies have provided conclusive evidence of a strong prognostic effect of RKIP expression in MMR-deficient CRC, an effect that was attributed to anti-apoptotic effects in the context of microsatellite deficiency (Minoo et al, 2007). However, a positive correlation between loss of RKIP expression and MMR proteins was not observed (Al-Mulla et al, 2006; Minoo et al, 2007). Silencing of the RKIP promoter through methylation of CpG islands has been identified as a possible mechanism leading to RKIP loss in MMR-proficient CRC (Al-Mulla et al, 2008). Further, we have previously characterised a significant relation between decreased RKIP expression and high CpG island methylator phenotype in CRC (Zlobec et al, 2011). As MMR-deficient cancers demonstrate the highest degree of promoter methylation (Bettington et al, 2013), the observed increased frequency of RKIP-loss may be indirect evidence of RKIP promoter-silencing through aberrant methylation. Interestingly, conflicting results were provided by a previous study of RKIP promoter methylation in CRC: In 28 selected cases with loss of RKIP, no methylation of the promoter region of RKIP could be identified by methylation-specific PCR independent of MMR status (Minoo et al, 2007). However, these results may be influenced by the small number of patients investigated, the resolution of the method employed as well as the selected promoter region, and should be further analysed.
This study must be interpreted in the context of its limitations. First, a differentiation between phosphorylated RKIP and unmodified RKIP protein could not be detected based on the immunohistochemical expression analysis performed. Experimental studies have provided conclusive evidence that RKIP can be phosporylated at serine 153 (Ser-153) by protein kinase C, which leads to dissociation from Raf-1 (Corbit et al, 2003). The analysis of the contribution of pRKIP to the prognostic impact of RKIP expression may be an interesting leverage point for further study. Further, mismatch repair status was determined using a four-marker panel for MMR-protein expression, which does not allow a specific differentiation between sporadic and hereditary (HNPCC-associated) microsatellite instable CRCs (Cicek et al, 2011). Owing to the low frequency of MMR-deficient cancers and limited patient material, a specific differentiation between HNPCC and sporadic microsatellite instable CRC patients was not attempted.
The present study provides a detailed characterisation of RKIP expression patterns in CRC in correlation with tumour morphology, molecular features and prognosis. By geographic protein expression analysis, we demonstrate that loss of RKIP expression in the tumour centre is strongly associated with both the biological and clinical behaviour of the cancer. First, loss of RKIP expression in the tumour centre correlates with the features of EMT and is most predictive of aggressive histopathological features including tumour budding, an infiltrative tumour border configuration, vascular invasion and distant metastasis. Second, RKIP expression in the tumour centre holds independent prognostic value that is maintained under chemotherapeutic treatment of advanced CRC. Third, molecular analysis demonstrated that RKIP expression is not influenced by B-Raf or KRAS mutations. We therefore recommend the evaluation of RKIP expression in the tumour centre of CRC as a biomarker for prospective validation in randomized multicenter trials.
Acknowledgments
We would like to thank Caroline Hammer, Mary Economou and Dr Dominique Müller, PhD, for their expert technical support.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Material
References
- Al-Mulla F, Bitar MS, Al-Maghrebi M, Behbehani AI, Al-Ali W, Rath O, Doyle B, Tan KY, Pitt A, Kolch W. Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3β. Cancer Res. 2011a;71:1334–1343. doi: 10.1158/0008-5472.CAN-10-3102. [DOI] [PubMed] [Google Scholar]
- Al-Mulla F, Bitar MS, Taqi Z, Rath O, Kolch W. RAF kinase inhibitory protein (RKIP) modulates cell cycle kinetics and motility. Mol Biosyst. 2011b;7:928–941. doi: 10.1039/c0mb00208a. [DOI] [PubMed] [Google Scholar]
- Al-Mulla F, Bitar MS, Feng J, Park S, Yeung KC. A new model for raf kinase inhibitory protein induced chemotherapeutic resistance. PLoS One. 2012;7:e29532. doi: 10.1371/journal.pone.0029532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mulla F, Hagan S, Behbehani AI, Bitar MS, George SS, Going JJ, García JJ, Scott L, Fyfe N, Murray GI, Kolch W. Raf kinase inhibitor protein expression in a survival analysis of colorectal cancer patients. J Clin Oncol. 2006;24:5672–5679. doi: 10.1200/JCO.2006.07.5499. [DOI] [PubMed] [Google Scholar]
- Al-Mulla F, Hagan S, Al-Ali W, Jacob SP, Behbehani AI, Bitar MS, Dallol A, Kolch W. Raf kinase inhibitor protein: mechanism of loss of expression and association with genomic instability. J Clin Pathol. 2008;61:524–529. doi: 10.1136/jcp.2007.046987. [DOI] [PubMed] [Google Scholar]
- Baritaki S, Katsman A, Chatterjee D, Yeung KC, Spandidos DA, Bonavida B. Regulation of tumour cell sensitivity to TRAIL-induced apoptosis by the metastatic suppressor Raf kinase inhibitor protein via Yin Yang 1 inhibition and death receptor 5 up-regulation. J Immunol. 2007;179:5441–5453. doi: 10.4049/jimmunol.179.8.5441. [DOI] [PubMed] [Google Scholar]
- Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. 2013;62:367–386. doi: 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
- Chatterjee D, Bai Y, Wang Z, Beach S, Mott S, Roy R, Braastad C, Sun Y, Mukhopadhyay A, Aggarwal BB, Darnowski J, Pantazis P, Wyche J, Fu Z, Kitagwa Y, Keller ET, Sedivy JM, Yeung KC. RKIP sensitizes prostate and breast cancer cells to drug-induced apoptosis. J Biol Chem. 2004;279:17515–17523. doi: 10.1074/jbc.M313816200. [DOI] [PubMed] [Google Scholar]
- Cicek MS, Lindor NM, Gallinger S, Bapat B, Hopper JL, Jenkins MA, Young J, Buchanan D, Walsh MD, Le Marchand L, Burnett T, Newcomb PA, Grady WM, Haile RW, Casey G, Plummer SJ, Krumroy LA, Baron JA, Thibodeau SN. Quality assessment and correlation of microsatellite instability and immunohistochemical markers among population- and clinic-based colorectal tumours results from the Colon Cancer Family Registry. J Mol Diagn. 2011;13:271–281. doi: 10.1016/j.jmoldx.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbit KC, Trakul N, Eves EM, Diaz B, Marshall M, Rosner MR. Activation of Raf-1 signaling by protein kinase C through a mechanism involving Raf kinase inhibitory protein. J Biol Chem. 2003;278:13061–13068. doi: 10.1074/jbc.M210015200. [DOI] [PubMed] [Google Scholar]
- Doyle B, Hagan S, Al-Mulla F, Scott L, Harden S, Paul J, Mulcahy H, Murray GI, Sheahan K, O'Sullivan J, Kolch W. Raf kinase inhibitor protein expression combined with peritoneal involvement and lymphovascular invasion predicts prognosis in Dukes' B colorectal cancer patients. Histopathology. 2013;62:505–510. doi: 10.1111/his.12014. [DOI] [PubMed] [Google Scholar]
- Fu Z, Smith PC, Zhang L, Rubin MA, Dunn RL, Yao Z, Keller ET. Effects of raf kinase inhibitor protein expression on suppression of prostate cancer metastasis. J Natl Cancer Inst. 2003;95:878–889. doi: 10.1093/jnci/95.12.878. [DOI] [PubMed] [Google Scholar]
- Fu Z, Kitagawa Y, Shen R, Shah R, Mehra R, Rhodes D, Keller PJ, Mizokami A, Dunn R, Chinnaiyan AM, Yao Z, Keller ET. Metastasis suppressor gene Raf kinase inhibitor protein (RKIP) is a novel prognostic marker in prostate cancer. Prostate. 2006;66:248–256. doi: 10.1002/pros.20319. [DOI] [PubMed] [Google Scholar]
- Fujimori Y, Inokuchi M, Takagi Y, Kato K, Kojima K, Sugihara K. Prognostic value of RKIP and p-ERK in gastric cancer. J Exp Clin Cancer Res. 2012;31:30. doi: 10.1186/1756-9966-31-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumour heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan S, Al-Mulla F, Mallon E, Oien K, Ferrier R, Gusterson B, García JJ, Kolch W. Reduction of Raf-1 kinase inhibitor protein expression correlates with breast cancer metastasis. Clin Cancer Res. 2005;11:7392–7397. doi: 10.1158/1078-0432.CCR-05-0283. [DOI] [PubMed] [Google Scholar]
- Huerta-Yepez S, Yoon NK, Hernandez-Cueto A, Mah V, Rivera-Pazos CM, Chatterjee D, Vega MI, Maresh EL, Horvath S, Chia D, Bonavida B, Goodglick L. Expression of phosphorylated raf kinase inhibitor protein (pRKIP) is a predictor of lung cancer survival. BMC Cancer. 2011;11:259. doi: 10.1186/1471-2407-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jass JR, Atkin WS, Cuzick J, Bussey HJ, Morson BC, Northover JM, Todd IP. The grading of rectal cancer: historical perspectives and a multivariate analysis of 447 cases. Histopathology. 1986;10:437–459. doi: 10.1111/j.1365-2559.1986.tb02497.x. [DOI] [PubMed] [Google Scholar]
- Karamitopoulou E, Lugli A, Panayiotides I, Karakitsos P, Peros G, Rallis G, Patsouris ES, Terracciano L, Zlobec I. Systematic assessment of protein phenotypes characterizing high-grade tumour budding in mismatch repair-proficient colorectal cancer. Histopathology. 2010;57:233–243. doi: 10.1111/j.1365-2559.2010.03615.x. [DOI] [PubMed] [Google Scholar]
- Karamitopoulou E, Zlobec I, Panayiotides I, Patsouris ES, Peros G, Rallis G, Lapas C, Karakitsos P, Terracciano LM, Lugli A. Systematic analysis of proteins from different signaling pathways in the tumour centre and the invasive front of colorectal cancer. Hum Pathol. 2011;42:1888–1896. doi: 10.1016/j.humpath.2010.06.020. [DOI] [PubMed] [Google Scholar]
- Karamitopoulou E, Zlobec I, Kölzer V, Kondi-Pafiti A, Patsouris ES, Gennatas K, Lugli A. Proposal for a 10-high-power-fields scoring method for the assessment of tumor budding in colorectal cancer. Mod Pathol. 2013;26:295–301. doi: 10.1038/modpathol.2012.155. [DOI] [PubMed] [Google Scholar]
- Kim HS, Kim GY, Lim SJ, Kim YW. Loss of Raf-1 kinase inhibitory protein in pancreatic ductal adenocarcinoma. Pathology. 2010;42:655–660. doi: 10.3109/00313025.2010.522172. [DOI] [PubMed] [Google Scholar]
- Kim HS, Won KY, Kim GY, Kim SC, Park YK, Kim YW. Reduced expression of Raf-1 kinase inhibitory protein predicts regional lymph node metastasis and shorter survival in esophageal squamous cell carcinoma. Pathol Res Pract. 2012;208:292–299. doi: 10.1016/j.prp.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Koelzer VH, Baker K, Kassahn D, Baumhoer D, Zlobec I. Prognostic impact of β-2-microglobulin expression in colorectal cancers stratified by mismatch repair status. J Clin Pathol. 2012;65:996–1002. doi: 10.1136/jclinpath-2012-200742. [DOI] [PubMed] [Google Scholar]
- Martinho O, Gouveia A, Silva P, Pimenta A, Reis RM, Lopes JM. Loss of RKIP expression is associated with poor survival in GISTs. Virchows Arch. 2009;455:277–284. doi: 10.1007/s00428-009-0821-z. [DOI] [PubMed] [Google Scholar]
- Martinho O, Granja S, Jaraquemada T, Caeiro C, Miranda-Gonçalves V, Honavar M, Costa P, Damasceno M, Rosner MR, Lopes JM, Reis RM. Downregulation of RKIP is associated with poor outcome and malignant progression in gliomas. PLoS One. 2012;7:e30769. doi: 10.1371/journal.pone.0030769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics REporting recommendations for tumour MARKer prognostic studies (REMARK) Br J Cancer. 2005;93:387–391. doi: 10.1038/sj.bjc.6602678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR, Lugli A. Loss of raf-1 kinase inhibitor protein expression is associated with tumour progression and metastasis in colorectal cancer. Am J Clin Pathol. 2007;127:820–827. doi: 10.1309/5D7MM22DAVGDT1R8. [DOI] [PubMed] [Google Scholar]
- Park S, Yeung ML, Beach S, Shields JM, Yeung KC. RKIP downregulates B-Raf kinase activity in melanoma cancer cells. Oncogene. 2005;24:3535–3540. doi: 10.1038/sj.onc.1208435. [DOI] [PubMed] [Google Scholar]
- Perneger TV. What's wrong with Bonferroni adjustments. BMJ. 1998;7139:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakul N, Menard RE, Schade GR, Qian Z, Rosner MR. Raf kinase inhibitory protein regulates Raf-1 but not B-Raf kinase activation. J Biol Chem. 2005;280:24931–24940. doi: 10.1074/jbc.M413929200. [DOI] [PubMed] [Google Scholar]
- Wu K, Bonavida B. The activated NF-kappaB-Snail-RKIP circuitry in cancer regulates both the metastatic cascade and resistance to apoptosis by cytotoxic drugs. Crit Rev Immunol. 2009;29:241–254. doi: 10.1615/critrevimmunol.v29.i3.40. [DOI] [PubMed] [Google Scholar]
- Wu Y, Deng J, Rychahou PG, Qiu S, Evers BM, Zhou BP. Stabilization of snail by NF-kappaB is required for inflammation-induced cell migration and invasion. Cancer Cell. 2009;15:416–428. doi: 10.1016/j.ccr.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung K, Seitz T, Li S, Janosch P, McFerran B, Kaiser C, Fee F, Katsanakis KD, Rose DW, Mischak H, Sedivy JM, Kolch W. Suppression of Raf-1 kinase activity and MAP kinase signalling by RKIP. Nature. 1999;401:173–177. doi: 10.1038/43686. [DOI] [PubMed] [Google Scholar]
- Zlobec I, Baker K, Minoo P, Jass JR, Terracciano L, Lugli A. Node-negative colorectal cancer at high risk of distant metastasis identified by combined analysis of lymph node status, vascular invasion, and Raf-1 kinase inhibitor protein expression. Clin Cancer Res. 2008;14:143–148. doi: 10.1158/1078-0432.CCR-07-1380. [DOI] [PubMed] [Google Scholar]
- Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 2011;225:336–343. doi: 10.1002/path.2879. [DOI] [PubMed] [Google Scholar]
- Zlobec I, Steele R, Terracciano L, Jass JR, Lugli A. Selecting immunohistochemical cut-off scores for novel biomarkers of progression and survival in colorectal cancer. J Clin Pathol. 2007;60:1112–1116. doi: 10.1136/jcp.2006.044537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.