Abstract
Objective
There is increasing evidence that variation in gene copy number (CN) influences clinical phenotype. The low-affinity Fcγ receptor 3B (FCGR3B) located in the FCGR gene cluster is a CN polymorphic gene involved in the recruitment to sites of inflammation and activation of polymorphonuclear neutrophils (PMNs). Given recent evidence that low FCGR3B CN is a risk factor for systemic but not organ-specific autoimmune disease and the potential importance of PMN in the pathophysiology of rheumatoid arthritis (RA), the authors hypothesised that FCGR3B gene dosage influences susceptibility to RA.
Methods
FCGR3B CN was measured in 643 cases of RA and 461 controls from New Zealand (NZ), with follow-up analysis in 768 cases and 702 controls from the Netherlands and 250 cases and 211 controls from the UK. All subjects were of Caucasian ancestry.
Results
Significant evidence for an association between CN <2 and RA was observed in the Dutch cohort (OR 2.01 (95% CI 1.37 to 2.94), p=3×10–4) but not in the two smaller cohorts (OR 1.45 (95% CI 0.92 to 2.26), p=0.11 and OR 1.33 (95% CI 0.58 to 3.02), p=0.50 for the NZ and UK populations, respectively). The association was evident in a meta-analysis which included a previously published Caucasian sample set (OR 1.67 (95% CI 1.28 to 2.17), p=1.2×10–4).
Conclusions
One possible mechanism to explain the association between reduced FCGR3B CN and RA is the reduced clearance of immune complex during infl ammation. However, it is not known whether the association between RA and FCGR3B CN is aetiological or acts as a proxy marker for another biologically relevant variant. More detailed examination of genetic variation within the FCGR gene cluster is required.
INTRODUCTION
Copy number variation (CNV) in immune-response genes has been implicated in several autoimmune diseases including CCL3L1 involvement in rheumatoid arthritis (RA)1 and systemic lupus erythematosus (SLE),2 and association of β-defensins with Crohn’s disease3 and psoriasis.4 Recently, CNV in the gene coding for the Fcγ receptor 3B (FCGR3B), the most abundant neutrophil binding site for polymeric IgG and immune complexes, has also been shown to be associated with immune-mediated glomerulonephritis,5 SLE6,7 and, albeit with a conflicting nature of association, antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis: CN <2 as risk factor6 or CN >2 as risk factor.7
The Fcγ receptor IIIb is constitutively expressed by polymorphonuclear leucocytes8 and is involved in many critical neutrophil functions including recruitment to sites of inflammation9,10 and activation.11 In addition, anti-FcγRIIIb antibodies and soluble FcγRIIIb (released from apoptotic polymorphonuclear neutrophils (PMNs) or shed from activated PMNs by cleavage of the glycosylphosphatidylinositol anchor) delay neutrophil apoptosis12,13 and may compete with IgG for binding sites to perpetuate local inflammation.12 The presence of autoantibodies and accumulation of activated neutrophils at sites of inflammation is a feature of RA and SLE, and there is evidence that these cells are functionally abnormal. For example, extended survival of PMN is observed in RA and SLE, which may reflect dysregulation of apoptosis, a major mechanism by which the activity of neutrophils is controlled.14 The pathogenesis of RA is mediated in part by PMN, which compose 80–90% of the cellular infiltrate during active RA.15–17 In addition to releasing proinflammatory cytokines including tumour necrosis factor (TNF), activated PMN produce large amounts of potentially damaging reactive oxygen species (ROS) (O2, H2O2 and HOCL) and granular enzymes such as proteases and permeability-inducing factors, all of which potentially contribute to tissue damage and joint erosion (see Edwards and Hallett17 for review). FcγRIIIb has been implicated in the release of these toxic products in response to soluble immune complexes such as those found in RA joints.11 Genetic variation other than CN variants in the FCGR locus has already been associated with RA.18 Furthermore, FcγR-mediated production of ROS and apoptosis is impaired in PMN isolated from patients with RA.11,19
There is clinical evidence implicating FcγR in the pathogenesis of RA,14–16 association of a haplotype encompassing FCGR3B with RA,20 and association of FCGR3B CN with SLE and other systemic but not organ-specific immune-mediated pathology.5–7 RA is a disease with both organ-specific and non-specific features which primarily affects the synovial membrane but also involves a number of other tissues. On this basis, we hypothesised that FCGR3B CN plays a role in the pathogenesis of RA. This was tested in a multicentre study by measuring FCGR3B CN in three Caucasian RA case–control cohorts.
METHODS
Study subjects
All study subjects were of European Caucasian descent. The New Zealand (NZ) RA cohort consisted of 643 patients recruited from outpatient clinics in Auckland, Bay of Plenty, Wellington, Canterbury, Otago and Southland in NZ. The control group (n=461) consisted of healthy subjects recruited from Otago. The UK RA cohort consisted of 250 patients with RA recruited at Lewisham and Guy’s and St Thomas’ Hospitals. One hundred and fifty-nine UK controls were purchased from the European Collection of Cell Cultures (ECACC) (http://www.hpacultures.org.uk/collections/ecacc.jsp) and genotyped by us, with the data augmented by a further 52 ECACC control samples genotyped by Hollox et al.21 The Dutch cases (n=768) were recruited from the central and eastern regions of the Netherlands.22,23 These were compared to 702 controls recruited from blood donor centres within Utrecht and Amsterdam. All the patients with RA were diagnosed according to the 1987 American College of Rheumatology criteria for RA.24 Demographic and clinical information on the cases and controls is given in table 1.
Table 1.
Demographic and clinical characteristics
| Age (mean, range, N*) |
Women % (N) |
RF % (N) |
CCP % (N) |
SE positivity % (N) |
|
|---|---|---|---|---|---|
| New Zealand | |||||
| Cases | 65.5, 20–88, 351† | 78.8 (643) | 82.3 (560) | 67.0 (367) | 80.1 (626) |
| Controls | 44.6, 17–95, 461† | 61.2 (461) | – | – | – |
| UK | |||||
| Cases | NA | 79.3 (241) | 63.1 (241) | NA | 79.1 (239) |
| Controls | NA | NA | – | – | – |
| The Netherlands | |||||
| Cases | 48.7, 4–79, 578‡ | 65.5 (586) | 78.5 (685) | NA | NA |
| Controls | NA | 49.2 (702) | – | – | – |
N is the number of individuals from which data were available.
Age at recruitment.
Age at disease onset.
CCP, cyclic citrullinated peptide; NA, not available; RF, rheumatoid factor.
For the NZ and Netherlands cohorts and the UK case cohort, DNA was prepared from whole blood by chloroform extraction. The ECACC genomic DNA was derived from lymphoblastoid cell lines. Recruitment and DNA extraction occurred over a period of more than 5 years for the NZ samples, whereas DNA extraction occurred over a period of less than 2 years for the Dutch samples and UK case samples. All TaqMan Real-Time PCR was done at the University of Otago in NZ. This necessitated shipping dried samples from the UK and the Netherlands to NZ where they were subsequently resuspended. All aqueous DNA was stored suspended in Tris-EDTA at −20°C.
Each of the three sample sets had ≥95% power (α=0.05) to detect an effect equivalent to that reported for FCGR3B CN <2 in SLE in a UK sample set (OR 2.21).6 At lower effect sizes, power decreased (eg, at OR 1.5 the power was 75% in the NZ sample set, 86% in the Netherlands sample set and 41% in the UK sample set). The combined sample set, however, was adequately powered to detect effects >1.3 (79% power at OR 1.3).
Measurement of FCGR3B CN
At the FCGR3 locus there are two patterns of CNV that include FCGR3B25—the major one also encompasses FCGR2C and the minor one also encompasses HSPA7/FCGR2C/FCGR3A/HSPA6 (deviation from CN=2 present in 11.2% and 0.8%, respectively, of a healthy sample set from the Netherlands25). The exact boundaries of CNV are not known.
FCGR3B CN was measured using TaqMan Real-Time PCR, as described in the online supplement, using an assay based on one previously described for CCL3L1.26 Variations in DNA quality increase the ‘noise’ of the assay system,21 which can prevent clear clustering of continuous values around whole integers and poor quality DNA increases the risk of false positive results.27 All NZ and Netherlands case genomic DNA samples were electrophoresed on an agarose gel and integrity scored from 1 (single high molecular weight band) to 4 (most degraded). Samples with scores of 3 and 4 were discarded from analysis. This was not done for the Netherlands control samples because they exhibited discrete CN bins (see fig 4 in online supplement), suggesting no blurring of CN calls owing to DNA degradation.27 Insufficient genomic DNA was available for the UK samples to enable testing by agarose gel. To minimise DNA degradation, NZ samples were freshly diluted from 200 μg/ml stock and used within 2 weeks.
Assignation of FCGR3B CN
In order to compensate for differences in mean and variance distribution between cases and controls, the a posteriori model fitting CNVtools algorithm28 was used to determine the cut-off CN value between 1 and 2 for each sample set and CN was assigned as described in more detail in the online supplement.
As additional evaluations of the qPCR assay, 72 NZ controls were first analysed by paralogue ratio testing (PRT)/restriction enzyme digest variant ratio using a previously developed and validated assay.21 Our data demonstrated good agreement between PRT (which has a first-pass error rate of 8%29) and TaqMan assays within the range of 1–3 copies (see table 2 in online supplement), and there was strong agreement between the assigned NA type by both methods. Eight of the PRT results differed by one copy compared with CNVtools assignment, with the PRT value uncertain for one of the differences. We then compared genotypes between the 30 overlapping UK ECACC controls genotyped by us and by Hollox et al21 (see table 3 in online supplement) and achieved 100% concordance in CN assignment.
Association analysis
The distributions of FCGR3B CN between patients and controls after CN assignment by CNVtools were compared using the χ2 test. Logistic regression analysis was used to measure the RA-associated risks of low FCGR3B CN as the OR relative to CN ≥2 using the STATA 7.0 statistical package (StataCorp, College Station, Texas, USA). Because a mixture of CNVtools fitting models28 were needed for our cohorts, combined (meta-) analysis of all three cohorts was performed using the Mantel–Haenszel test, comparing CN <2 to ≥2 after testing for heterogeneity by the Breslow–Day test.
RESULTS
The distribution of FCGR3B CN as a continuous variable is shown in figure 1. CN assignment showed that 93.6% of the combined control cohort had two or more copies of FCGR3B and 6.4% carried single copy deletions (table 2). No control individuals lacked both alleles. One patient in each of the Dutch and NZ case cohorts had a null FCGR3B genotype. The FCGR3B CN allele distribution among our controls was very similar to that observed previously based on measurement by the multiplex ligand-dependent probe amplification technique in a cohort of 129 healthy individuals of Northern European ancestry (CN 0, 0%; 1, 7%; 2, 83%; 3, 10%),24 and similar to that observed using an integrated ;approach in UK samples (CN 0, 0%; CN 1, 4%; 2, 87%; >3, 9%).21
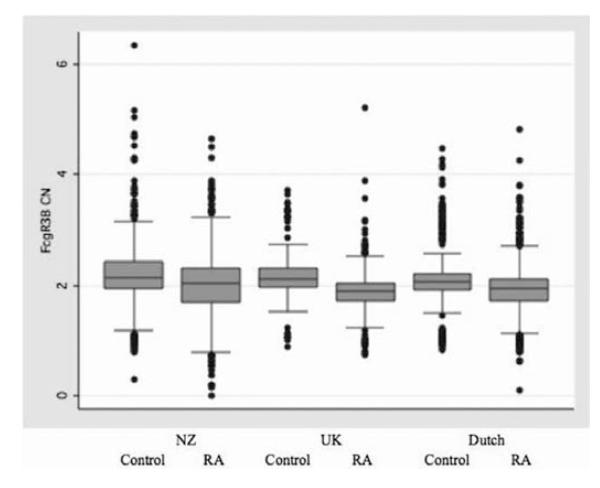
Figure 1.
Distribution of Fcγ receptor 3B copy number (CN) as a continuous variable in the three case and control cohorts. Data are shown without removal of samples. Boxes and lines encompass samples within 1 and 2 SD from the mean, respectively. Mean (SD) values for the six sample sets are (from left to right): 2.23 (0.45), 2.04 (0.39), 2.18 (0.25), 1.89 (0.20), 2.14 (0.24) and 1.92 (0.26).
Table 2.
Fcγ receptor 3B (FCGR3B) copy number (CN) and risk for rheumatoid arthritis (RA)
| FCGR3B CN | Cases, N (frequency) | Controls, N (frequency) | OR (95% CI) | p Value | |
|---|---|---|---|---|---|
|
|
|||||
| New Zealand RA | <2 | 60 (0.099) | 32 (0.071) | 1.45 (0.92 to 2.26) | 0.11 |
| ≥2 | 546 (0.901) | 421 (0.929) | 1.00 | ||
| UK RA | <2 | 15 (0.062) | 10 (0.047) | 1.33 (0.58 to 3.02) | 0.50 |
| ≥2 | 228 (0.938) | 202 (0.953) | 1.00 | ||
| Dutch RA | <2 | 86 (0.116) | 43 (0.061) | 2.01 (1.37 to 2.94) | 3.0×10−4 |
| ≥2 | 655 (0.884) | 658 (0.939) | 1.00 | ||
| Breunis RA25 | <2 | 10 (0.089) | 9 (0.070) | 1.31 (0.51 to 3.34) | 0.57 |
| ≥2 | 102 (0.901) | 120 (0.930) | 1.00 | ||
| Combined RA* | <2 | 171 (0.100) | 94 (0.063) | 1.67 (1.28 to 2.17) | 1.2×10−4 |
| ≥2 | 1531 (0.900) | 1401 (0.937) | 1.00 | ||
p=0.60, Breslow–Day.
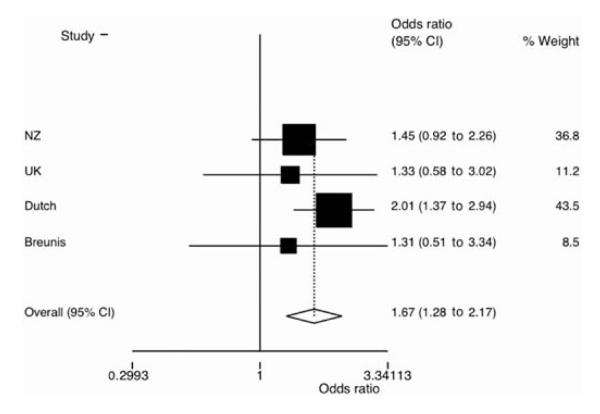
To test the hypothesis that FCGR3B CN influences susceptibility to RA, the influence on disease risk of CN <2 of FCGR3B was tested under the prediction that any association of FCGR3B with RA would be similar to that evident in SLE where CN <2 increases the risk of disease by slowing the rate of immune complex clearance during transient inflammation.5–7 Table 2 shows the disease risk for CN 0–1 relative to CN ≥2. Significant evidence for an association between CN <2 and RA was observed in the Dutch cohort (OR 2.01 (95% CI 1.37 to 2.94), p=3×10−4) but not in the two smaller cohorts (OR 1.45 (95% CI 0.92 to 2.26), p=0.11 and OR 1.33 (95% CI 0.58 to 3.02), p=0.50 for the NZ and UK populations, respectively). In a combined (meta) analysis that included previously published data from a small case–control sample set of Northern European ancestry (table 2; OR 1.31 (95% CI 0.51 to 3.34)), there was strong evidence for CN <2 FCGR3B increasing the risk of disease (figure 2; OR 1.67 (95% CI 1.28 to 2.17), p=1.2×10−4).
Figure 2.
Meta-analysis of the association between low Fcγ receptor 3B (FCGR3B) copy number and rheumatoid arthritis for the four sample sets using Metan (STATA Version 7.0) with a fixed Mantel–Haenszel pooling model. FCGR3B CN of 0–1 was designated as the minor allele and copy number ≥2 as the major allele.
Given that rheumatoid factor (RF) is an anti-FCG, we tested for an association between FCGR3B CN and the presence or absence of RF (table 3). There was some evidence for an association of low CN with RF-negative RA in the NZ and UK cohorts (p=0.08 and p=0.02, respectively), but not in the Dutch cohort (p=0.69). In the combined cohorts there was no evidence for differential association between FCGR3B CN <2 and RF status, nor was there any evidence for an association between NA type and RA for NZ and UK samples with CN 1 or 2 (see table 4 in online supplement).
Table 3.
Fcγ receptor 3B (FCGR3B) distribution in rheumatoid factor (RF) positive and negative patients
| FCGR3B CN | RF negative, N (frequency) |
RF positive, N (frequency) |
OR (95% CI) | p Value | |
|---|---|---|---|---|---|
| New Zealand (RA) | <2 | 14 (0.156) | 41 (0.094) | 0.56 (0.29 to 1.08) | 0.08 |
| 2 | 76 (0.844) | 396 (0.896) | 1.00 | ||
| UK RA | <2 | 10 (0.115) | 5 (0.034) | 0.27 (0.09 to 0.82) | 0.02 |
| 2 | 77 (0.885) | 142 (0.966) | 1.00 | ||
| Dutch RA | <2 | 16 (0.112) | 64 (0.124) | 1.13 (0.63 to 2.02) | 0.69 |
| 2 | 127 (0.888) | 451 (0.876) | 1.00 | ||
| Combined RA* | <2 | 40 (0.125) | 110 (0.100) | 0.78 (0.53 to 1.14) | 0.20 |
| 2 | 280 (0.875) | 989 (0.900) | 1.00 |
p=0.055, Breslow–Day.
CN, copy number; RA, rheumatoid arthritis.
DISCUSSION
Our results provide evidence that CNV in FCGR3B is associated with RA (combined analysis of four Caucasian RA case–control sample sets: OR 1.67 (95% CI 1.28 to 2.17), p=1.2×10−4). While only the largest sample set exhibited a significant association with RA, the other three included in the meta-analysis consistently showed an increased risk for RA (OR >1) in the presence of FCGR3B deletion, and there was no significant intercohort heterogeneity (table 2; p=0.60, Breslow–Day). While the ANCA-associated systemic vasculitis data require further clarification,6,7 our results are consistent with previous reports of an association between low CN and systemic autoimmunity in other diseases including glomerulonephritis and SLE.5–7 This suggests a common pathogenic role for FCGR3B (or an associated variant in the FCGR locus) in systemic autoimmune diseases.
Confirmed genetic associations have defined aberration in the activation, regulation and migration of T cells as central to the aetiology of RA in Caucasian populations, including HLA-DRB1,30 protein tyrosine phosphatase N22,31 a locus near the TNF-induced protein 3 gene,32,33 the signal transducer and activator of transcription 4 gene,34 a locus between the TNF receptor-associated factor 1 and C5 genes35,36 and the CD40, chemokine ligand 21, CD122, KIF5A and protein kinase chloroquine genes.37,38 The association of FCGR3B CN with RA reported here still requires further replication in other populations before it can be considered confirmed. Nonetheless, if this association is validated, it would implicate genetic regulation of immune complex deposition and clearance in the pathophysiology of RA. Although there was no association between FCGR3B CN and RF status, we had no information on the presence or absence of anti-cyclic citrullinated peptides (anti-CCPs) in all of our populations and therefore could not analyse the relationship between anti-CCP and FCGR3B CN. Antibodies to citrullinated proteins are now considered a more specific marker of disease than RF status,39 and although there is a large overlap between anti-CCP and the presence/absence of RF, it is possible that FCGR3B deletion is more strongly associated with anti-CCP negative RA. This needs to be addressed in future studies.
Although quantitative PCR (qPCR) has been a popular method for measuring CNV owing to suitability for genotyping a large number of samples, and is a relatively accurate method of detecting single-copy deletions and duplications,29 there are a number of factors that can influence the accuracy of assays. Because CN is measured relative to a reference gene, variation in amplification efficiency and experimental conditions can alter the ratio of the two products and thus apparent CN.28 DNA quality also influences assay efficiency and can introduce false positives and batch effects when comparing cohorts collected and stored under different conditions.27,28 Because the CN values are (rarely) integer values, arbitrary ‘binning’ of samples to whole numbers can introduce false associations if the distribution of two populations differs in mean and/or variance.28 To compensate for batch effects and systematic errors, cases and controls were assayed concurrently. The modelling algorithm CNVtools was used to determine the cut-off between CNs 1 and 2 and those with values ±0.05 of the cut-off between CNs 1 and 2 were removed. Independent verification of the qPCR technique was carried out using PRT (see Materials and methods). Because degraded DNA samples can yield spurious CNV results,27 samples exhibiting excessive degradation were removed from analysis in this study.
Genotyping within the FCGR locus is a complex undertaking owing to the extensive homology that exists between the FCGR genes. Because there is FCGR3B duplication and deletion on multiple haplotypes,21,25 there are no known single nucleotide polymorphisms (SNPs) that can be used to accurately predict CN.21 A major conclusion from the comprehensive analysis of the FCGR locus by Hollox et al21 was that direct measurement of CN by an integrated suite of methods combined with association analysis of SNPs is required to fully evaluate the contribution of FCGR3B to RA. Here we focused on direct measurement of FCGR3B CN by a single method (TaqMan) that is suited to larger-scale genotyping. Future work on this locus in RA should include data on association of SNP haplotypes.
FCGR3B CN correlates with the level of receptor expression (both surface and soluble forms), neutrophil adherence to IgG and internalisation of opsonised particles.7 While our results appear counterintuitive to the possibility that FcγRIIIB deletion has a role in RA through this process, they highlight the fact that the differential roles of PMN activation and immune complex clearance in the immune response (and the exact function of FcγRIIIB) require further investigation. It is also relevant to emphasise that our data are consistent with the findings observed in SLE.6,7 There are at least three possible explanations for how reduced FcγRIIIb levels could increase the risk of RA.
First, FcγRIIIb can bind intravascular immune complexes in the absence of any other inflammatory mediators but fails to generate ROS, whereas FcγRIIa (predominantly expressed by macrophages and natural killer (NK) cells) is responsible for ROS-mediated tissue damage.40 The reduced adherence and phagocytic capabilities of PMN in the case of low FCGR3B CN may delay clearance of immune complexes during episodes of transient inflammation and prolong the inflammatory response either to initial joint damage or to an exogenous infection that subsequently triggers an autoimmune response. This would allow the development of an inflammatory milieu where FcγRIIa activity predominates, as has been suggested for glomerulonephritis and SLE.6,7,41
Second, changes in FCGR3B CN may alter PMN function indirectly by shifting the balance of activating receptors and potentially ‘unmasking’ the effects of functional polymorphisms in other FCG receptors. Furthermore, PMN are an important source of proinflammatory chemokines, which are partly mediated through FcγRIIa and FcγRIIIa.8 Both are FCG receptors that have been implicated in RA and SLE.41
A third possibility is that the increased susceptibility to RA associated with low FCGR3B CN is due not to reduced FCGR3B dosage but to deletion in FCGR2C, since CNV in these two genes is strongly related.25,42 FcγRIIc is expressed on macrophages, PMN43 and NK cells44 where it triggers antibody-dependent cellular toxicity (ADCC).45 ADCC may play an important role in regulating autoimmune responses by lysing autologous immature dendritic cells.45,46 Defective NK cell cytotoxicity has been reported in a number of autoimmune disorders including RA, juvenile RA, SLE and macrophage activation syndrome.47,48 An SNP within exon 5 of FCGR2C is functionally equivalent to a deletion of the gene. The null allele of this polymorphism occurs more often in patients with RA than in controls and results in an apparent shift towards increased expression of the inhibitory FcγRIIb.49 Whether this is also the case in the context of FCGR3B/FCGR2C deletion is unknown. It should be noted that rare (~1%) deletion events encompassing FCGR3B/FCGR2C/FCGR3A have been observed in Caucasian populations.25
In this paper we provide evidence for an association between a low FCGR3B CN and susceptibility to RA. Whether this is due to biological effects of FcγRIIIb itself or is a proxy marker for another biologically relevant polymorphism will require further study, validation in other sample sets and better characterisation of structural variation within the FCGR locus.
Supplementary Material
Acknowledgements
The authors would like to thank NZ research nurses Gael Hewett and Sue Yeoman for assistance in recruiting patients, and Chris Barnes for his assistance with CNVtools.
Funding This work was supported by the Health Research Council of New Zealand, Arthritis New Zealand and the NZ Child Health Research Foundation, Medical Research Council (UK) intramural funding from the Clinical Sciences Centre and Wellcome Trust project grant 083167. BZA was supported by a grant from the Netherlands Organization for Health Research and Development (ZonMw; grant number 016.096.121).
Footnotes
Competing interests None.
Ethics approval Ethical approval for the study in New Zealand was given by the MultiRegion (cases) and Lower South Ethics Committees (controls), in the UK by the local research ethics committees of Lewisham Hospital and Guy’s and St Thomas’ Hospitals and in The Netherlands by the local ethics committee. All subjects gave written informed consent.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.McKinney C, Merriman ME, Chapman PT, et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67:409–13. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 2.Mamtani M, Rovin B, Brey R, et al. CCL3L1 gene-containing segmental duplications and polymorphisms in CCR5 affect risk of systemic lupus erythaematosus. Ann Rheum Dis. 2008;67:1076–83. doi: 10.1136/ard.2007.078048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley RW, Pearson J, Gearry RB, et al. Association of higher DEFB4 genomic copy number with Crohn’s disease. Am J Gastroenterol. 2010;105:354–9. doi: 10.1038/ajg.2009.582. [DOI] [PubMed] [Google Scholar]
- 4.Hollox EJ, Huffmeier U, Zeeuwen PL, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–5. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aitman TJ, Dong R, Vyse TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–5. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 6.Fanciulli M, Norsworthy PJ, Petretto E, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–3. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willcocks LC, Lyons PA, Clatworthy MR, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–82. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fanger MW, Shen L, Graziano RF, et al. Cytotoxicity mediated by human Fc receptors for IgG. Immunol Today. 1989;10:92–9. doi: 10.1016/0167-5699(89)90234-X. [DOI] [PubMed] [Google Scholar]
- 9.Kocher M, Siegel ME, Edberg JC, et al. Cross-linking of Fc gamma receptor IIa and Fc gamma receptor IIIb induces different proadhesive phenotypes on human neutrophils. J Immunol. 1997;159:3940–8. [PubMed] [Google Scholar]
- 10.Coxon A, Cullere X, Knight S, et al. Fc gamma RIII mediates neutrophil recruitment to immune complexes. A mechanism for neutrophil accumulation in immune-mediated inflammation. Immunity. 2001;14:693–704. doi: 10.1016/s1074-7613(01)00150-9. [DOI] [PubMed] [Google Scholar]
- 11.Fossati G, Moots RJ, Bucknall RC, et al. Differential role of neutrophil Fcgamma receptor IIIB (CD16) in phagocytosis, bacterial killing, and responses to immune complexes. Arthritis Rheum. 2002;46:1351–61. doi: 10.1002/art.10230. [DOI] [PubMed] [Google Scholar]
- 12.Durand V, Pers JO, Renaudineau Y, et al. Soluble Fcgamma receptor IIIb alters the function of polymorphonuclear neutrophils but extends their survival. Eur J Immunol. 2001;31:1952–61. doi: 10.1002/1521-4141(200107)31:7<1952::aid-immu1952>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 13.Youinou P, Durand V, Renaudineau Y, et al. Pathogenic effects of anti-Fc gamma receptor IIIb (CD16) on polymorphonuclear neutrophils in non-organ-specific autoimmune diseases. Autoimmun Rev. 2002;1:13–19. doi: 10.1016/s1568-9972(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 14.Peng SL. Neutrophil apoptosis in autoimmunity. J Mol Med. 2006;84:122–5. doi: 10.1007/s00109-005-0007-3. [DOI] [PubMed] [Google Scholar]
- 15.Wipke BT, Allen PM. Essential role of neutrophils in the initiation and progression of a murine model of rheumatoid arthritis. J Immunol. 2001;167:1601–8. doi: 10.4049/jimmunol.167.3.1601. [DOI] [PubMed] [Google Scholar]
- 16.Mandik-Nayak L, Allen PM. Initiation of an autoimmune response: insights from a transgenic model of rheumatoid arthritis. Immunol Res. 2005;32:5–13. doi: 10.1385/IR:32:1-3:005. [DOI] [PubMed] [Google Scholar]
- 17.Edwards SW, Hallett MB. Seeing the wood for the trees: the forgotten role of neutrophils in rheumatoid arthritis. Immunol Today. 1997;18:320–4. doi: 10.1016/s0167-5699(97)01087-6. [DOI] [PubMed] [Google Scholar]
- 18.Lee YH, Ji JD, Song GG. Associations between FCGR3A polymorphisms and susceptibility to rheumatoid arthritis: a metaanalysis. J Rheumatol. 2008;35:2129–35. doi: 10.3899/jrheum.080186. [DOI] [PubMed] [Google Scholar]
- 19.Fairhurst AM, Wallace PK, Jawad AS, et al. Rheumatoid peripheral blood phagocytes are primed for activation but have impaired Fc-mediated generation of reactive oxygen species. Arthritis Res Ther. 2007;9:R29. doi: 10.1186/ar2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morgan AW, Barrett JH, Griffiths B, et al. Analysis of Fcgamma receptor haplotypes in rheumatoid arthritis: FCGR3A remains a major susceptibility gene at this locus, with an additional contribution from FCGR3B. Arthritis Res Ther. 2006;8:R5. doi: 10.1186/ar1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollox EJ, Detering JC, Dehnugara T. An integrated approach for measuring copy number variation at the FCGR3 (CD16) locus. Hum Mutat. 2009;30:477–84. doi: 10.1002/humu.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welsing PM, van Riel PL. The Nijmegen inception cohort of early rheumatoid arthritis. J Rheumatol Suppl. 2004;69:14–21. [PubMed] [Google Scholar]
- 23.Kievit W, Fransen J, Oerlemans AJ, et al. The efficacy of anti-TNF in rheumatoid arthritis, a comparison between randomised controlled trials and clinical practice. Ann Rheum Dis. 2007;66:1473–8. doi: 10.1136/ard.2007.072447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnett FC, Edworthy SM, Bloch DA, et al. The American Rheumatism Association 1987 revised criteria for the classifi cation of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 25.Breunis WB, van Mirre E, Geissler J, et al. Copy number variation at the FCGR locus includes FCGR3A, FCGR2C and FCGR3B but not FCGR2A and FCGR2B. Hum Mutat. 2009;30:E640–50. doi: 10.1002/humu.20997. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 genecontaining segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 27.Cukier HN, Pericak-Vance MA, Gilbert JR, et al. Sample degradation leads to false-positive copy number variation calls in multiplex real-time polymerase chain reaction assays. Anal Biochem. 2009;386:288–90. doi: 10.1016/j.ab.2008.11.040. [DOI] [PubMed] [Google Scholar]
- 28.Barnes C, Plagnol V, Fitzgerald T, et al. A robust statistical method for case-control association testing with copy number variation. Nat Genet. 2008;40:1245–52. doi: 10.1038/ng.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Armour JA, Palla R, Zeeuwen PL, et al. Accurate, high-throughput typing of copy number variation using paralogue ratios from dispersed repeats. Nucleic Acids Res. 2007;35:e19. doi: 10.1093/nar/gkl1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wordsworth BP, Lanchbury JS, Sakkas LI, et al. HLA-DR4 subtype frequencies in rheumatoid arthritis indicate that DRB1 is the major susceptibility locus within the HLA class II region. Proc Natl Acad Sci USA. 1989;86:10049–53. doi: 10.1073/pnas.86.24.10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YH, Rho YH, Choi SJ, et al. The PTPN22 C1858T functional polymorphism and autoimmune diseases – a meta-analysis. Rheumatology (Oxford) 2007;46:49–56. doi: 10.1093/rheumatology/kel170. [DOI] [PubMed] [Google Scholar]
- 32.Plenge RM, Cotsapas C, Davies L, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomson W, Barton A, Ke X, et al. Rheumatoid arthritis association at 6q23. Nat Genet. 2007;39:1431–3. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Remmers EF, Plenge RM, Lee AT, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plenge RM, Seielstad M, Padyukov L, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis – a genomewide study. N Engl J Med. 2007;357:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kurreeman FA, Rocha D, Houwing-Duistermaat J, et al. Replication of the tumor necrosis factor receptor-associated factor 1/complement component 5 region as a susceptibility locus for rheumatoid arthritis in a European family-based study. Arthritis Rheum. 2008;58:2670–4. doi: 10.1002/art.23793. [DOI] [PubMed] [Google Scholar]
- 37.Barton A, Thomson W, Ke X, et al. Rheumatoid arthritis susceptibility loci at chromosomes 10p15, 12q13 and 22q13. Nat Genet. 2008;40:1156–9. doi: 10.1038/ng.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raychaudhuri S, Remmers EF, Lee AT, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–23. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishimura K, Sugiyama D, Kogata Y, et al. Meta-analysis: diagnostic accuracy of anti-cyclic citrullinated peptide antibody and rheumatoid factor for rheumatoid arthritis. Ann Intern Med. 2007;146:797–808. doi: 10.7326/0003-4819-146-11-200706050-00008. [DOI] [PubMed] [Google Scholar]
- 40.Tsuboi N, Asano K, Lauterbach M, et al. Human neutrophil Fcgamma receptors initiate and play specialized nonredundant roles in antibody-mediated inflammatory diseases. Immunity. 2008;28:833–46. doi: 10.1016/j.immuni.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuchiya N, Kyogoku C, Miyashita R, et al. Diversity of human immune system multigene families and its implication in the genetic background of rheumatic diseases. Curr Med Chem. 2007;14:431–9. doi: 10.2174/092986707779941041. [DOI] [PubMed] [Google Scholar]
- 42.de Haas M, Kleijer M, van Zwieten R, et al. Neutrophil Fc gamma RIIIb deficiency, nature, and clinical consequences: a study of 21 individuals from 14 families. Blood. 1995;86:2403–13. [PubMed] [Google Scholar]
- 43.Cassel DL, Keller MA, Surrey S, et al. Differential expression of Fc gamma RIIA, Fc gamma RIIB and Fc gamma RIIC in hematopoietic cells: analysis of transcripts. Mol Immunol. 1993;30:451–60. doi: 10.1016/0161-5890(93)90113-p. [DOI] [PubMed] [Google Scholar]
- 44.Metes D, Galatiuc C, Moldovan I, et al. Expression and function of Fc gamma RII on human natural killer cells. Nat Immun. 1994;13:289–300. [PubMed] [Google Scholar]
- 45.Ernst LK, Metes D, Herberman RB, et al. Allelic polymorphisms in the FcgammaRIIC gene can influence its function on normal human natural killer cells. J Mol Med. 2002;80:248–57. doi: 10.1007/s00109-001-0294-2. [DOI] [PubMed] [Google Scholar]
- 46.Pazmany L. Do NK cells regulate human autoimmunity? Cytokine. 2005;32:76–80. doi: 10.1016/j.cyto.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Dalbeth N, Callan MF. A subset of natural killer cells is greatly expanded within inflamed joints. Arthritis Rheum. 2002;46:1763–72. doi: 10.1002/art.10410. [DOI] [PubMed] [Google Scholar]
- 48.Villanueva J, Lee S, Giannini EH, et al. Natural killer cell dysfunction is a distinguishing feature of systemic onset juvenile rheumatoid arthritis and macrophage activation syndrome. Arthritis Res Ther. 2005;7:R30–7. doi: 10.1186/ar1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stewart-Akers AM, Cunningham A, Wasko MC, et al. Fc gamma R expression on NK cells influences disease severity in rheumatoid arthritis. Genes Immun. 2004;5:521–9. doi: 10.1038/sj.gene.6364121. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.