Abstract
Objectives
To evaluate FCGR3B copy number variation (CNV) in African and European populations and to determine if FCGR3B copy number is associated with SLE and SLE nephritis risk in Afro-Caribbeans, adjusting for African genetic ancestry.
Methods
We estimated FCGR3B to determine if there were ethnic variations in CNV (unrelated unadmixed Europeans and Africans). We then examined CNV at FCGR3B in relation to SLE and SLE nephritis within a case–control collection of 134 cases of SLE (37 with SLE nephritis) and 589 population controls of mainly Afro-Caribbean descent resident in Trinidad.
Results
We found a significant difference in copy number FCGR3B distribution between unadmixed African and European UK cohorts, with 27 (29%) vs 3 (5%) for those with low (0 or 1) copy FCGR3B, respectively, P = 0.002. In a Trinidadian SLE case–control study, low FCGR3B CNV was associated with SLE risk 1.7 (95% CI 1.1, 2.8), P = 0.02, which remained after adjustment for African genetic ancestry; odds ratios (ORs) 1.7 (95% CI 1.0, 2.8), P = 0.04.
Conclusion
Our studies suggest that FCGR3B low copy number is associated with SLE risk in Afro-Caribbean populations independently of CNV due to African ancestry.
Keywords: SLE, Admixture, FCGR3B, Copy number variation, African
Introduction
Emerging analyses of copy number variation (CNV) across the genome has highlighted the role that structural variation may play in the aetiology of genetic disease susceptibility. CNV has been reported in a number of recent studies [1], with increasing reports of individual gene CNV and association of CNV with specific diseases [2–13], including an association between low FCGR3B CNV and systemic immunity in a number of European cohorts [14–16] and C4 gene deletions and SLE risk [13]. There is also evidence for regional CNV variation between different ethnic groups [1]. However, association of low FCGR3B CNV was not confirmed in a Chinese lupus nephritis cohort of 202 individuals, compared with 146 controls [17]. Further studies on Columbian patients of Spanish (Paisa) origin have confirmed association of low and high FCGR3B CNV in 146 subjects with SLE, and identified possible epistasis with CCL3L1 (CC chemokine ligand 3-like 1) [18], which is encoded by a variable copy number gene that may additionally enhance inflammatory responses and increase the chance of autoimmune disease [10].
SLE and SLE nephritis show large ethnic differences in disease risk, with prevalence rates 6–8 times higher in African-Americans and in Caribbean migrants of West African descent than in Europeans, but little is known about FCGR3B CNV in ethnically diverse African cohorts with SLE. We decided to investigate the association of low CNV in unadmixed Africans and Europeans, and subsequently in a case–control study of SLE within a Trinidadian Afro-Caribbean population.
Methods
CNV was measured in this study by paralogue ratio test (PRT) assay as previously described and validated [19]. First, we established FCGR3B CNV in a population-based control sample of unrelated unadmixed Europeans and Africans from the UK with no known renal disease. Individuals were selected via primary care clinics if they claimed all four grandparents were of either European or African descent. Additionally, data were validated from data collected on country of origin and mother’s main language. Finally, these individuals were genotyped using independent genome-wide ancestry informative markers (AIMs) as described previously [20], five-way admixture estimated using ADMIXMAP software (admixture mapping software; open source software developed at London School of Hygiene & Tropical Medicine) and those included with >97.5% African ancestry. All study participants gave written informed consent. The study was approved by London School of Hygiene & Tropical Medicine ethics board and the Trinidad and Tobago Ministry of Health.
We then examined CNV at FCGR3B (as described earlier by PRT methods) in relation to SLE and SLE nephritis within a case–control collection of 134 cases of SLE (37 with SLE nephritis) and 589 population controls of mainly Afro-Caribbean descent resident in Trinidad [21]. Novel aspects of this study included the use of five-way admixture in this population to examine separately the effects of admixture on SLE and CNV. Analysis of admixture was based on a panel of AIMs chosen to have large allele frequency differentials between West African, European, Chinese, Indian and Native American populations. A Bayesian model for population admixture, individual admixture and locus ancestry was fitted by Markov chain simulation [20].
All analyses were carried out using Stata statistical software (release 10.0, 2008; Stata Corporation, College Station, TX, USA), and adjusted for five-way admixture, age and gender. The association between the relevant outcome and CNV status at time of survey was explored using logistic regression analysis–effects on risk being estimated by odds ratios (ORs) with 95% CIs.
Results
We were able to obtain FCGR3B CNV results on ~96% of the samples using the PRT assay, which comprised: (i) UK unadmixed European and African control cohort; (ii) SLE Afro-Caribbean Trinidad cohort; and (iii) non-SLE Afro-Caribbean Trinidad controls.
FCGR3B
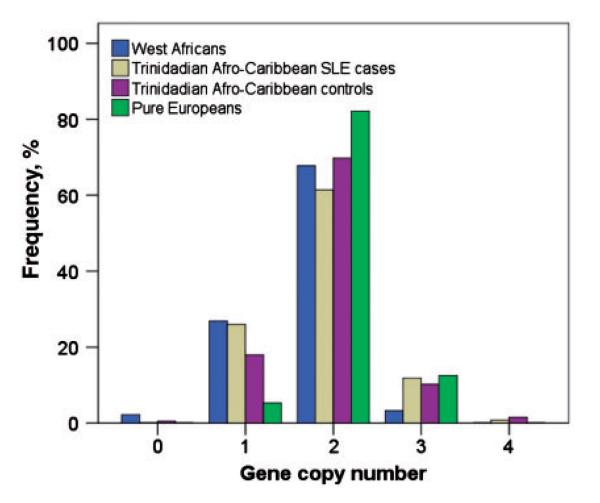
In the West African unadmixed control population, 27/93 (29%) also had significantly higher proportions of low (0, 1) FCGR3B CNV compared with European control 3/56 (5%) populations, with P = 0.002. Similar results were found for low FCGR3B in a Trinidadian Afro-Caribbean control population, P < 0.05 compared with European controls. Prevalence of FCGR3B CNV in European controls was similar when validated with results from a secondary European cohort (MRC 1946 European cohort). In Fig. 1, One Trinidadian Afro-Caribbean control (0 cases), two African controls living in the UK and no Europeans had zero copies of FCGR3B. Numbers of low FCGR3B CNV were significantly higher in SLE cases compared with Afro-Caribbean controls, P = 0.01. The Afro-Caribbean control with zero FCGR3B CNV had no evidence of other autoimmune disease or high ANA status identified from medical records. Trinidadian Afro-Caribbean SLE cases had significantly higher proportions of low (0, 1) FCGR3B CNV [32/126 (25%)] compared with non-SLE Afro-Caribbean control [79/479 (16%)] populations, with P = 0.01.
Fig. 1.
Distribution of FCGR3B by PRT CNV in West African (unadmixed), Afro-Caribbean and European populations. Totals (N): Trinidadian Afro-Caribbean SLE cases (n = 126); Trinidadian Afro-Caribbean non-SLE controls (n = 479); West Africans (n = 93); and Europeans (n = 56).
Table 1 shows summary characteristics across three groups of Afro-Caribbean patients in Trinidad: (i) Afro-Caribbean non-SLE controls; (ii) Afro-Caribbean SLE cases; and (iii) Afro-Caribbean SLE nephritis cases. Estimation of African (P = 0.0004) and European ancestry (P = 0.002) significantly differed between SLE cases and non-SLE controls. Estimates of Chinese (P = 0.0001) and Native American ancestry (P = 0.02) were significantly higher in SLE cases than in non-SLE controls. Of those with low FCGR3B CNV with genetic estimates of ancestry (admixture), SLE cases did not have significantly lower mean African ancestry than non-SLE controls (67 vs 68%, respectively), P = 0.7.
Table 1.
SLE and SLE nephritis in relation to FCGR3B copy number and African ancestry: characteristics at survey of participants
| Risk factor | Afro-Caribbean controls, n (%) n = 589 |
Afro-Caribbean SLE cases, n (%) n = 134 |
Afro-Caribbean SLE nephritis subset of SLE cases, n (%) n = 37 |
P-value All SLE cases vs non-SLE controls |
|---|---|---|---|---|
| Age at survey, yearsa | 0.2 | |||
| 15–24 | 16 (3) | 1 (1) | 0 (0) | |
| 25–34 | 76 (13) | 17 (13) | 7 (19) | |
| 35–44 | 163 (28) | 43 (32) | 12 (32) | |
| 45–54 | 148 (25) | 43 (32) | 12 (32) | |
| 55–64 | 117 (20) | 20 (15) | 3 (8) | |
| >65 | 64 (11) | 10 (7) | 3 (8) | |
| Education after 18 years | NA | 25 (19) | 7 (19) | |
| Lack of running water at 12 years | NA | 50 (37) | 15 (41) |
| Genetic ancestry Mean (SD) |
n = 558 (five-way admixture) |
n = 119 (five-way admixture) |
n = 32 (five-way admixture) |
|
|---|---|---|---|---|
| African | 0.68 (0.17) | 0.62 (0.24) | 0.61 (0.26) | 0.0004 |
| European | 0.12 (0.11) | 0.10 (0.07) | 0.09 (0.07) | 0.002 |
| Native American | 0.05 (0.05) | 0.07 (0.05) | 0.07 (0.06) | 0.02 |
| Indian | 0.08 (0.08) | 0.09 (0.07) | 0.09 (0.08) | 0.12 |
| Chinese | 0.08 (0.07) | 0.12 (0.12) | 0.14 (0.14) | 0.0001 |
| Proportions of African ancestry per copy category |
n = 450 | n = 113 | n = 31 | |
|---|---|---|---|---|
| 0, 1 copies FCGR3B | 75 (17) | 28 (25) | 10 (32) | |
| Mean African ancestry (s.d.), % | 0.68 (0.18) | 0.67 (0.25) | 0.69 (0.21) | 0.7 |
| 2 copies FCGR3B | 342 (76) | 70 (62) | 19 (61) | |
| Mean African ancestry (s.d.), % | 0.69 (0.18) | 0.59 (0.25) | 0.54 (0.29) | 0.001 |
| ≥3 copies FCGR3B | 33 (7) | 15 (13) | 2 (6) | |
| Mean African ancestry (s.d.), % | 0.65 (0.22) | 0.65 (0.21) | 0.85 (0.02) | 1.0 |
Five missing age data. N/A: not available.
Table 2 shows the association of low (0 or 1) CNV at FCGR3B with SLE from a case–control study of SLE and SLE nephritis in Trinidad Afro-Caribbeans. The crude OR adjusted for age group and gender was 1.7 (95% CI 1.1, 2.8), P = 0.02. The OR for low FCGR3B CNV associated with SLE risk remained similar after adjustment for five-way ancestry including African admixture; OR 1.7 (95% CI 1.0, 2.8), P = 0.04. For SLE nephritis, the crude OR for low FCGR3B adjusted for age group and gender was 2.3 (95% CI 1.1, 4.8), and adjusted for five-way ancestry including African admixture OR was 2.4 (95% CI 1.1, 5.5), P = 0.03.
Table 2.
Adjusted regression models for SLE and SLE nephritis risk with FCGR3B; controlling for African genetic ancestry
| SLE cases vs non-SLE controls |
SLE nephritis vs non-SLE controls |
SLE nephritis vs non-renal SLE |
||||
|---|---|---|---|---|---|---|
|
n = 562 (prevalence %), P-value |
Adjusted ORa (95% CI) genotypic |
n = 480, P-value |
Adjusted ORa (95% CI) genotypic |
n = 113, P-value |
Adjusted ORa (95% CI) genotypic |
|
| ≥2 copies FCGR3B | 1.0 (95% CI) | 1.0 (95% CI) | 1.0 (95% CI) | |||
| 0, 1 copies FCGR3B | 0.04 | 1.7 (1.0, 2.8) | 0.03 | 2.4 (1.1, 5.5) | 0.3 | 1.7 (0.7, 4.5) |
OR adjusted for age, gender and African genetic ancestry.
We confirmed that the association with low CNV was independent of five-way admixture for SLE and SLE nephritis. There was no additional risk for SLE nephritis compared with SLE patients without nephritis in either the adjusted or unadjusted models. ANA status was strongly associated with SLE diagnosis but was not associated with African admixture.
Discussion
The most significant results are the distributions of FCGR3B number between African and European populations. This is statistically significant and present in several populations of West African descent (West Africans in the UK, and Trinidadian Afro-Caribbeans).
This study suggests an association of low FCGR3B CNV with SLE and SLE nephritis in an Afro-Caribbean cohort, independent of ethnic background and is the first study to include five-way admixture. Calculations based on the actual numbers genotyped indicate our study was 80% powered to detect ORs of 1.7 and 2.4, respectively, for association of FCGR3B with SLE and lupus nephritis. Recent studies have identified that FCGR3B may be involved in immune complex clearance, as low FCGR3B is associated with SLE [12, 14, 15]. Willcocks et al. [12] showed that both in a family with Fc gamma RIIIb deficiency and in the normal population, FCGR3B CNV correlates with protein expression, with neutrophil uptake of and adherence to immune complexes, and with soluble serum Fc gamma RIIIb. The authors suggested that reduced Fc gamma RIIIb expression may contribute to the impaired clearance of immune complexes, a recognized feature of SLE, which may support the association between low FCGR3B CNV and SLE found in European (and other) populations [12]. Further recent studies have identified association between high and low FCGR3B CNV with SLE (and primary SS) in a Spanish Paisa Columbian population, stated as 85% European and 15% Amerindian with possible epistatic interactions in those SLE individuals with two copies of CCL3L1, although no formal adjustment for admixture was undertaken [18]. No association of FCGR3B was found with SLE nephritis in 202 Chinese individuals lupus nephritis cohort [17].
Further examination showed FCGR3B was not in linkage disequilibrium (LD) with any of the AIMs selected to estimate admixture. Although Afro-Caribbeans are at substantially higher risk of SLE and SLE nephritis, the risk of low FCGR3B CNV with SLE remains after adjusting for African ancestry. As in previous studies, there was no additional risk for low CNV with organ-specific immunity such as SLE nephritis, within SLE cases. Further studies will determine if the effect of FCGR3B CNV is associated with SLE risk in other ethnic groups.
Rheumatology key messages.
Our studies suggest FCGR3B low copy number is associated with SLE risk in Afro-Caribbean populations independently of CNV due to African ancestry.
We found significant differences in distribution of FCGR3B CNV between African and European populations.
Acknowledgements
We are grateful to all patients and staff who contributed to the case–control collection in Trinidad and the UK. M.M., M.F., E.P. and T.J.A designed this study. M.F. and A.L.R. performed PCR-based experiments. M.M. and M.F. performed statistical and data analysis. M.M., A.L.P. and P.M. provided material and contributed to the collection of the case–control studies. P.M. provided the admixture analyses. The manuscript was written by M.M., M.F., E.P., T.J.V. and T.J.A.
Funding: Work for this study was supported with help from the ARC (M0600 & M0651). M.M. is supported by a NIHR post-doctoral award.
Footnotes
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brasch-Andersen C, Christiansen L, Tan Q, Haagerup A, Vestbo J, Kruse TA. Possible gene dosage effect of glutathione-S-transferases on atopic asthma: using real-time PCR for quantification of GSTM1 and GSTT1 gene copy numbers. Hum Mutat. 2004;24:208–14. doi: 10.1002/humu.20074. [DOI] [PubMed] [Google Scholar]
- 3.Burns JC, Shimizu C, Gonzalez E, et al. Genetic variations in the receptor-ligand pair CCR5 and CCL3L1 are important determinants of susceptibility to Kawasaki disease. J Infect Dis. 2005;192:344–9. doi: 10.1086/430953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cook EH, Jr, Scherer SW. Copy-number variations associated with neuropsychiatric conditions. Nature. 2008;455:919–23. doi: 10.1038/nature07458. [DOI] [PubMed] [Google Scholar]
- 5.Fellermann K, Stange DE, Schaeffeler E, et al. A chromosome 8 gene-cluster polymorphism with low human beta-defensin 2 gene copy number predisposes to Crohn disease of the colon. Am J Hum Genet. 2006;79:439–48. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 7.Hollox EJ, Huffmeier U, Zeeuwen PL, et al. Psoriasis is associated with increased beta-defensin genomic copy number. Nat Genet. 2008;40:23–5. doi: 10.1038/ng.2007.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivaschenko TE, Sideleva OG, Baranov VS. Glutathione-S-transferase micro and theta gene polymorphisms as new risk factors of atopic bronchial asthma. J Mol Med. 2002;80:39–43. doi: 10.1007/s001090100274. [DOI] [PubMed] [Google Scholar]
- 9.McCarroll SA, Huett A, Kuballa P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McKinney C, Merriman ME, Chapman PT, et al. Evidence for an influence of chemokine ligand 3-like 1 (CCL3L1) gene copy number on susceptibility to rheumatoid arthritis. Ann Rheum Dis. 2008;67:409–13. doi: 10.1136/ard.2007.075028. [DOI] [PubMed] [Google Scholar]
- 11.Piirila P, Wikman H, Luukkonen R, et al. Glutathione S-transferase genotypes and allergic responses to diisocyanate exposure. Pharmacogenetics. 2001;11:437–45. doi: 10.1097/00008571-200107000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Willcocks LC, Lyons PA, Clatworthy MR, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–82. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, Chung EK, Wu YL, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am J Hum Genet. 2007;80:1037–54. doi: 10.1086/518257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aitman TJ, Dong R, Vyse TJ, et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–5. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 15.Fanciulli M, Norsworthy PJ, Petretto E, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39:721–3. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaschl H, Aitman TJ, Vyse TJ. Copy number variation in the human genome and its implication in autoimmunity. Clin Exp Immunol. 2009;156:12–6. doi: 10.1111/j.1365-2249.2008.03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lv J, Yang Y, Zhou X, et al. FCGR3B copy number variation is not associated with lupus nephritis in a Chinese population. Lupus. 2010;19:158–61. doi: 10.1177/0961203309350319. [DOI] [PubMed] [Google Scholar]
- 18.Mamtani M, Anaya JM, He W, Ahuja SK. Association of copy number variation in the FCGR3B gene with risk of autoimmune diseases. Genes Immun. 2010;11:155–60. doi: 10.1038/gene.2009.71. [DOI] [PubMed] [Google Scholar]
- 19.Hollox EJ, Detering JC, Dehnugara T. An integrated approach for measuring copy number variation at the FCGR3 (CD16) locus. Hum Mutat. 2009;30:477–84. doi: 10.1002/humu.20911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKeigue PM, Carpenter JR, Parra EJ, Shriver MD. Estimation of admixture and detection of linkage in admixed populations by a Bayesian approach: application to African-American populations. Ann Hum Genet. 2000;64:171–86. doi: 10.1017/S0003480000008022. [DOI] [PubMed] [Google Scholar]
- 21.Molokhia M, Hoggart C, Patrick AL, et al. Relation of risk of systemic lupus erythematosus to west African admixture in a Caribbean population. Hum Genet. 2003;112:310–8. doi: 10.1007/s00439-002-0883-3. [DOI] [PubMed] [Google Scholar]