Abstract
Repulsive guidance molecule (RGM) family members RGMa, RGMb/Dragon and RGMc/hemojuvelin were recently found to act as BMP co-receptors that enhance BMP signaling activity. While our previous studies have shown that hemojuvelin regulates hepcidin expression and iron metabolism through the BMP pathway, the role of the BMP signaling mediated by Dragon remains largely unknown. We have previously shown that Dragon is expressed in neural cells, germ cells and renal epithelial cells. Here, we demonstrate that Dragon is highly expressed in macrophages. Studies with RAW264.7 and J774 macrophage cell lines reveal that Dragon negatively regulates IL-6 expression in a BMP ligand dependent manner, via the p38 MAPK and Erk1/2 pathways but not the Smad1/5/8 pathway. We also generated Dragon knockout mice, and found that IL-6 is upregulated in macrophages and dendritic cells derived from whole lung tissue of these mice compared to respective cells derived from wild type littermates. These results indicate that Dragon is an important negative regulator of IL-6 expression in immune cells, and that Dragon-deficient mice may be a useful model for studying immune and inflammatory disorders.
Introduction
Bone morphogenetic proteins (BMPs) represent a large subfamily of the transforming growth factor β (TGF-β) superfamily of ligands that transduce their signals through type I and II serine/threonine kinase receptors and intracellular Smad proteins. TGF-β superfamily members play numerous roles in physiologic and pathologic processes including cell proliferation, differentiation, apoptosis, and specification of developmental fate during embryogenesis and in adult tissues (1). TGF-β signaling also regulates immune function as demonstrated by the targeted inactivation of TGF-β1 in mice, which led to a mixed inflammatory cell response and to tissue necrosis (2). Subsequent studies revealed that activins (3,4) and BMPs (5–11) also regulate inflammatory cytokines and chemokines in various cell types including macrophages, monocytes and osteoblastic cells.
Dragon (RGMb), along with two other members of the RGM (repulsive guidance molecule) family, RGMa and RGMc (hemojuvelin), are glycophosphatidylinositol (GPI)-linked membrane-associated proteins. Recently, we showed that the three RGM proteins are all co-receptors that enhance BMP signaling through increased utilization of BMP type II receptor ActRIIA by BMP2 and BMP4 (12–18). Dragon is expressed in neural tissues, where it may promote cell-cell adhesion by homophilic interactions (19). Dragon is also expressed in other organs including the ovary, testis and kidneys (13,16). In the kidney, Dragon is expressed in the epithelium of renal tubules, where it may facilitate the formation of tight junctions via the BMP/Smad signaling pathway (16). However, the expression and function of Dragon in other cells and organs have not been characterized.
Since BMP signaling can regulate macrophage function, we investigated whether Dragon plays a role in this process. We found that Dragon is highly expressed in macrophages, and is directly involved in the suppression of IL-6 expression through the p38 MAPK and Erk1/2 pathways. Through the generation of Dragon knockout mice, we report a central role of Dragon in controlling IL-6 expression in lung macrophages in vivo. This is the first BMP signaling function in vivo that has been identified for the Dragon protein.
Materials and Methods
Reverse Transcription (RT)-PCR
Total RNA was isolated from RAW264.7 macrophages using an RNeasy mini kit (Qiagen Inc.) according to the manufacturer’s instructions. First-strand cDNA synthesis was performed using an iScript cDNA synthesis kit (Bio-Rad). Transcripts of mouse BMP2, BMP4, BMP5-7, and RGMb were amplified using the primers previously described (14, 16).
siRNA knockdown
Mouse Dragon and BMPRII siRNAs were purchased from Ambion and the sequences were described previously (14). SMARTpool siRNAs against mouse Smad4 were purchased from Dharmacon. siRNA duplexes (100 nM) were added to subconfluent RAW264.7 or J774 macrophages using Lipofectamine 2000 (Invitrogen) or DharmaFectI (Dharmacon). Cells were then incubated with or without BMP4 (50 ng/ml; R&D Systems), LPS (10 ng/ml, Sigma), the p38 MAPK inhibitor SB203580 (2.5 μM), or the Erk1/2 MAPK inhibitor PD98059 (2.5 μM). Assays to measure mRNA levels of IL-6, MCP-1, TNF-α, IL-1β, IFN-γ, RGMb, Id1 and RPL19, or phosphorylation levels of Smad1/5/8, p38 MAPK or Erk1/2 MAPK were performed 46 h after transfection. For the experiments with PASMC, HUVEC, IMCD3 and C2C12 cells, Dragon siRNA duplexes were used at 60–80 nM.
Dragon cDNA transfection
Mouse Dragon cDNA (200 ng/ml) were transfected into RAW264.7 macrophages using Lipofectamine 2000. Trancfected cells were then incubated with Noggin (500 ng/ml; R&D Systems) or LDN-193189 (0, 40 and 400 ng/ml, Shanghai United Pharmatech Company, Shanghai, China). Assays to measure mRNA levels of IL-6 and RPL19 were performed 46 h after transfection.
Measurement of Gene Expression
Real-time quantification of mRNA transcripts was performed as previously described (Xia JBC and Blood). First-strand cDNA was amplified with the primers as previously described (9, 14, 20, 21). Results are expressed as a ratio of the gene of interest to RPL19.
Western blotting
Lung tissues or RAW264.7 cells were lysed in TBS (Tris.HCl, 50 mM, NaCl 150 mM, 1% Triton X-100, pH 7.4) containing protease inhibitor mixture (Pierce) and phosphatase inhibitor mixture (Pierce) for 30 min on ice. After centrifugation for 10 min at 4 °C, the supernatant was assayed for protein concentration by colorimetric assay (BCA kit, Pierce). 20–40 μg of protein was separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes. Membranes were probed with rabbit anti-phospho-Smad1/5/8, anti-phospho-MAPK p38, anti-phospho-Erk1/2 polyclonal antibodies (1:1000 dilution; Cell Signaling Technology, Beverly, MA), or goat anti-mIL-6 (R&D Systems). Membranes were stripped in 0.2 m glycine (pH 2.5) and 0.5% Tween 20 for 10 min and reprobed with rabbit anti-Smad1, anti-total MAPK p38, anti-total Erk1/2 antibodies (Cell Signaling), or anti-actin (Sigma).
Dragon knockout mice
To generate Dragon knockout mice (C57/B6/129), a mouse genomic library was screened using a mRGMb specific probe (Incyte Genomics, Palo Alto, CA). The second coding exon of mRGMb was disrupted by inserting a cassette containing an eGFP, followed by an IRES-NLS-LacZ-pA and a thymidine kinase (TK)-neomycin cassette using homologous recombination in embryonic stem (ES) cells. ES cell recombinants were screened by Southern blotting analyses. The genotyping of mRGMb mutant mice was performed by Southern blotting (primers for the probe: 5′-gtt cct agg gag aat agc gtc tcc-3′/5′-aca ggc acg ttc gtc act tga acc-3′)(Supplemental Figure 4) or PCR (5′-gtc aat ccg ccg ttt gtt ccc acg g-3′/5′-gcg tgt acc aca gcg gat ggt tcg g-3′ for the knockout allele; 5′-aca ggc acg ttc gtc act tga acc-3′/5′-gtt cct agg gag aat agc gtc tcc-3′ for the wild-type allele). Northern blot and RT-PCR analyses of mRGMb KO mice confirmed the absence of RGMb mRNA in the brain and lung respectively (Supplemental Figure 4). A sequence previously described for in situ hybridization was used for the probe for Northern blotting (22).
Mice were housed under specific pathogen-free conditions with a light/dark cycle of 12h/12h and ad lib access to food and water. Heterozygous animals were bred to obtain homozygous Dragon-null mice. All procedures were performed in accordance with Massachusetts General Hospital animal care regulations.
Flow cytometry
Mice were sacrificed 10–12 days after birth. Lungs were digested with collagenase I (Sigma-Aldrich) at 37° C for 1 h. Red blood cells were lysed with ACK lysis buffer. The cells were washed and resuspended in HBSS supplemented with 0.2% (wt/vol) BSA and 1% (wt/vol) FCS. Total number of living cells was determined with Trypan blue (Mediatech, Inc.).
Cell suspensions were incubated with a cocktail of mAbs against T cells (CD90-PE, 53-2.1), B cells (B220-PE, RA3-6B2), NK cells (CD49b-PE, DX5 and NK1.1-PE, PK136), granulocytes (Ly-6G-PE, 1A8), myeloid cells (CD11b-APC, M1/70), and monocyte subsets (Ly-6C-FITC, AL-21) (BD Biosciences). Monocytes were identified as CD11bhi (CD90/B220/CD49b/NK1.1/Ly-6G)lo (F4/80/CD11c)lo Ly-6Chi/lo; Macrophages as F4/80hi (CD11blo); dendritic cells as CD11chi CD11bhi; and neutrophils as CD11bhi Ly-6Ghi. Reported cell numbers were calculated as the product of total living cells and percent cells within their respective gates. Data were acquired on a LSRII (BD Biosciences) and analyzed with FlowJo v.8.5.2 (Tree Star, Inc.). For gene expression analysis, the cells mentioned above were sorted with a FACS Aria II (BD Biosciences) into 250 μl RLT buffer and total RNA was extracted using an RNeasy mini kit.
Data analysis
The results are presented as the mean ± SD of three determinations from two or three independent experiments for the in vitro studies, or the mean ± SD of 6–7 mice (Figure 6A, B &C), 6 mice (Figure 7A), 3–4 mice (Figure 7B) or 4–6 mice (Supplemental Figures 7) for the in vivo studies. Dragon expression levels in different cell populations in the lung of wild type mice were analyzed by one-way ANOVA. Other differences were assessed by Student t test. Differences of p < 0.05 were considered significant.
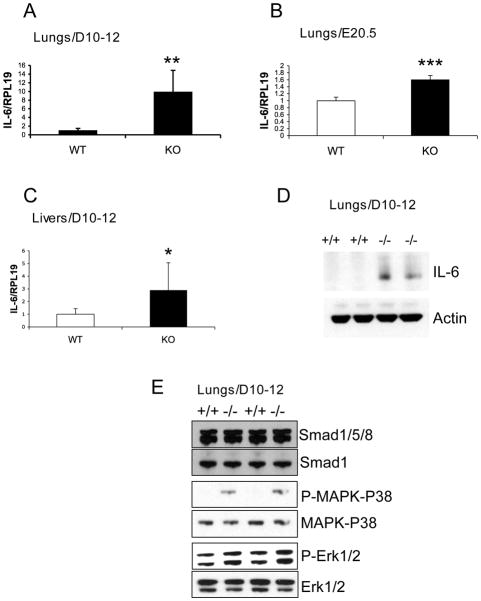
Figure 6.
IL-6 mRNA expression is up regulated in the lung and liver of Dragon knockout mice and embryos. (A-C) Total RNA was extracted from lungs (A) and livers (C) of wild type (WT) and Dragon knockout (KO) mice at ages of day 10–12 or from lungs (B) of E20.5 wild type and Dragon fetuses. mRNA levels were measured by quantitative real-time PCR, were normalized to RPL19 mRNA levels, and are expressed as a fraction of values from WT samples. (D) IL-6 protein is up regulated in the lung of Dragon knockout mice. Lysates from lungs of 10–12 days old wild type (+/+) and Dragon knockout (−/−) mice were subjected to Western blotting for IL-6. The membrane was stripped and probed with Actin antibodies. (E) Changes in phosphorylation levels of Smad1/5/8, and p38 and Erk1/2 MAPK in Dragon knockout lungs. Lysates from lungs of 10–12 days old wild type (+/+) and Dragon knockout (−/−) mice were subjected to Western blotting for phospho-Smad1/5/8, phospho-p38 MAPK and phospho-Erk1/2. The membranes were stripped and probed with total Smad1, p38-MAPK and Erk1/2 antibodies respectively. *, p < 0.05; **, p < 0.01; *** p < 0.001 versus wild type. n=6–7.
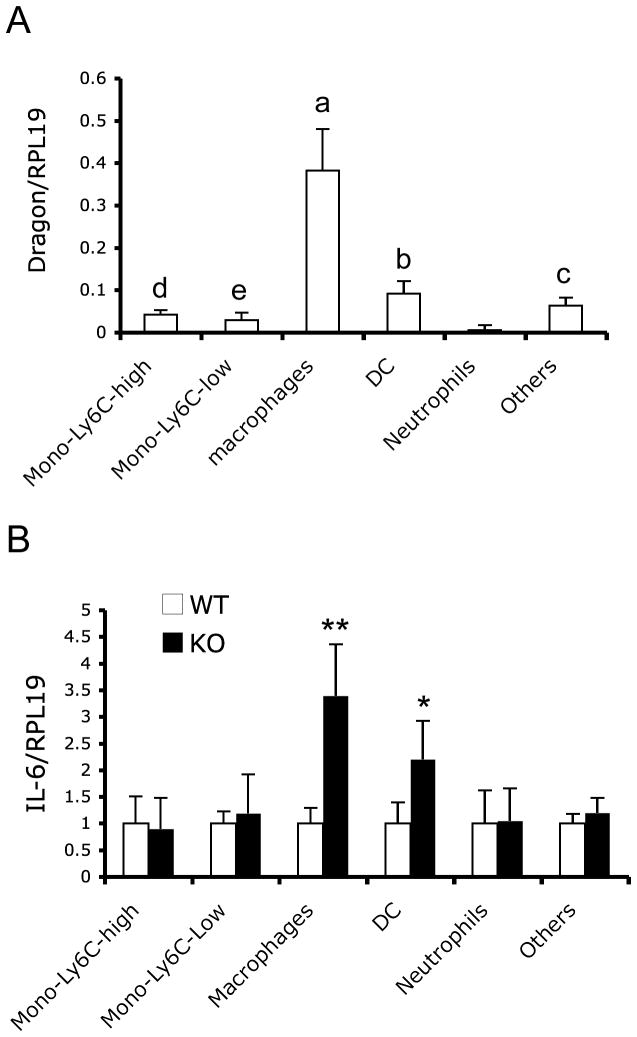
Figure 7.
Dragon mRNA expression in Ly-6Chigh and Ly-6Clow monocytes, macrophages, dendritic cells (DC) and neutrophils in lungs of 10–12 days old wild type mice (A), and IL-6 mRNA expression in the five cell populations and the rest of the cells (Others) in wild type and knockout mice (B). Total RNA were extracted from sorted Ly-6Chigh and Ly-6Clow monocytes, macrophages, DC, neutrophils and other cells in lungs of 10–12 days old wild type and knockout mice. Real time PCR analyses were performed to quantify Dragon and IL-6 mRNA levels. RPL-19 was used as an internal control. abcdeVaules with different superscripts differ significant; *, p < 0.05; **, p < 0.01 (B). n=6 (A), or 3–4 (B).
Results
Dragon suppresses IL-6 expression through a BMP ligand dependent pathway
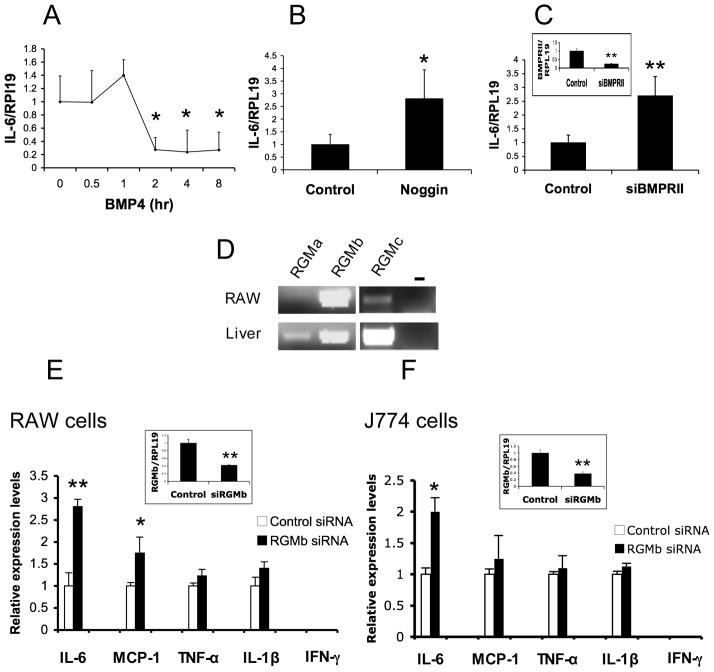
It has been shown that BMP signaling inhibits IL-6 expression in a number of cell types including macrophages (5–7, 9). Consistent with these previous findings, BMP4 inhibited IL-6 mRNA expression in RAW264.7 macrophages (Figure 1A). The inhibition was seen as early as 2 hrs after exogenous BMP4 treatment (Figure 1A). Incubation with noggin, an extracellular antagonist of BMP ligands (Figure 1B) or inhibition of BMP type II receptor BMPRII expression by siRNA increased IL-6 mRNA expression (Figure 1C). These results confirm the previously published findings that BMP signaling inhibits IL-6 expression in macrophages in vitro (5, 6).
Figure 1.
IL-6 is a target of Dragon in RAW264.7 and J774 macrophages. (A, B and C) IL-6 mRNA expression is inhibited by BMP4 (A), and increased by noggin (B) or by inhibition of BMPRII expression (C). RAW264.7 macrophages were incubated with BMP4 (50 ng/ml) for 0–8 hrs, or incubated with noggin (500 ng/ml) overnight, or transfected with control or BMPRII siRNA. Cells were then harvested for RNA extraction and real time PCR for IL-6 and RPL19. IL-6 expression levels of the treated cells were expressed as a fraction of values from the controls. (D) Expression of RGMa, RGMb and RGMc mRNAs in RAW264.7 macrophages. Total RNA from RAW cells was extracted for RT-PCR to determine the expression of RGMa, RGMb and RGMc. Total RNA from the mouse liver was used in PCR analyses as positive controls. (E and F) IL-6 mRNA expression is up regulated by inhibition of Dragon expression in RAW264.7 (E) and J774 (F) macrophages. RAW264.7 (E) or J774 (F) cells were transfected with control or Dragon siRNA, and analysed for mRNA levels of IL-6, MCP-1, TNF-α, IL-1β, IFN-γ and Dragon. The expression levels of these factors are normalized to RPL19 and expressed as a fraction of values from the respective controls. The efficacy of BMPRII siRNA or Dragon siRNA was shown in the insets. *, p < 0.05; **, p < 0.01.
We have previously shown that Dragon is a co-receptor that enhances BMP signaling (12, 13, 16). To examine whether Dragon regulates IL-6 expression in macrophages, we first examined the expression of Dragon in RAW264.7 cells. As shown by RT-PCR, Dragon mRNA was highly expressed in RAW264.7 cells while the expression of two other members of the RGM family was either weak (RGMc) or undetectable (RGMa) (Figure 1D). Inhibition of Dragon expression by siRNA increased basal IL-6 mRNA levels by 2.8 fold (Figure 1E). Inhibition of Dragon expression only modestly increased MCP-1 expression, and did not alter the expression of TNF-α, and IL-1β (Figure 1E). A selective increase in IL-6 expression by inhibition of Dragon was also observed in J774 macrophages (Figure 1F). In addition, inhibition of Dragon expression in RAW264.7 cells increased LPS-stimulated IL-6 mRNA levels by 1.8 fold while MCP-1, TNF-α, and IL-1β mRNA levels did not change (Supplemental Figure 1). These results suggest that IL-6 is an important cytokine target of Dragon action in macrophages.
IL-6 and Dragon mRNAs were also expressed in mouse pulmonary artery smooth muscle cells (PASMC), human umbilical venous endothelial cells (HUVEC), mouse inner medullar collecting duct (IMCD3) cells, and C2C12 mouse myoblasts. Inhibition of Dragon mRNA by 70%-90% did not alter IL-6 expression in PASMC, HUVEC and IMCD3 cells, and slightly increased IL-6 expression in C2C12 cells (Supplemental Figure 2). These results suggest that the action of Dragon in IL-6 expression is specific for macrophage cell types.
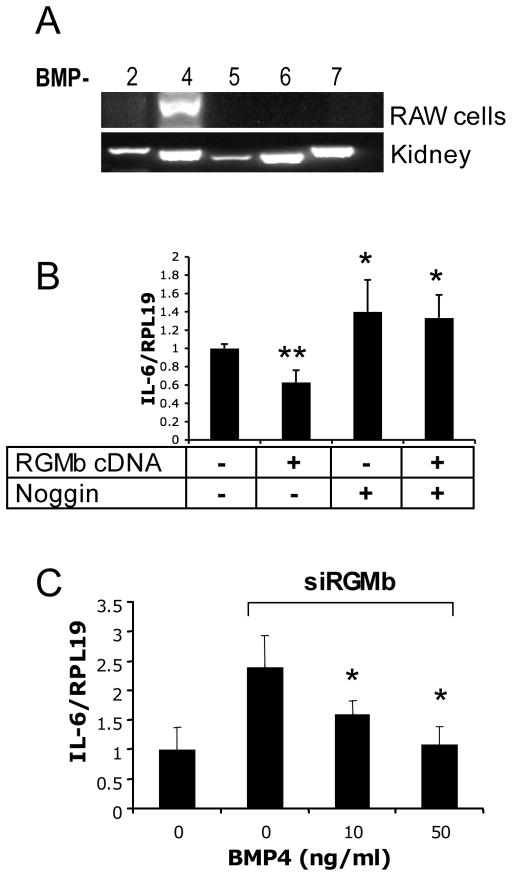
Our previous studies have shown that Dragon. Fc binds to radiolabeled BMP2 and BMP4, and that BMP signaling induced by transfected Dragon was reduced after inhibition of BMP4 expression in IMCD3 cells, suggesting that BMP2 and BMP4 are the endogenous BMP ligand for the Dragon co-receptor (12, 16). To examine whether the ability of Dragon to inhibit IL-6 expression is dependent on BMP ligands, we examined the expression of BMP2, BMP4 and the closely related BMP5, BMP6, and BMP7 (Figure 2A) in RAW264.7 macrophages. BMP4 mRNA was detected while BMP2, -5, -6 and -7 were not. Transfection of RAW264.7 cells with Dragon cDNA inhibited IL-6 expression, while addition of noggin abolished this inhibition (Figure 2B). Silencing of Dragon induced IL-6 expression, while exogenous BMP4 addition abolished this induction (Figure 2C). These results suggest that endogenous BMP ligand(s) is required for Dragon to inhibit IL-6 expression.
Figure 2.
Dragon action on IL-6 expression is BMP ligand dependent. (A) Expression of BMP ligands BMP2 and BMP4–BMP7 in RAW264.7 macrophages. Total RNA from RAW cells was extracted for RT-PCR to determine the expression of BMP2 and BMP4–BMP7 mRNAs. Total RNA from the mouse kidney was used in PCR analyses as positive controls. (B) Inactivation of BMP ligands by noggin abolished inhibition of IL-6 expression induced by RGMb overexpression. RGMb is RAW264.7 cells were transfected with and without Dragon cDNA, followed by incubation with and without noggin (500 ng/ml). The cells were then harvested for real time PCR analysis of IL-6 and RPL19 mRNA levels. (C) Induction of IL-6 expression by inhibition of RGMb expression was attenuated by exogenous BMP4. RAW264.7 cells were transfected with and without Dragon siRNA, and the cells transfected with Dragon siRNA were incubated with increasing amounts of BMP4 (0–50 ng/ml). The cells were then harvested for real time PCR analysis for IL-6 and RPL19 mRNA levels. *, p < 0.05; **, p < 0.01 versus the controls (bar 1).
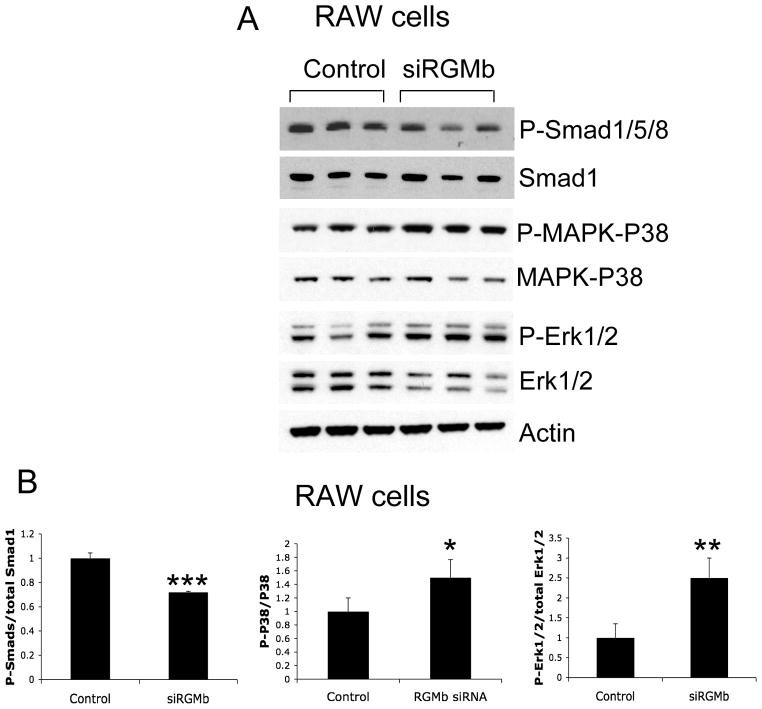
The action of Dragon in IL-6 expression is mediated by the p38 MAPK and Erk1/2 pathways but not the Smad1/5/8 pathway
To explore the mechanisms involved in the regulation of IL-6 expression by Dragon, we examined the effects on the Smad1/5/8, p38 MAPK and Erk1/2 pathways, which well documented pathways that mediate BMP actions (1). As shown in Figures 3A and 3B, phospho-Smad1/5/8 levels were decreased, but phospho-p38-MAPK and phospho-Erk1/2 levels were increased, after inhibition of Dragon expression by siRNA. To examine whether the Smad pathway plays a role in the regulation of IL-6 expression by BMP4 and Dragon, we used LDN-193189, a small molecule BMP inhibitor, which inhibits Smad1/5/8 phosphorylation but does not affect p38 MAPK and Erk1/2 activity (16, 23). LDN-193189 treatment did not affect the inhibition of IL-6 expression induced by BMP4 (Figure 4A). In contrast, the induction of phospho-Smad1/5/8 levels and Id1 expression by BMP4 was dramatically reduced by LDN-193189 (Figure 4B & C). LDN-193189 treatment also did not affect the inhibition of IL-6 expression induced by Dragon (Figure 4D). Finally, inhibition of Smad4 expression by siRNA did not affect IL-6 mRNA levels while it significantly reduced Id1 mRNA expression in RAW264.7 cells (Supplemental Figure 3). These results suggest that regulation of IL-6 by Dragon/BMP does not involve the Smad pathway.
Figure 3.
Changes in phosphorylation levels of Smad1/5/8, and p38 and Erk1/2 MAPK in Dragon knockdown RAW264.7 cells. (A) RAW264.7 cells were transfected with control (three replicates) and Dragon siRNA (three replicates). 46 hrs after transfection, the cells were lysed and analyzed by Western blotting for phospho-Smad1/5/8 and total Smad1; phospho-p38 and total p38; phospho-Erk1/2 and total Erk1/2; and actin. (B) Western blot chemiluminescence from experiments in (A) was quantified using IPLab Spectrum software for phospho-Smad1/5/8 relative to Smad1, phospho-p38 relative to total p38, and phospho-Erk1/2 relative to total Erk1/2. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Figure 4.
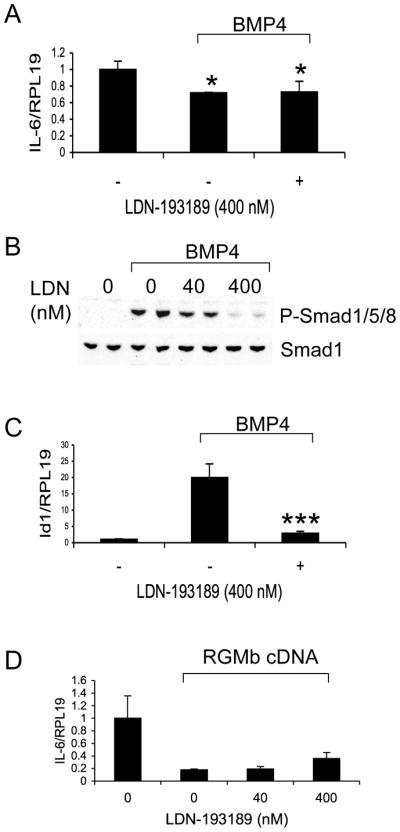
Inhibition of IL-6 expression by BMP4 and Dragon is not affected by LDN-193189. (A) RAW264.7 cells were treated with or without BMP4 (50 ng/ml) in the presence or absence of LDN-193189 (400 nM). The cells were then harvested for real time PCR analysis for IL-6 and RPL19 mRNA levels. (B) RAW264.7 cells were treated with or without BMP4 (50 ng/ml) in the presence of increasing amounts of LDN-193189 (0–400 nM). The cells were lyzed and analyzed by Western blotting for phospho-Smad1/5/8 and total Smad1. (C) RAW264.7 cells were treated with or without BMP4 (50 ng/ml) in the presence or absence of LDN-193189 (400 nM). The cells were then harvested for real time PCR analysis for Id1 mRNA levels. (D) RAW264.7 cells were transfected with or without Dragon cDNA. The cells transfected with Dragon cDNA were then treated with increasing amounts of LDN-193189 (0–400 nM). The cells were harvested for real time PCR analysis for IL-6 and RPL19 mRNA levels. *, p < 0.05; ***, p < 0.001 versus cells treated with BMP4 in the absence of LDN-193189 (bar 2, C).
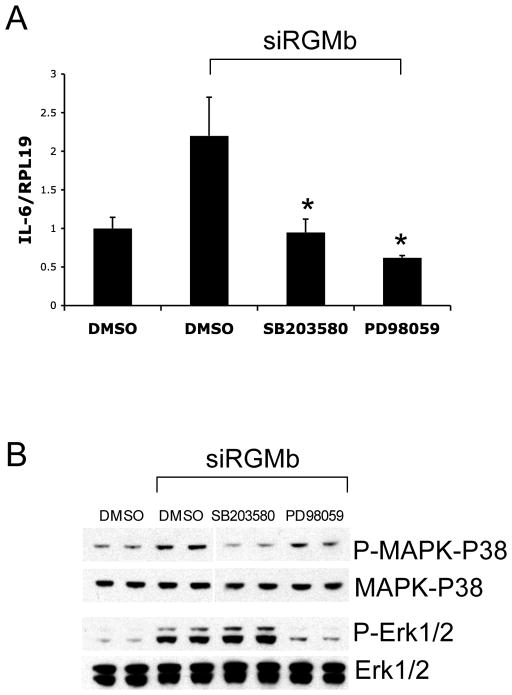
To examine whether the p38 and Erk1/2 pathways are involved in the regulation of IL-6 expression by Dragon, we used p38 inhibitor SB203580 or Erk1/2 inhibitor PD98059. IL-6 expression induced by Dragon siRNA was abolished by treatment with SB203580 or PD98059 (Figure 5A), which respectively reduced phospho-p38-MAPK and phospho-Erk1/2 protein levels that were induced when Dragon was inhibited (Figure 5B). These results suggest that p38-MAPK and Erk1/2 mediate the action of Dragon on lowering IL-6 mRNA levels.
Figure 5.
Induction of IL-6 expression by Dragon siRNA is abolished by p38 inhibitor SB203580 or Erk1/2 inhibitor PD98059. (A) RAW264.7 cells were transfected with control or Dragon siRNA, followed by incubation with DMSO, SB203580 (2.5 μM), or PD98059 (2.5 μM). The cells were analyzed by real time PCR for IL-6 and RPL19 mRNA levels. (B) RAW264.7 cells were transfected with control or Dragon siRNA, followed by incubation with DMSO, SB203580 (2.5 μM), or PD98059 (2.5 μM). The cells were lyzed and analyzed by Western blotting for phospho-p38 and total p38; and phospho-Erk1/2 and total Erk1/2. *, p < 0.05 versus cells transfected with Dragon siRNA in the absence of any inbibitors (bar 2).
IL-6 expression is up regulated in the lungs and livers of Dragon knockout mice
To examine if Dragon plays a role in regulating IL-6 expression in vivo, we analyzed IL-6 expression in the lungs, livers, spleens, colons and muscles of Dragon knockout (KO) mice. Dragon KO mice were generated using gene targeting in ES cells by homologous recombination (Supplemental Figure 4). Since Dragon knockout mice die 2 to 3 weeks after birth (cause of mortality under investigation), we collected samples from Dragon KO and wild type (WT) littermates at day 10–12 of age. As shown in Figure 6A, IL-6 mRNA expression was up regulated by 10-fold in the lungs of KO mice compared to the wild type. IL-6 protein was also up regulated as shown by Western blotting analysis (Figure 6D). IL-6 mRNA levels were significantly higher in E20.5 KO lungs than in E20.5 WT lungs (Figure 6B). IL-6 mRNA levels were 2.9-fold higher in KO livers than in WT livers of 10–12 days old mice (Figure 6C). IL-6 mRNA expression was 16-fold lower in the spleen, colon and muscle compared to the IL-6 levels in the lung of 10–12 day old WT mice (data not shown), and IL-6 mRNA levels in these organs were not altered between KO and WT (Supplemental Figure 5). In addition to IL-6, a number of other inflammatory cytokines and chemokines including MCP-1, TNF-α, IL-1β and IFN- γ were also up regulated in the lung of Dragon KO mice at day 10–12 after birth (Supplemental Figure 6). These results suggest that Dragon is a negative regulator of IL-6 and other inflammatory factors in the lung and liver in vivo.
In the lungs of Dragon KO mice, phospho-Smad1/5/8 levels did not change, while phospho-p38-MAPK and phospho-Erk1/2 levels in KO lungs were increased compared to WT lungs (Figure 6E). These results are consistent with the in vitro observation in cultured RAW264.7 cells that Dragon acts through the p38 MAPK and Erk1/2 pathways, but not the Smad1/5/8 pathway, to regulate IL-6 expression.
IL-6 mRNA is up regulated in macrophages in the lungs of Dragon knockout mice
Next, we sought to characterize which cells increase IL-6 expression in the lungs of Dragon KO mice. To this end, we purified and analyzed diverse populations of the tissue immune cell repertoire from 10–12 days old WT and Dragon KO mice (Supplemental Figure 7 and Figure 7). The total number of cells in the lung was similar between WT and Dragon KO mice. The numbers of Ly-6Chigh and Ly-6Clow monocytes, macrophages, DCs and neutrophils were not significantly different between the two genotypes (Supplemental Figure 7).
Dragon mRNA was expressed in Ly-6Chigh and Ly-6Clow monocytes, macrophages, and DCs, while it is barely detectable in neutrophils collected from lungs of 10–12 day old WT mice (Figure 7A). Dragon expression was much higher in macrophages than in Ly-6Chigh and Ly-6Clow monocytes, DCs, neutrophils and other cells, with Dragon expression being higher in DCs than in monocytes, neutrophils and other cells (Figure 7A). IL-6 mRNA expression was increased by 3.4-fold in macrophages and 2.2-fold in dendritic cells due to Dragon deletion, while no changes in IL-6 mRNA levels were observed in Ly-6Chigh and Ly-6Clow monocytes, neutrophils and other cells (Figure 7B). These results suggest that high levels of Dragon in lung macrophages (and dendritic cells) serve to suppress IL-6 expression in vivo, which are consistent with our in vitro data.
Discussion
The three RGM family members RGMa, RGMb (Dragon), and RGMc (hemojuvelin) are BMP coreceptors that enhance BMP signaling. Our published studies have shown that RGMc/hemojuvelin acts through BMP pathway to regulate hepcidin expression and iron metabolism, but the biological actions of the BMP signaling function of RGMa and RGMb/Dragon are poorly characterized. Previous studies have shown that BMPs (5, 6, 9) regulate inflammatory cytokines and chemokines including IL-6 in various cell types including macrophages and monocytes. In the present study, we confirmed that BMP4 inhibited IL-6 expression in RAW264.7 macrophages. Interestingly, inhibition of Dragon expression in RAW264.7 or J774 macrophages also increased IL-6 expression while Dragon overexpression inhibited IL-6 expression in a BMP ligand dependent manner. These results suggest that Dragon regulates IL-6 expression via the BMP pathway in RAW264.7 macrophages. Inhibition of Dragon expression did not have significant effects on IL-6 expression in PASMC, HUVEC, mIMCD3 and C2C12 cells. These cells have been shown be responsive to BMP ligands by ourselves and others (9, 14, 16, 24, 25, 26). For example, inhibition of BMPRII expression increased IL-6 expression in PASMC derived from mouse (data not shown) and humans (9). Thus, the IL-6 effect of Dragon appears to be specific for macrophages cells. The reason for this cell type specific action is unclear.
Our study also demonstrated that Dragon action on IL-6 expression is mediated by the p38 MAPK and Erk1/2 pathways but not by the Smad1/5/8 pathway. Our data show 1) phosphorylation levels of p38 MAPK and Erk1/2 were increased by inhibition of Dragon expression in vitro and by deletion of Dragon in vivo, while phosphorylation levels of Smad1/5/8 were reduced by inhibition of Dragon in vitro but not affected by deletion of Dragon in vivo; 2) the induction of IL-6 expression by inhibition of Dragon expression is blocked by p38 MAPK or Erk1/2 specific inhibitor; 3) inhibition of IL-6 expression by exogenous Dragon or BMP4 was not affected by LDN-193189, and IL-6 expression was not affected by inhibition of Smad4 expression. A study by Hagen et al. showed that inhibition of BMPRII in mouse PASMC also led to increased p38 MAPK activity resulting in increased IL-6 expression. These results suggest an inhibitory effect of BMP2 signaling on p38 MAPK and Erk1/2 activation in these cells. In contrast, previous studies have shown a stimulatory effect of BMP2 on p38 MAPK and Erk1/2 activation in other cell types (27). To add more complexity, a recent study demonstrated that BMP6 and BMP7 suppressed estrogen-induced p38 and Erk1/2 activation, while BMP2 and BMP4 enhanced this stimulation in MCF-7 cells (28). Therefore, whether BMP signaling leads to stimulation or inhibition of the p38 MAPK and Erk1/2 pathways appear to be dependent on the specific BMP ligands and cell types studied.
Previous studies have shown that BMP receptors oligomerize in at least two different manners. BMP type I and type II receptors can form hetero-oligomeric complexes prior to ligand binding. BMPs bind to these preformed hetero-oligomeric complexes (PFC), and then activate the Smad signaling pathway. The other alternative is that BMP ligand binds to BMP type I receptors and then recruits BMP type II receptors into a complex (BMP-induced signaling complexes: BISC), thus activating XIAP-Tak1-Tab1 complex and downstream p38 and Erk1/2 pathways (27, 29). BMP type I and type II receptors partially reside in lipid rafts. The non-Smad signaling induced by BMP2 depends on the association of BMP receptors with rafts and caveolae, while the Smad pathway is independent of lipid raft association (27). Interestingly, Dragon is also localized in lipid rafts (13). Whether Dragon is involved in regulating the formation of BISC and activation of XIAP and TAK1 remains to be investigated.
We found that IL-6 expression was dramatically increased in lung macrophages and dendritic cells, as well as in whole lung and liver tissues in the Dragon knockout mice compared with wild-type mice. These results revealed an important physiological role of Dragon in regulating IL-6 expression. The magnitude of the increase in IL-6 mRNA levels in the whole lungs (10-fold) was larger than those in macrophages (3.4-fold) and in dendritic cells (2.2-fold). The reason for the differences between the whole organ and specific cell populations remains unclear, but the cells were analyzed for mRNA levels a few hours after the cells were collected due to the preparation time required for FACS procedures, and IL-6 mRNA expression may have declined during this period of time. IL-6 expression in cells other than monocytes, macrophages, dendritic cells and neutrophils in the lung was not altered by Dragon deletion. This result is consistent with our in vitro data showing that inhibition of Dragon did not change IL-6 expression in PASMC, a cell type in the lung, which is involved in mechanisms of disorders such as pulmonary arterial hypertension (PAH) with elevated IL-6 expression.
Expression levels of other factors including MCP-1, TNF-α, IL-1β and IFN- γ were also elevated in the lungs of Dragon KO mice. Our in vitro data using RAW264.7 and J774 cells show that IL-6 is a direct target of Dragon/BMP pathway while expression of MCP-1, TNF-α and IL-1β were not appreciably affected by inhibition of Dragon expression. Therefore, the changes in these factors in Dragon KO mice might be due to secondary effects of Dragon/BMP action.
We characterized the Dragon expression profile in five immune cell populations isolated from the lungs of day 10–12 mice, and found that Dragon is most highly expressed in macrophages and DC, and lower in other cell types. This increased expression of Dragon in macrophages and DC likely accounts for why these cell types dramatically increased their IL-6 expression in the Dragon KO mice compared to wild type mice, while there was no change in IL-6 expression in Ly-6Chigh and Ly-6Clow monocytes, neutrophils and other cells.
The growth of Dragon KO mice was retarded (data not shown), and the mice die between 2–3 weeks after birth. Our preliminary results show that the Dragon KO mice developed growth hormone (GH) resistance in the liver and skeletal muscle (data not shown). GH resistance has been reported in other mice injected with exogenous IL-6 expression (30) or in mice with transgenic over-expression of IL-6 (31, 32). Therefore, in the future, it will be interesting to investigate whether abnormal expression of IL-6 and other inflammatory factors contribute to the retarded growth and even the mortality of Dragon KO mice.
Dragon has an ability to promote cell-cell adhesion by homophilic interactions (19). Whether this property of Dragon contributes to the difference in migration and other functions among different immune cell populations has not yet been addressed.
In summary, we have demonstrated the first in vivo biological role for the BMP signaling function of Dragon, which is that Dragon suppresses IL-6 expression in macrophages. Dragon is most highly expressed in macrophages compared to other immune cells in the lung, and deletion of Dragon in mice leads to increased expression of IL-6 and other inflammatory factors in macrophages and dendritic cells as well as in the lung and the liver. These results suggest that Dragon plays an important role in regulating the immune system.
Supplementary Material
Acknowledgments
The authors thank Drs. Paul Yu, and Hideyuki Beppu for the mouse PASMC. The authors also thank Dr. Chia Chi Sun for reading the manuscript, and Drs. Jinzhong Qin and Fang Wang for helpful discussions.
Grant support
H.Y.L. was supported by National Institutes of Health (NIH) grants RO1 DK-071837 and RO1 DK-069533. Y.X. was supported in part by the NIH grants R03HD60641 and K01DK084081. V.C.R., M.J.P., and R.W. were supported by NIH grants U01 HL080731, P50 CA86355, and U54 CA126515.
References
- 1.Shi Y, Massague J. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 2.Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, Annunziata N, Doetschman T. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xia Y, Schneyer AL. The biology of activin: recent advances in structure, regulation and function. J Endocrinol. 2009;202:1–12. doi: 10.1677/JOE-08-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vallet S, Mukherjee S, Vaghela N, Hideshima T, Fulciniti M, Pozzi S, Santo L, Cirstea D, Patel K, Sohani AR, Guimaraes A, Xie W, Chauhan D, Schoonmaker JA, Attar E, Churchill M, Weller E, Munshi N, Seehra JS, Weissleder R, Anderson KC, Scadden DT, Raje N. Activin A promotes multiple myeloma-induced osteolysis and is a promising target for myeloma bone disease. Proc Natl Acad Sci U S A. 2010;107:5124–5129. doi: 10.1073/pnas.0911929107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould SE, Day M, Jones SS, Dorai H. BMP-7 regulates chemokine, cytokine, and hemodynamic gene expression in proximal tubule cells1. Kidney Int. 2002;61:51–60. doi: 10.1046/j.1523-1755.2002.00103.x. [DOI] [PubMed] [Google Scholar]
- 6.Lee MJ, Yang CW, Jin DC, Chang YS, Bang BK, Kim YS. Bone morphogenetic protein-7 inhibits constitutive and interleukin-1β-induced monocyte chemoattractant protein-1 expression in human mesangial cells: role for JNK/AP-1 pathway. J Immunol. 2003;170:2557–2563. doi: 10.4049/jimmunol.170.5.2557. [DOI] [PubMed] [Google Scholar]
- 7.Maric I, Poljak L, Zoricic S, Bobinac D, Bosukonda D, Sampath KT, Vukicevic S. Bone morphogenetic protein-7 reduces the severity of colon tissue damage and accelerates the healing of inflammatory bowel disease in rats. J Cell Physiol. 2003;196:258–264. doi: 10.1002/jcp.10275. [DOI] [PubMed] [Google Scholar]
- 8.Hori M, Sawai H, Tsuji Y, Okamura H, Koyama K. Bone morphogenetic protein-2 counterregulates interleukin-18 mRNA and protein in MC3T3-E1 mouse osteoblastic cells. Connect Tissue Res. 2006;47:124–132. doi: 10.1080/03008200600685350. [DOI] [PubMed] [Google Scholar]
- 9.Hagen M, Fagan K, Steudel W, Carr M, Lane K, Rodman DM, West J. Interaction of interleukin-6 and the BMP pathway in pulmonary smooth muscle. Am J Physiol Lung Cell Mol Physiol. 2007;292:L1473–1479. doi: 10.1152/ajplung.00197.2006. [DOI] [PubMed] [Google Scholar]
- 10.Kwon SJ, Lee GT, Lee JH, Kim WJ, Kim IY. Bone morphogenetic protein-6 induces the expression of inducible nitric oxide synthase in macrophages. Immunology. 2009;128(1 Suppl):e758–765. doi: 10.1111/j.1365-2567.2009.03079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hong JH, Lee GT, Lee JH, Kwon SJ, Park SH, Kim SJ, Kim IY. Effect of bone morphogenetic protein-6 on macrophages. Immunology. 2009;128(1 Suppl):e442–50. doi: 10.1111/j.1365-2567.2008.02998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong SJ, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, Lin HY, Brivanlou AH, Attisano L, Woolf CJ. DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- 13.Xia Y, Sidis Y, Mukherjee A, Samad T, Brenner G, Woolf CJ, Lin HY, Schneyer A. Localization and action of Dragon (repulsive guidance molecule b), a novel bone morphogenetic protein coreceptor, throughout the reproductive axis. Endocrinology. 2005;146:3614–3621. doi: 10.1210/en.2004-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xia Y, Yu PB, Sidis Y, Beppu H, Bloch KD, Schneyer AL, Lin HY. Repulsive guidance molecule (RGMa) alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J Biol Chem. 2007;282:18129–18140. doi: 10.1074/jbc.M701679200. [DOI] [PubMed] [Google Scholar]
- 15.Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 2008;111:5195–5204. doi: 10.1182/blood-2007-09-111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia Y, Babitt JL, Bouley R, Zhang Y, Da Silva N, Chen S, Zhuang Z, Samad TA, Brenner GJ, Anderson JL, Hong CC, Schneyer AL, Brown D, Lin HY. Dragon enhances BMP signaling and increases transepithelial resistance in kidney epithelial cells. J Am Soc Nephrol. 2010;21:666–677. doi: 10.1681/ASN.2009050511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Babitt JL, Zhang Y, Samad TA, Xia Y, Tang J, Campagna JA, Schneyer AL, Woolf CJ, Lin HY. Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:29820–29827. doi: 10.1074/jbc.M503511200. [DOI] [PubMed] [Google Scholar]
- 18.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- 19.Samad TA, Srinivasan A, Karchewski LA, Jeong SJ, Campagna JA, Ji RR, Fabrizio DA, Zhang Y, Lin HY, Bell E, Woolf CJ. DRAGON: A Member of the Repulsive Guidance Molecule-Related Family of Neuronal- and Muscle-Expressed Membrane Proteins Is Regulated by DRG11 and Has Neuronal Adhesive Properties. J Neurosci. 2004;24:2027–2036. doi: 10.1523/JNEUROSCI.4115-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaPensee CR, Hugo ER, Ben-Jonathan N. Insulin stimulates interleukin-6 expression and release in LS14 human adipocytes through multiple signaling pathways. Endocrinology. 2008;149:5415–5422. doi: 10.1210/en.2008-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, Babitt JL. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niederkofler, Salie VR, Sigrist M, Arber S. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J Neurosci. 2004;24:808–818. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg Med Chem Lett. 2008;18:4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu PB, Beppu H, Kawai N, Li E, Bloch KD. Bone morphogenetic protein (BMP) type II receptor deletion reveals BMP ligand-specific gain of signaling in pulmonary artery smooth muscle cells. J Biol Chem. 2005;280:24443–24450. doi: 10.1074/jbc.M502825200. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Q, Heinke J, Vargas A, Winnik S, Krauss T, Bode C, Patterson C, Moser M. ERK signaling is a central regulator for BMP-4 dependent capillary sprouting. Cardiovasc Res. 2007;76:390–399. doi: 10.1016/j.cardiores.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Nojima J, Kanomata K, Takada Y, Fukuda T, Kokabu S, Ohte S, Takada T, Tsukui T, Yamamoto TS, Sasanuma H, Yoneyama K, Ueno N, Okazaki Y, Kamijo R, Yoda T, Katagiri T. Dual roles of smad proteins in the conversion from myoblasts to osteoblastic cells by bone morphogenetic proteins. J Biol Chem. 2010;285:15577–15586. doi: 10.1074/jbc.M109.028019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sieber C, Kopf J, Hiepen C, Knaus P. Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 2009;20:343–355. doi: 10.1016/j.cytogfr.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi M, Otsuka F, Miyoshi T, Otani H, Goto J, Yamashita M, Ogura T, Makino H, Doihara H. Bone morphogenetic protein 6 (BMP6) and BMP7 inhibit estrogen-induced proliferation of breast cancer cells by suppressing p38 mitogen-activated protein kinase activation. J Endocrinol. 2008;199:445–55. doi: 10.1677/JOE-08-0226. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi K, Nagai S, Ninomiya-Tsuji J, Nishita M, Tamai K, Irie K, Ueno N, Nishida E, Shibuya H, Matsumoto K. XIAP, a cellular member of the inhibitor of apoptosis protein family, links the receptors to TAB1-TAK1 in the BMP signaling pathway. EMBO J. 1999;18:179–87. doi: 10.1093/emboj/18.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang P, Li N, Li JS, Li WQ. The role of endotoxin, TNF-alpha, and IL-6 in inducing the state of growth hormone insensitivity. World J Gastroenterol. 2002;8:531–536. doi: 10.3748/wjg.v8.i3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieskovska J, Guo D, Derman E. IL-6-overexpression brings about growth impairment potentially through a GH receptor defect. Growth Horm IGF Res. 2002;12:388–398. doi: 10.1016/s1096-6374(02)00079-5. [DOI] [PubMed] [Google Scholar]
- 32.Lieskovska J, Guo D, Derman E. Growth impairment in IL-6-overexpressing transgenic mice is associated with induction of SOCS3 mRNA. Growth Horm IGF Res. 2003;13:26–35. doi: 10.1016/s1096-6374(02)00135-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.