Abstract
Endoplasmic reticulum (ER) stress activates a set of signaling pathways, collectively termed the unfolded protein response (UPR). The three UPR branches (IRE1, PERK, and ATF6) promote cell survival by reducing misfolded protein levels. UPR signaling also promotes apoptotic cell death if ER stress is not alleviated. How the UPR integrates its cytoprotective and proapoptotic outputs to select between life or death cell fates is unknown. We found that IRE1 and ATF6 activities were attenuated by persistent ER stress in human cells. By contrast, PERK signaling, including translational inhibition and proapoptotic transcription regulator Chop induction, was maintained. When IRE1 activity was sustained artificially, cell survival was enhanced, suggesting a causal link between the duration of UPR branch signaling and life or death cell fate after ER stress. Key findings from our studies in cell culture were recapitulated in photoreceptors expressing mutant rhodopsin in animal models of retinitis pigmentosa.
The UPR comprises a set of signaling pathways that collectively adjust the cell’s ER protein folding capacity according to need. As such, UPR signaling reestablishes homeostasis in the face of changing developmental and environmental conditions, thereby preserving ER protein folding fidelity. Upon unmitigated ER stress, the UPR also triggers apoptosis. Thus, rather than produce misfolded or malfunctioning proteins, cells are eliminated, perhaps to protect the organism from rogue cells that do not receive or relay signals properly.
Physiologic or pathologic processes that create an imbalance between protein folding load and capacity induce the UPR through ER-resident transmembrane proteins—IRE1, PERK, and ATF6—that act as sensors in the ER lumen and transmit the information to the rest of the cell (1). IRE1 is a transmembrane kinase/endoribonuclease (RNAse) that, upon activation, initiates the nonconventional splicing of Xbp-1 mRNA (2, 3). Spliced Xbp-1 mRNA encodes a transcription activator that drives transcription of genes such as ER chaperones, whose products directly participate in ER protein folding (4). PERK is a transmembrane kinase that phosphorylates the eukaryotic translation initiation factor 2 subunit α (eIF2α), thereby reducing protein synthesis and counteracting ER protein overload (5). eIF2α phosphorylation also allows the selective translation of some mRNAs that contain small open reading frames in their 5′ untranslated regions, thereby leading to the production of transcription activators such as ATF4 (6). ATF6 is a transcription factor that is made initially as an ER-resident transmembrane protein. Upon protein misfolding, the ATF6 cytoplasmic domain (ATF6f) is liberated from its membrane anchor by regulated proteolysis (7, 8). The transcription factors thus produced (i.e., XBP1, ATF4, and ATF6f) collaborate to activate UPR target genes, thereby controlling the cell’s response to ER stress.
Genetic studies have begun to assign cytoprotective or proapoptotic functions to individual UPR target genes. For instance, expression of the ER chaperone BiP protects cells from ER stress (9), whereas CHOP, a B-ZIP transcription factor induced by the PERK branch of the UPR, promotes cell death (10). Paradoxically, all known ER stresses simultaneously elicit protective and toxic outputs from the UPR, and it has remained unclear how the UPR integrates these opposing outputs to arrive at a life or death decision.
IRE1 signaling is attenuated during persistent ER stress
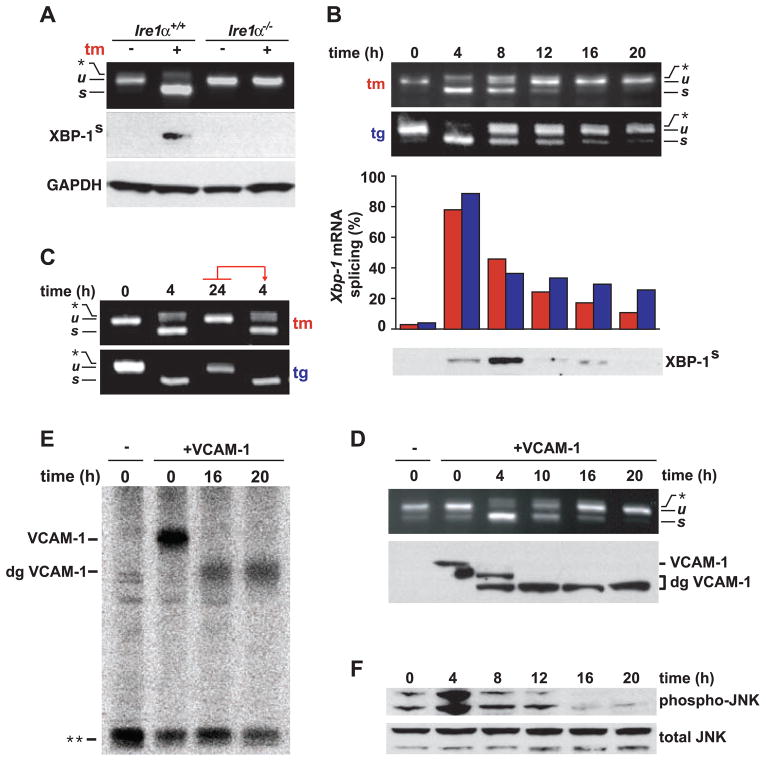
To determine the activation status of IRE1 after ER stress, we examined Xbp-1 mRNA splicing by reverse transcription polymerase chain reaction (RT-PCR) in human embryonic kidney (HEK) 293 cells (Fig. 1A). We observed the appearance of spliced Xbp-1 mRNA after treating the cells with tunicamycin or thapsigargin: agents that elicit ER stress by blocking N-linked glycosylation or inhibiting the ER Ca2+ pump, respectively (Fig. 1B). Unexpectedly, we found a strong diminution in Xbp-1 mRNA splicing with prolonged exposure to either drug (Fig. 1B). Consistent with the mRNA splicing data, XBP-1s protein levels (the form of XBP-1 derived from its spliced mRNA) also decreased with prolonged drug treatment (Fig. 1B). Other human cell lines showed qualitatively similar effects, although they differed in the observed timing of onset and shutoff, as well as in the degree of Xbp-1 mRNA splicing (fig. S1).
Fig. 1.
Kinetics of IRE1 signaling with persistent ER stress. (A) WT Ire1α+/+ or Ire1α−/− mouse embryo fibroblasts were treated with tunicamycin (tm) (5 μg/ml), and Xbp-1 mRNA splicing was determined by RT-PCR. Unspliced (u) and spliced (s) Xbp-1 mRNA products are indicated. The asterisk indicates the position of a hybrid amplicon (27). XBP-1s was detected by immunoblotting. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels served as a protein loading control. (B) HEK293 cells were treated with tm (5 μg/ml) or thapsigargin (tg) (500 nM) for the indicated times. tm, red bars; tg, blue bars. Results are representative of five independent experiments. (C) HEK293 cells were treated with agents for the indicated times. At 24 hours, media containing drug were transferred to fresh cells. After 4 additional hours, Xbp-1 mRNA splicing was determined by RT-PCR. (D) HEK293 cells, transfected with VCAM-1, were treated with tm (5 μg/ml) for the indicated times. Xbp-1 mRNA splicing was determined by RT-PCR. Mature and deglycosylated (dg) VCAM-1 species were determined by immunoblotting. (E) HEK293 cells, transfected with VCAM-1, were treated with tm for the indicated times and were pulse-labeled, and radiolabeled VCAM-1 was detected after immunoprecipitation. The double asterisk indicates the position of a nonspecific band used as a loading control. (F) HEK293 cells were treated with tg for the indicated times, and phospho-JNK protein levels were assessed by immunoblotting. Total JNK protein levels served as a loading control.
To ascertain that loss of Xbp-1 mRNA splicing at the late time points was not due to inactivation of the ER stress–inducing agents, we transferred media from HEK293 cells that had been treated with tunicamycin or thapsigargin for 24 hours (a time when little Xbp-1 mRNA splicing was seen) to plates of fresh, untreated cells. After 4 hours of incubation in conditioned media, Xbp-1 mRNA splicing was induced to a degree indistinguishable to that seen in cells treated with fresh agents (Fig. 1C).
We next tested if, at late time points, cells continued to accumulate misfolded proteins in their ER or had acquired means of neutralizing the effects of the ER stress–inducing agents. To this end, we examined the glycosylation status of vascular cell adhesion molecule–1 (VCAM-1), a transmembrane protein that is cotranslationally inserted into the ER membrane where it becomes N-glycosylated (11). We transfected HEK293 cells with VCAM-1, added tunicamycin, and compared IRE1 activity with the glycosylation status of VCAM-1 (Fig. 1D). HEK293 cells expressing VCAM-1 spliced Xbp-1 mRNA in a manner indistinguishable from that of wild-type (WT) cells after tunicamycin treatment, and, as in untransfected cells, Xbp-1 mRNA splicing progressively decayed back to baseline levels with prolonged treatment (Fig. 1D). VCAM-1 was fully glycosylated before tunicamycin addition and first became partially glycosylated and then unglycosylated (Fig. 1D). Not only the steady-state pool but also all newly synthesized VCAM-1 was unglycosylated at later time points (Fig. 1E).
To determine if other IRE1-dependent functions were attenuated in a manner akin to Xbp-1 mRNA splicing, we examined the activation status of c-Jun N-terminal kinase (JNK). In response to ER stress, IRE1 initiates a signal transduction pathway that activates JNK (12). In HEK293 cells, JNK was rapidly phosphorylated with ER stress (Fig. 1F). The initial burst in phosphorylation was followed by a progressive decrease in phospho-JNK levels, a trend that paralleled Xbp-1 mRNA splicing (compare the time points from 4 hours to 20 hours in Fig. 1F with the equivalent time points in Fig. 1B). Thus, cells responded to unmitigated ER stress by first activating and then attenuating IRE1 activity.
Behavior of ATF6 and PERK signaling with persistent ER stress
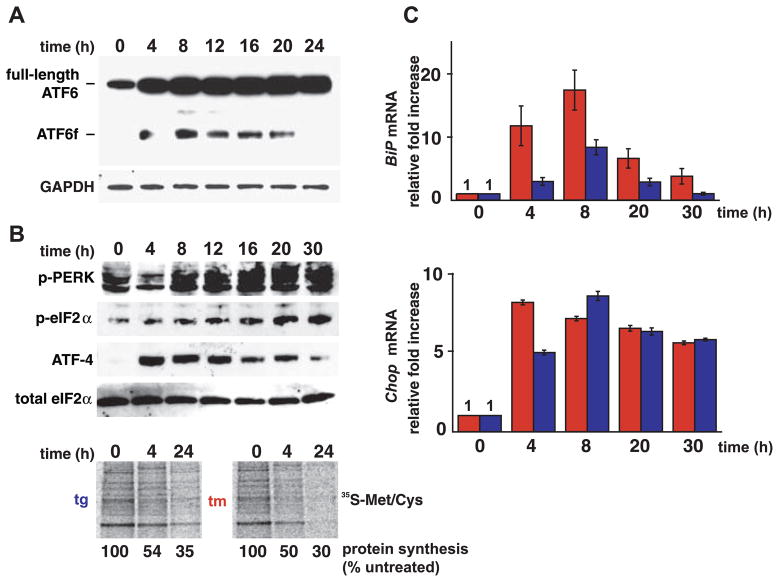
To assess whether other branches of the UPR elicit kinetic behavior similar to that elicited by IRE1, we monitored ATF6 and PERK activities over time (after UPR induction). To measure ATF6 activation, we followed the liberation of its cleaved cytosolic fragment, ATF6f, using a FLAG-tagged ATF6 reporter that recapitulated ATF6 processing upon induction of ER stress (13). We saw rapid production of ATF6f after exposure of HEK293 cells to ER stress (Fig. 2A). With prolonged ER stress, ATF6f levels diminished and ultimately disappeared (Fig. 2A). Thus, like IRE1 signaling, ATF6 activation also diminished after prolonged ER stress, albeit with different kinetics: Although IRE1 signaling decayed within 8 hours, cessation of ATF6f production was not apparent until after at least 20 hours of continuous stress.
Fig. 2.
Kinetics of ATF6 and PERK with persistent ER stress. (A) HEK293 cells, expressing FLAG-tagged ATF6α, were treated with tg (500 nM) for the indicated times, and ATF6f was detected by immunoblotting. GAPDH levels served as a protein loading control. (B) HEK293 cells were treated with tg, and phospho-PERK, phospho-eIF2α, and ATF-4 levels were determined by immunoblotting. Total eIF2α levels served as a protein loading control. In the bottom panels, cells were treated with drug for the indicated times and were pulse-labeled, and radioisotope incorporation was measured via phosphoimaging. 35S-Met/Cys, 35S-labeled methionine/cysteine. (C) Cells were treated with agents (tm, red bars; tg, blue bars) for the indicated hours, and normalized BiP (top panel) and Chop (bottom panel) mRNA levels were measured by quantitative PCR and shown relative to levels in untreated cells. Error bars represent SDs from five independent experiments.
The dynamics of the transcriptional targets of IRE1 and ATF6 were consistent with the induction kinetics of these ER-proximal UPR signal transducers. Induction of BiP mRNA, encoding an HSP70-class ER chaperone that is transcriptionally regulated by both ATF6f and XBP-1s (4, 7), peaked at 8 hours after drug treatment and then declined to near preinduction levels (Fig. 2C).
To assess the activation kinetics of the PERK branch of the UPR, we monitored the accumulation of phosphorylated PERK and its downstream product: phosphorylated eIF2α. PERK activation did not diminish even after prolonged ER stress (Fig. 2B). Consistent with this finding, the translational capacity of cells remained attenuated at all times after the imposition of ER stress (Fig. 2B). Similarly, we observed continuous production of ATF4 (Fig. 2B) and its transcriptional target Chop after ER stress (Fig. 2C), although there was some gradual diminution at the later time points. Thus, by contrast to IRE1 and ATF6, PERK branch activation is largely sustained with unmitigated ER stress.
Chemical-genetic control of IRE1 signaling in human cells
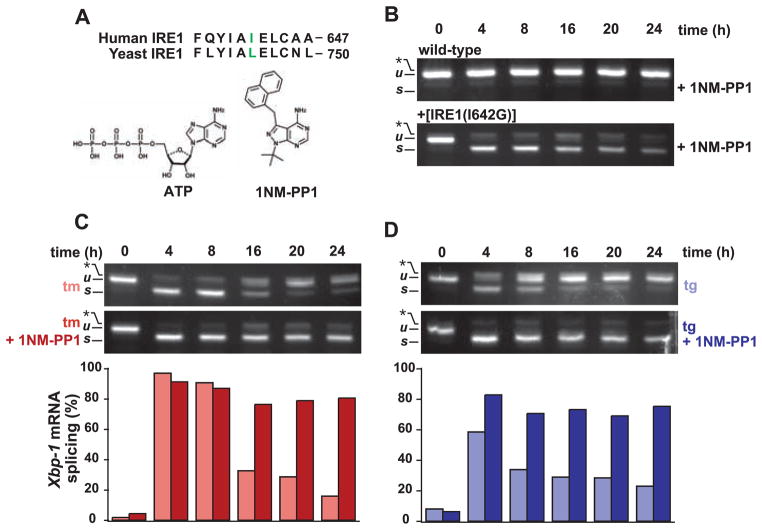
To ask whether the attenuation of IRE1 activity had physiological consequences, we sought to selectively control IRE1 activity. We followed a strategy that permitted chemical regulation of IRE1’s RNase activity using the adenosine triphosphate (ATP) analog 4-amino-1-tert-butyl-3-(1′-naphthylmethyl)pyrazolo[3,4-d] pyrimidine (1NM-PP1) (Fig. 3A), which binds selectively to the kinase domain of IRE1 mutants bearing an enlarged ATP-binding site (14). A leucine-to-glycine [Leu745→Gly745 (L745G)] mutation rendered yeast IRE1 sensitive to 1NM-PP1. We therefore constructed an allele of human IRE1 in which the orthologous amino acid [isoleucine 642 (Ile642)] was changed to glycine [Fig. 3A, “IRE1(I642G)”]. Because conventional transfection or transduction gene expression activated the UPR constitutively and caused cell death (15, 16), we used flippase-mediated, site-specific DNA recombination to introduce the drug-sensitized IRE1(I642G) mutant allele directly into the genome of HEK293 cells bearing a defined frt site (17). No deleterious growth defects were observed in the transgenic cells expressing the IRE1(I642G) allele. The UPR was not constitutively induced in these cells, as indicated by the absence of spliced Xbp-1 mRNA (Fig. 3B, 0-hours time point). Application of 1NM-PP1 alone induced robust splicing of Xbp-1 mRNA in IRE1(I642G)-expressing cells but had no effect on Xbp-1 mRNA in the parental cells (Fig. 3B). By contrast, the corresponding 1NM-PP1–sensitized allele in yeast required both ER protein misfolding and 1NM-PP1 to activate HAC1 mRNA splicing. The human allele, thus, provided a molecular switch that could be toggled by 1NM-PP1 regardless of ER protein folding status.
Fig. 3.
Chemical-genetic control of human IRE1. (A) Alignments of a portion of the ATP-binding domains of yeast and human IRE1 are shown. The residue mutated to glycine is shown in color (28). The structure of the ATP analog, 1NM-PP1, is shown below. (B) Parental WT and transgenic HEK293 cells expressing 1NM-PP1–sensitized IRE1 were treated for the indicated times with 1NM-PP1 (5 μM), and Xbp-1 mRNA splicing was determined by RT-PCR. (C) Transgenic HEK293 cells were treated with tm (5 μg/ml) ± 1NM-PP1 (5 μM). Xbp-1 mRNA splicing was assessed by RT-PCR and quantified. Results are representative of five independent experiments. (D) Transgenic HEK293 cells were treated with tg (300 nM) ± 1NM-PP1 (5 μM). Xbp-1 mRNA splicing was assessed by RT-PCR and quantified. Results are representative of five independent experiments.
We used IRE1(I642G)-expressing cells to test if we could manipulate Xbp-1 mRNA splicing during prolonged ER stress. As in WT cells, in the absence of 1NM-PP1, robust Xbp-1 mRNA splicing occurred in the transgenic cells after drug treatment at early time points and then diminished at later time points (Fig. 3, C and D; compare top panels to Fig. 1A). Thus, the expression of the mutant allele had no deleterious effects on IRE1 activation or attenuation by prolonged ER stress. By contrast, in the presence of 1NM-PP1, Xbp-1 mRNA splicing in transgenic cells was induced and remained elevated (Fig. 3, C and D). Thus, artificial activation of IRE1(I642G) by 1NM-PP1 overcame the attenuation of IRE1 activity seen upon prolonged ER stress, sustaining Xbp-1 mRNA splicing at levels approaching those seen at early time points (Fig. 3, C and D, bottom panels). Although 1NM-PP1 activated IRE1(I642G)’s RNAse activity, we detected no comparable activation of JNK signaling (fig. S2), indicating that the mRNA splicing function of IRE1(I642G) is selectively activated by 1NM-PP1.
IRE1 activity enhances cell viability
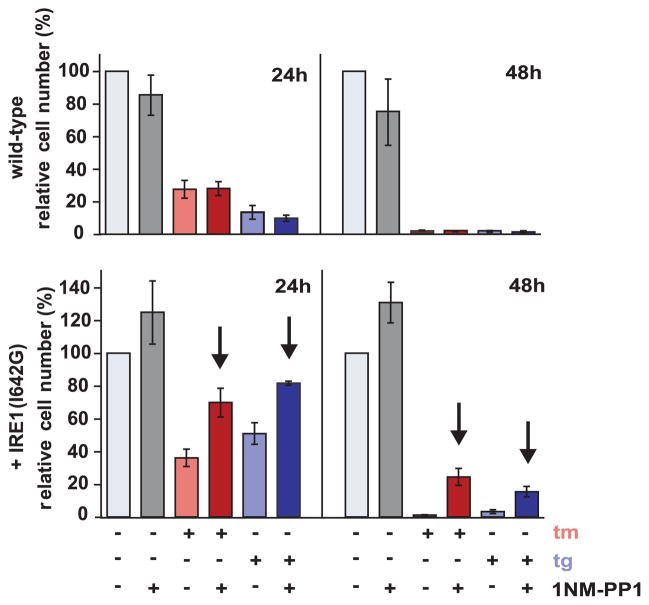
1NM-PP1 control of IRE1 allowed us to assess the physiological consequences of IRE1 activation and attenuation during the UPR. In particular, we asked whether extended IRE1 activation would have a beneficial effect on cell viability upon prolonged ER stress. ER stress induced by both tunicamycin and thapsigargin is toxic to HEK293 cells. Forty-eight hours after treatment, less than 2% of the WT cells survived (Fig. 4 and fig. S3). The addition of 1NM-PP1 had no substantial effect, although it diminished the viability of WT cells slightly (~25% reduction in viable cell number).
Fig. 4.
IRE1 signaling enhances cell viability. Parental WT and transgenic HEK293 cells were treated with the indicated agents, and adherent, cresyl violet–stained positive cells were counted at the indicated times and are shown relative to counts of mock-treated cells. Error bars represent SDs from three independent experiments. The arrows indicate P value < 0.01 when the corresponding samples ± 1NM-PP1 were compared (Student’s t test).
By contrast, 1NM-PP1 treatment of HEK293 cells expressing IRE1(I642G) significantly improved their survival. At the 48-hours time point, cell numbers after thapsigargin and tunicamycin treatment were 5 and 20 times higher, respectively, in the presence of 1NM-PP1 than in its absence (Fig. 4). Even in the absence of experimental induction of ER stress, 1NM-PP1 proved beneficial in cells expressing IRE1(I642G), affording a ~30% enhancement of cell growth (Fig. 4 and fig. S3). Thus, IRE1 signaling directly enhanced cell viability in the face of ER stress.
UPR behavior in models of retinitis pigmentosa
The UPR has been postulated to play a role in the pathogenesis of protein misfolding diseases (18). Autosomal dominant retinitis pigmentosa (adRP) is a human protein misfolding disease most commonly caused by a proline-to-histidine mutation at position 23 of rhodopsin (P23H rhodopsin) that leads to its retention within the ER (19–21). Retinal photoreceptors expressing P23H rhodopsin ultimately die, leading to blindness, but the molecular pathways linking rhodopsin misfolding in the ER to cell death are unclear (22). To explore whether the UPR is instrumental in retinal cell death, we examined its activation status in cells expressing P23H rhodopsin.
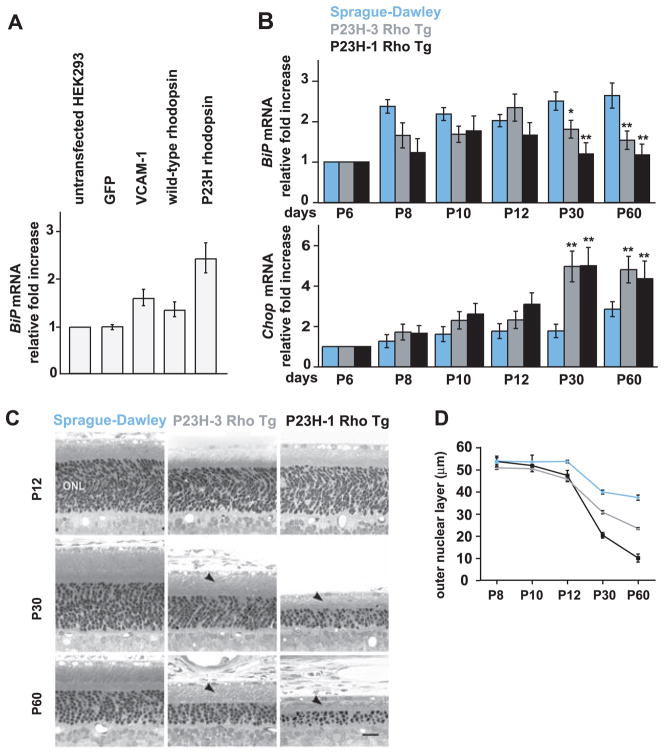
We assessed P23H rhodopsin’s ability to induce ER stress in transfected HEK293 cells. Increased BiP mRNA levels were detected in cells expressing two control ER-targeted proteins, VCAM-1 and WT rhodopsin (Fig. 5A), but not in cells expressing cytosolic green fluorescent protein (GFP), indicating that increasing the protein folding load of the ER induced the UPR. BiP mRNA expression was significantly higher in cells expressing P23H rhodopsin as compared with cells expressing WT rhodopsin (Fig. 5A). The rhodopsin mRNA levels were identical in cells expressing WT and mutant forms of the protein. Thus, P23H rhodopsin is a more potent UPR inducer than WT rhodopsin, presumably because of its folding defect.
Fig. 5.
BiP and Chop expression in animals expressing P23H rhodopsin. (A) HEK293 cells were transfected as indicated, and normalized BiP mRNA levels were measured by quantitative PCR and are shown relative to levels in untreated cells. Values represent the means ± SDs from five independent experiments. (B) Normalized BiP and Chop mRNA levels were measured in retinas [from WT Sprague-Dawley or transgenic rats expressing P23H rhodopsin at high (P23H-1 Rho Tg) or low (P23H-3 Rho Tg) levels] by quantitative PCR and are shown relative to levels at PND 6 (P6): a time when >95% of the mature complement of retinal photoreceptors has been generated. Error bars represent SDs from three animals at each age. *, P = 0.003; **, P < 0.001 [as compared with age-matched WT animals (Student’s t test)]. (C) Light micrographs of representative retinal sections from the inferior posterior retinas of WT, P23H-1, and P23H-3 rats at the indicated ages. The outer nuclear layer (ONL), which is proportional to photoreceptor nuclei numbers, thins as photoreceptors degenerate, and rhodopsin-containing outer segments (arrowheads) shorten. Scale bar, 25 μm. (D) The mean ONL thickness was measured in retinal cross sections from transgenic and WT rats at the indicated ages. Error bars represent SDs of ONL thickness from three to six animals at each age. The onset of ONL thinning in both transgenes was at P12 (P < 0.05), with progressive thinning at later ages (P < 0.0001) (Student’s t test).
We next examined BiP and Chop mRNA levels in retinas from transgenic rat models of adRP that express mouse P23H rhodopsin at low (P23H-3 Rho Tg) or high (P23H-1 Rho Tg) levels (23, 24). In WT Sprague-Dawley rats and those expressing the P23H rhodopsin transgene at either level, BiP mRNA increased after the birth of photoreceptor neurons [postnatal day 6 (PND 6)] and increased up to PND 10 or 12 (Fig. 5B). Thereafter, BiP mRNA levels selectively dropped in both transgenic lines expressing P23H rhodopsin (Fig. 5B, top panel). By contrast, Chop mRNA levels concomitantly increased in animals expressing P23H rhodopsin but remained low in WT animals (Fig. 5B, bottom panel). The time course of BiP mRNA decline and Chop mRNA rise tightly matched the rate of retinal degeneration in P23H rhodopsin transgenic animals (Fig. 5, C and D). Furthermore, the changes in BiP and Chop mRNA levels in retinas expressing misfolded rhodopsin mirrored the results seen in cell culture after prolonged ER stress (compare Figs. 5B and 2C). Thus, selective attenuation of cytoprotective UPR output coupled with sustained CHOP production—seen after drug-induced protein misfolding in vitro or, in the case of P23H rhodopsin, constitutive mis-folded protein production in vivo—contributed to cell death.
Discussion
The UPR elicits paradoxical outputs, inducing cytoprotective functions that reestablish homeostasis and cell destructive functions that promote apoptosis. We found that the switch between cytoprotective and proapoptotic output lies in part in the duration of individual UPR branch activity. After rapid initial activation of all UPR branches by ER stress, IRE1 signaling was selectively attenuated in human cells, even though stress persisted. ATF6 signaling also declined with slower kinetics, yet PERK signaling persisted much longer in the presence of unmitigated ER stress. We observed enhanced cell survival after experimentally prolonging IRE1 signaling, thereby demonstrating a causal link between IRE1 activity and cell survival. Thus, IRE1 signaling attenuation by persistent ER stress emerges as a key step in making the life or death decision after UPR induction.
Our results suggest a model by which distinct combinations of individual UPR signaling pathways determine a cell’s fate after ER stress. The initial combined activation of IRE1, PERK, and ATF6 produces cytoprotective outputs such as reduced translation, enhanced ER protein folding capacity, and clearance of misfolded ER proteins, along with proapoptotic outputs such as CHOP production. Cytoprotective outputs would outweigh proapoptotic factors at this point, which would be helped by the relatively longer mRNA and protein half-lives of factors such as BiP (25). This phase of predominantly beneficial UPR output would thus provide a “window of opportunity” for cells to readjust their ER to cope with stress. If these steps fail to reestablish homeostasis, IRE1 signaling and then ATF6 signaling are attenuated, creating an imbalance in which unchecked proapoptotic output guides the cell toward its demise. The variation in IRE1 signaling kinetics across different human cell types (fig. S1) may reflect differential susceptibility to ER stress–induced cell death among different cells and organs, reinforcing growing evidence that the metazoan UPR is tailored toward the physiologic functions of particular organs and cell types (26).
The experimental conditions used in our cell culture studies induce irreparable protein mis-folding and resemble pathological processes in which inherited mutations produce misfolded proteins. It was unexpected that in retinal degeneration models, rhodopsin molecules bearing the causative mutation of the disease induced changes in UPR activity that resembled those observed with unmitigated stress after drug treatment. Whereas WT retinal cells induced BiP concomitant with the developmental need to fold large amounts of rhodopsin, retinal cells expressing mutant rhodopsin selectively shut down BiP production and increased CHOP production, suggesting that down-regulation of IRE1, coupled with maintenance of PERK signaling, may drive the cell death seen with the P23H rhodopsin mutation. Similarly, insufficient or imbalanced UPR output could also trigger cell loss in other diseases that arise from persistent ER stress.
Supplementary Material
AUTHORS’ SUMMARY.
Organism health depends on the accuracy of the signals sent and received by constituent cells. Proteins, either secreted from the cell or embedded in the plasma membrane to monitor the environment, transmit much of this information. On the basis of these signals, cells make vital decisions—when and where to divide, migrate or change shape, differentiate, or die.
Cells have evolved elaborate mechanisms to ensure the accuracy with which proteins are folded and assembled before export or transport to the cell surface. Stringent quality control is imposed by the endoplasmic reticulum (ER), a membrane-bound labyrinth of tubes and sacs, where virtually all plasma membrane and secreted proteins begin their journey to the surface. Only properly folded proteins are allowed to leave the ER; misfolded proteins are degraded. In this way, cells display or release only high-quality, functional proteins.
To maintain fidelity, the cell needs to fold proteins as they are made, and this system needs to adapt to changing conditions. This feat is achieved by a set of intracellular signaling pathways, collectively termed the “unfolded protein response” (UPR), which senses when the ER has accumulated too many unfolded proteins. The UPR then activates transcription of certain genes that serve to increase the ER’s protein folding capacity as needed. UPR signaling can protect cells from such ER stress by expanding the amount of ER in the cell, enhancing the degradation of misfolded proteins, and reducing the synthesis of new proteins. If homeostasis cannot be reestablished, however, UPR signaling eventually induces cell death by apoptosis, an effective means of protecting the organism from rogue cells expressing dysfunctional signaling molecules. How does the UPR switch between these mutually incompatible life and death fates for the cell?
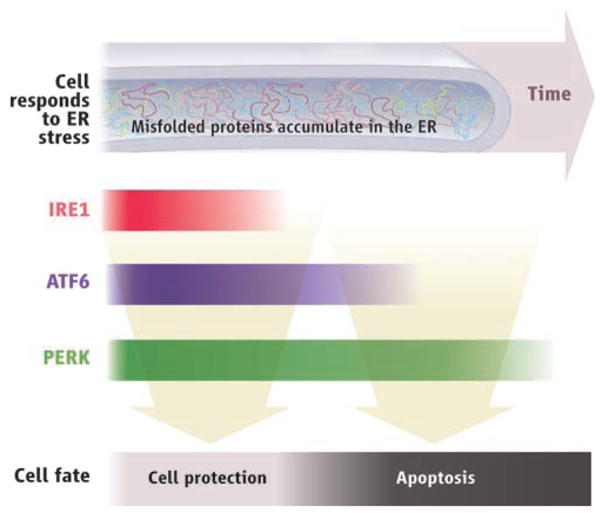
No trigger for ER stress has been identified that selectively elicits only protective responses or only apoptosis. Instead, ER stress activates all UPR signaling pathways, thereby simultaneously producing antagonistic outputs. To address this paradox, we developed assays to examine the molecular behavior of three parallel branches of the UPR—governed by the ER-stress sensors IRE1, PERK, and ATF6, respectively—in human cells exposed to persistent, pharmacologically induced ER stress. As expected, all three branches were activated upon induction of ER stress but, unexpectedly, the behavior of individual signaling pathways varied markedly with time after the onset of stress. The responses set in motion via IRE1 quickly attenuated within 8 hours despite the persistence of the stress; the ATF6 responses showed somewhat delayed attenuation (see figure). By contrast, the responses mediated by PERK persisted under prolonged ER stress and were still evident 30 hours after stress onset.
These findings suggested that the varied time courses of the individual UPR branches influence the cell’s ultimate fate in response to ER stress. To test this, we developed a chemical-genetic strategy to control IRE1 and its downstream targets, independent of ER stress. We created isogenic human cells that expressed an artificial, drug-inducible mutant IRE1, which allowed us to sustain prolonged IRE1 signaling in the face of persistent ER stress. Cell survival was significantly enhanced under these conditions, demonstrating that the termination of IRE1 activity is an important factor in allowing cell death after UPR activation.
Last, we found similar time-related switches in the endpoint of the UPR (from cell protection to apoptosis) in animal models of a heritable degenerative proteinopathy—retinitis pigmentosa—where photoreceptor cells die as a result of the expression of misfolded rhodopsin molecules, leading to blindness. Our findings thus provide a molecular rationale for how cells control whether to live or die when confronted with ER stress.
Pressing next questions include identifying the mechanisms by which different UPR branches can be selectively controlled. Phosphatases modulate PERK branch activity (1) and could also influence IRE1 signaling. Other transcriptional, translational, and posttranslational regulatory mechanisms may also contribute. Furthermore, different cell types may tailor the UPR for their own needs. Which parts of the model apply to all cells and which are tailored to allow different cell types to “dial-in” physiologically appropriate tolerance levels to ER stress? Do tissues with inherently different IRE1, ATF6, or PERK activities display greater or lesser resistance to cell death upon induction of ER stress? In addition to transcriptional output of the UPR branches, direct protein-protein interactions may also contribute to the control of apoptosis. Mammalian IRE1, for example, associates with the apoptosis-regulating BCL-2 protein family members BAK and BAX (2). IRE1 signaling activates the JNK pathway, possibly by directly phosphorylating downstream targets (3). Finally, the association of ER stress with diverse human diseases—cancer, diabetes, proteinopathies, and viral infections—raises the possibility of altering pathogenesis by manipulating the UPR.
The unfolded protein response. Accumulation of misfolded proteins (ER stress) triggers both cell protective and cell death responses, but with different time courses.
Acknowledgments
We thank the Walter lab, B. Farese, J. L. Garrison, R. J. Kaufman, R. Locksley, M. Matthes, D. Morgan, J. Nathans, R. Prywes, F. Sanchez, and B. Yen for comments, provision of reagents, or technical advice. This work was supported by the Amyotrophic Lateral Sclerosis Association, U.S. Department of Defense, Foundation Fighting Blindness, John Douglas French Alzheimer’s Foundation, National Eye Institute, NIH, and Research to Prevent Blindness. P.W. and K.M.S. are Howard Hughes Medical Institute Investigators.
Footnotes
Summary References
- 1.Novoa I, Zeng H, Harding HP, Ron D. J Cell Biol. 2001;153:1011. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hetz C, et al. Science. 2006;312:572. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 3.Urano F, et al. Science. 2000;287:664. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
References and Notes
- 1.Ron D, Walter P. Nat Rev Mol Cell Biol. 2007;8:519. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 2.Calfon M, et al. Nature. 2002;415:92. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 3.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. Cell. 2001;107:881. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 4.Lee AH, Iwakoshi NN, Glimcher LH. Mol Cell Biol. 2003;23:7448. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding HP, Zhang Y, Ron D. Nature. 1999;397:271. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 6.Harding HP, et al. Mol Cell. 2000;6:1099. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 7.Haze K, Yoshida H, Yanagi H, Yura T, Mori K. Mol Biol Cell. 1999;10:3787. doi: 10.1091/mbc.10.11.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye J, et al. Mol Cell. 2000;6:1355. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 9.Morris JA, Dorner AJ, Edwards CA, Hendershot LM, Kaufman RJ. J Biol Chem. 1997;272:4327. doi: 10.1074/jbc.272.7.4327. [DOI] [PubMed] [Google Scholar]
- 10.Zinszner H, et al. Genes Dev. 1998;12:982. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrison JL, Kunkel EJ, Hegde RS, Taunton J. Nature. 2005;436:285. doi: 10.1038/nature03821. [DOI] [PubMed] [Google Scholar]
- 12.Urano F, et al. Science. 2000;287:664. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 13.Shen J, Prywes R. Methods. 2005;35:382. doi: 10.1016/j.ymeth.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Papa FR, Zhang C, Shokat K, Walter P. Science. 2003;302:1533. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- 15.Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. Genes Dev. 2000;14:2725. doi: 10.1101/gad.839400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XZ, et al. EMBO J. 1998;17:5708. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen HR, Panning B. Chromosoma. 2007;116:373. doi: 10.1007/s00412-007-0100-1. [DOI] [PubMed] [Google Scholar]
- 18.Schroder M, Kaufman RJ. Annu Rev Biochem. 2005;74:739. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 19.Sohocki MM, et al. Hum Mutat. 2001;17:42. doi: 10.1002/1098-1004(2001)17:1<42::AID-HUMU5>3.0.CO;2-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sung CH, Davenport CM, Nathans J. J Biol Chem. 1993;268:26645. [PubMed] [Google Scholar]
- 21.Kaushal S, Khorana HG. Biochemistry. 1994;33:6121. doi: 10.1021/bi00186a011. [DOI] [PubMed] [Google Scholar]
- 22.Mendes HF, van der Spuy J, Chapple JP, Cheetham ME. Trends Mol Med. 2005;11:177. doi: 10.1016/j.molmed.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Steinberg RH, et al. Invest Ophthalmol Visual Sci. Vol. 37. Association for Research in Vision and Ophthalmology (ARVO); 1996. p. S698. Abstract 3190. [Google Scholar]
- 24.Machida S, et al. Invest Ophthalmol Visual Sci. 2000;41:3200. [PubMed] [Google Scholar]
- 25.Rutkowski DT, et al. PLoS Biol. 2006;4:374. doi: 10.1371/journal.pbio.0040374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Acosta-Alvear D, et al. Mol Cell. 2007;27:53. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 27.Back SH, Schroder M, Lee K, Zhang K, Kaufman RJ. Methods. 2005;35:395. doi: 10.1016/j.ymeth.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.