Abstract
DNA double-strand breaks (DSBs) can arise from multiple sources, including exposure to ionizing radiation. The repair of DSBs involves both post-translational modification of nucleosomes and concentration of DNA repair proteins at the site of damage. Consequently, nucleosome packing and chromatin architecture surrounding the DSB may limit the ability of the DNA damage response to access and repair the break. Here, we review early chromatin-based events that promote the formation of open, relaxed chromatin structures at DSBs and which allow the DNA repair machinery to access the spatially confined region surrounding the DSB, thereby facilitating mammalian DSB repair.
Keywords: Chromatin remodeling, DNA repair, H2AX, acetylation, nucleosome
DNA Double Strand Breaks and Cancer
Maintaining the integrity of genetic information is critical for both normal cellular functions and for suppressing mutagenic events that can lead to cancer. Damage to DNA can arise from external sources, such as exposure to ionizing radiation (IR), ultraviolet radiation (UV) or environmental toxins, or from endogenous sources such as reactive oxygen species or errors during DNA replication. These events can generate a wide range of DNA lesions, including modified bases or sugar residues, the formation of DNA adducts, cross-linking of the DNA strands and production of single and double strand breaks. Consequently, cells have evolved at least six different DNA repair pathways to deal with these distinct types of DNA damage (Kennedy and D’Andrea, 2006). Among these lesions, DNA double strand breaks (DSBs) are particularly lethal because they result in physical cleavage of the DNA backbone. DSBs can occur through replication fork collapse, during the processing of interstrand crosslinks, or following exposure to ionizing radiation (IR) (Ciccia and Elledge, 2010; Jackson and Bartek, 2009; Kennedy and D’Andrea, 2006). Because IR (radiation therapy) is widely used to treat cancer, understanding how cells repair DSBs created by IR, and how this process is altered in tumors, is of high significance.
Chromatin Structure and DSB repair
DSB repair takes place within the complex organization of the chromatin, and it is clear from work in many model systems that chromatin structure and nucleosome organization represent a significant barrier to the efficient detection and repair of DSBs. Mammalian cells contain a diverse array of specialized chromatin structures, such as active genes, telomeres, replication forks, intergenic regions and compact heterochromatin. These structures are distinguished by specific patterns of histone modifications, unique histone variants, arrays of chromatin binding proteins and by the density of nucleosome packing (de Wit and van Steensel, 2009; Grewal and Jia, 2007; Peng and Karpen, 2008). This complexity and diversity in chromatin organization presents a series of challenges to the DSB repair machinery. The impact of chromatin on DNA repair was first described in the “access-repair-restore” model ((Smerdon, 1991); reviewed in (Soria et al., 2012)). This model proposed the minimal steps needed to reorganize the chromatin and repair DNA damage. Broadly, the DSB repair machinery must be able to: (i) detect DNA damage in different chromatin structures; (ii) remodel the local chromatin architecture to provide access to the site of damage; (iii) reorganize the nucleosome-DNA template for processing and repair of the damage; and, importantly (iv) restore the local chromatin organization after repair has been completed. Since this model was first put forward in 1991, we now know many of the remodeling factors and histone modifying enzymes that act to create open chromatin structures and promote DNA repair, as well as factors such as histone chaperones, deacetylases and phosphatases that reassemble the chromatin after repair is complete. Here, we will focus on the “access” component of the “access-repair-restore” model, reviewing some of the early (seconds-minutes) remodeling events that occur after DNA damage and are required to create open chromatin structures. Although the “access-repair-restore” model is likely applicable to the repair of all types of DNA damage, we will focus our discussion specifically on the repair of DNA double-strand breaks (DSBs). In particular, we will examine 3 broad chromatin based events that occur during the first seconds-to-minutes after production of DSBs – the formation of open chromatin structures at DSBs through acetylation of histone H4; (ii) the importance of kap-1 in promoting chromatin relaxation in heterochromatin and (iii) the rapid polyADP-ribosylation (PARylation) of the chromatin by the polyADP-ribose polymerase (Parp) family, which promotes the transient recruitment of chromatin remodeling enzymes and heterochromatin factors to the DSB.
DSB Repair in Mammalian Cells
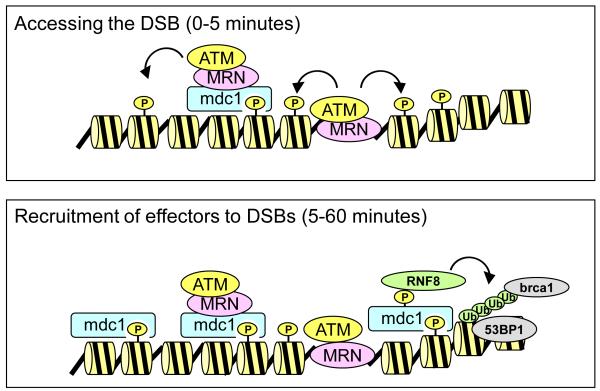
The mammalian DSB repair pathway is a complex signaling mechanism which regulates the two key responses to DSBs – the rapid activation of cell cycle checkpoints and the recruitment DNA repair proteins onto the chromatin at the DSB (figure 1). The MRN complex, consisting of the mre11, rad50 and nbs1 proteins, is first recruited to DSBs, where it functions to recruit and activate the ATM protein kinase (Lavin, 2008; Sun et al., 2010). Activated ATM has been shown to phosphorylate 100s of proteins (Matsuoka et al., 2007), including proteins involved in checkpoint activation (e.g. p53 and chk2) and DNA repair proteins such as brca1 and 53BP1 (Ciccia and Elledge, 2010; Jackson and Bartek, 2009; Kennedy and D’Andrea, 2006). A critical target for ATM is phosphorylation of the c-terminal of the histone variant H2AX. Phosphorylated H2AX (referred to as γH2AX) creates a binding site for the BRCT domains of the mdc1 protein (Lou et al., 2006; Stucki et al., 2005) (figure 1). Positioning of mdc1 at the DSB creates a docking site for additional DSB repair proteins, including the MRN-ATM complex (Chapman and Jackson, 2008; Melander et al., 2008). Consequently, phosphorylation of H2AX by ATM spreads away from the DSB, creating γH2AX domains that extend for 100’s of kilobases along the chromatin from the DSB (Bonner et al., 2008; Rogakou et al., 1999). The mdc1 protein also recruits late acting effector proteins, including the RNF8 and RNF168 ubiquitin ligases, which ubiquitinate the chromatin and promote loading of the brca1 and 53BP1 proteins (Doil et al., 2009; Kolas et al., 2007). Similar to γH2AX spreading, chromatin ubiquitination can also spread for 10s of kilobases from the DSB (Xu et al., 2010). This extension of chromatin ubiquitination is opposed by the activity of the two E3 ligases, TRIP12 and UBR5, which promote the ubiquitin-dependent degradation of RNF168 (Gudjonsson et al, 2012). DSB repair therefore involves the sequential recruitment and concentration of 1000s of copies of individual DSB repair proteins onto the chromatin, as well as extensive post-translational modification of the nucleosomes.
Figure 1. The mechanism of double strand break repair.
(Top) ATM phosphorylates H2AX at double strand breaks (−DSBs), creating a binding site for the mdc1 protein. ATM-MRN (mre11-rad50-nbs1) complexes then associate with mdc1, promoting the spreading of γH2AX along the chromatin for hundreds of kilobases. (Bottom) mdc1 recruits multiple DSB repair proteins to sites of damage, including the RNF8/RNF168 ubiquitin ligases. Chromatin ubiquitination (Ub) then facilitates loading of the brca1 complex and 53BP1 DSB repair proteins. Abbreviation: P, phosphorylation.
DSB Repair by HR and NHEJ
The actual repair of DSBs can proceed through two distinct mechanisms: the error-prone non-homologous end joining (NHEJ) pathway and the error-free homologous recombination (HR) pathway (Huertas, 2010; Jackson and Bartek, 2009). NHEJ involves minimal processing of the damaged DNA by nucleases, followed by direct re-ligation of the DNA ends. NHEJ requires the Ku70/80 DNA binding complex and the DNA-PKcs kinase. In contrast, HR requires the generation of single stranded DNA (ssDNA) intermediates, which are used for homology searching within adjacent sister chromatids. The production of ssDNA requires the initial nuclease activity of the CtIP-MRN complex (Sartori et al., 2007), followed by further end processing by additional nucleases to produce ssDNA intermediates (Symington and Gautier, 2011). This ssDNA is then used for homology searching in sister chromatids, which then provide the template for accurate repair of DSBs by HR. Importantly, because sister chromatids are only present during the S and G2 phases of the cell cycle, HR repair is restricted to this part of the cell cycle. Consequently, NHEJ predominates in G1 and HR in S phase and G2. However, how cells regulate the choice between HR and NHEJ repair pathways is not well understood, although both the 53BP1 and brca1 proteins can play a key role in this choice (Bothmer et al., 2010; Bunting et al., 2010).
Influence of Chromatin Organization on Genomic Stability
The nucleosome is the basic functional unit of chromatin and consists of 147bp of DNA wrapped around a histone octamer (Campos and Reinberg, 2009). Nucleosomes form linear 10nm beads-on-a-string structures which pack together to form 30nm arrays and other higher order structures. The core of each nucleosome contains two H3-H4 dimers and two H2A-H2B dimers. The n-terminal tails of histones extend out from the nucleosome and contain conserved lysine residues which can be modified by acetylation, methylation or ubiquitination. These modifications can function to attract specific chromatin complexes that can then alter nucleosome function. In addition to histone post-translational modifications, chromatin organization is also regulated by multi-subunit remodeling complexes built around a large motor ATPase. Four major ATPase families, including the SWI/SNF, CHD, INO80 and ISWI families have been identified in eukaryotes (Clapier and Cairns, 2009). These remodeling complexes utilize the energy from ATP hydrolysis to: (i) remove nucleosomes from the chromatin and create open DNA sequences; (ii) to shift the position of the nucleosome relative to the DNA by exposing (or burying) a DNA sequence (nucleosome sliding); or (iii) exchange pre-existing histones for specialized histone variants. Chromatin remodeling complexes and histone modifications can alter the interaction within or between adjacent nucleosomes and recruit chromatin binding proteins to specific regions (Cairns, 2005; Campos and Reinberg, 2009). Nucleosomes can therefore be envisaged as dynamic hubs to which chromatin modifying proteins and specific modifications attach, and which regulate the function and packing of the DNA in the chromatin.
The importance of chromatin organization in maintaining genomic stability is underscored by studies demonstrating that mutations rates are not even across the human genome. Sequencing of multiple cancer genomes has revealed that mutations accumulate at much higher levels in compact, H3K9me3 rich heterochromatin domains (Schuster-Bockler and Lehner, 2012), consistent with the slower rates of DNA repair reported in heterochromatin (Goodarzi et al., 2008; Noon et al., 2010). Further, inserts and deletions are depleted around nucleosomes, whereas mutations tend to cluster on the nucleosomal DNA (Chen et al., 2012; Sasaki et al., 2009; Tolstorukov et al., 2011), and can be influenced by the presence of specific epigenetic modifications on the nucleosome (Schuster-Bockler and Lehner, 2012; Tolstorukov et al., 2011). Some of these difference in mutation rates may accrue by negative selection (for example, selection against mutations in coding regions) or through protection of the DNA from mutagens by association with nucleosomes. However, the elevated mutation rates in compact, transcriptionally-silent heterochromatin domains (Schuster-Bockler and Lehner, 2012) implies that chromatin packing may impact the detection or repair of damage by the DNA repair machinery. That is, the ability of the DNA repair machinery to access the DNA can have a significant impact on genomic stability within specific regions.
DSBs Promote Rapid Histone H4 Acetylation
One of the best of the best characterized changes in chromatin organization is the rapid formation of open chromatin structures at DSBs. Several groups have demonstrated that this process is associated with increased acetylation of histones H2A and H4 on nucleosomes at DSBs (Downs et al., 2004; Jha et al., 2008; Kusch et al., 2004; Murr et al., 2006). This acetylation extends for hundreds of kilobases away from the break (Downs et al., 2004; Murr et al., 2006; Xu et al., 2010), similar to the spreading of γH2AX (figure 1). The acetylation of histone H4 at DSBs is dependent on the Tip60 acetyltransferase, a haplo-insufficient tumor suppressor protein which is required for the repair of DSBs (Doyon and Cote, 2004; Gorrini et al., 2007; Sun et al., 2010). Tip60 is rapidly recruited to DSBs where it can acetylate multiple DDR proteins, including histones H2A and H4, the ATM kinase, p53 and other repair proteins (Bird et al., 2002; Ikura et al., 2007; Jha et al., 2008; Sun et al., 2005; Sun et al., 2010; Sykes et al., 2006). Tip60 functions in DSB repair as a subunit of the human NuA4 (hNuA4) remodeling complex. hNuA4 contains at least 16 subunits (Doyon and Cote, 2004), of which 4 posses catalytic activity - the Tip60 acetyltransferase, the p400 motor ATPase and the Ruvbl1 and Ruvbl2 helicase-like proteins. Multiple sub-units of hNuA4, including Tip60 (Sun et al., 2009), Trrap (Downs et al., 2004; Kusch et al., 2004; Murr et al., 2006), p400 (Xu et al., 2010) and ruvbl1 and ruvbl2 (Jha et al., 2008) are co-recruited to DSBs, suggesting that these proteins are recruited together as components of hNuA4.
Interestingly, hNuA4 is a fusion of two separate yeast complexes – the smaller yeast NuA4 (yNuA4) complex, which contains the Tip60 homolog esa1, and ySWR1 complex, which contains the Swr1 ATPase and the yeast Ruvbl1 and Ruvbl2 homologs (Clapier and Cairns, 2009; Doyon and Cote, 2004). Both yNuA4 (Downs et al., 2004) and ySWR1 complexes (Papamichos-Chronakis et al., 2006; van Attikum et al., 2007) are recruited to enzymatically-generated DSBs in yeast. However, whereas yNuA4 and SWR1 are recruited to DSBs through direct interaction with γH2AX (Downs et al., 2004; van Attikum et al., 2007), hNuA4 is loaded onto chromatin through interaction with the mdc1 protein (Xu and Price, 2011; Xu et al., 2010). However, in both yeast and mammalian cells, loading of either yNuA4 or hNuA4 at DSBs leads to the rapid acetylation of the N-terminal tail of histone H4 by Tip60 (Downs et al., 2004; Ikura et al., 2007; Murr et al., 2006; Sun et al., 2009; Xu et al., 2010). Further, inactivation of Tip60 (Bird et al., 2002; Downs et al., 2004; Ikura et al., 2000; Murr et al., 2006) prevents H4 acetylation and leads to a significant increase in sensitivity to DSBs. Finally, mutation of the Tip60 acetylation sites on H4 in yeast increases sensitivity to DNA damage similar to that seen following Tip60 inactivation (Bird et al., 2002; Downs et al., 2004). Although mutation of the N-terminal of H4 is not possible in mammalian cells, the results from both yeast and mammalian systems indicate that the rapid recruitment of NuA4 complexes containing Tip60 to DSBs leads to the increased acetylation of histone H4 and H2A adjacent to the DSB.
Histone Acetylation Creates Open Chromatin Structures
It is well-established that open chromatin conformations at actively transcribed genes are associated with acetylation of histone H4 (Campos and Reinberg, 2009; de Wit and van Steensel, 2009). The N-terminal tail of histone H4 can interact with the acidic patch on the surface of H2A-H2B dimers of adjacent nucleosomes (Luger et al., 2012). Disruption of this interaction by acetylation of H4 on lysine 16 (Robinson et al., 2008; Shogren-Knaak et al., 2006) inhibits packing of 30nm fibers and leads to chromatin decompaction. The increase in acetylation of histones H2A and H4 at DSBs may therefore promote chromatin unpacking and direct the formation of open, relaxed chromatin structures detected at DSBs (Kruhlak et al., 2006). In fact, several studies have demonstrated that chromatin at DSBs undergoes a transition to a more open, less compact conformation. For example, the sensitivity of DNA to nuclease digestion increases after DNA damage (Smerdon et al., 1978; Ziv et al., 2006), indicating that linker DNA between nucleosomes is more accessible. Depletion of histone H1, which binds to linker DNA and promotes nucleosome packing, promotes chromatin relaxation, and facilitates DSB repair (Murga et al., 2007). Histones at DSBs are susceptible to extraction in low salt (Xu et al., 2010), implying a weaker interaction between DNA and histones at DSBs. Further, biophysical studies demonstrate that DSBs lead to a localized chromatin expansion at DSBs (Kruhlak et al., 2006). Finally, experiments in which Tip60 is inactivated (Murr et al., 2006; Xu et al., 2012; Xu et al., 2010), blocks the shift from a compact to a more open chromatin structure at DSBs, consistent with the Tip60-dependent acetylation of histone H4 creating open, flexible chromatin structures at DSBs.
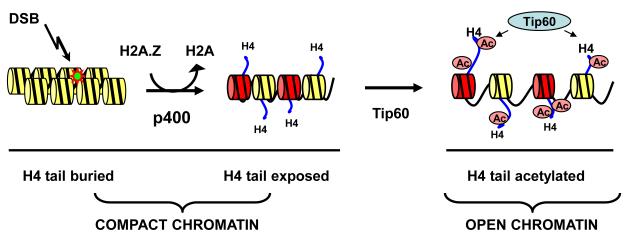
The p400 ATPase of hNuA4 Catalyzes H2A.Z Exchange at DSBs
In addition to Tip60, the hNuA4 complex also contains the p400 motor ATPase. p400 is a member of the Ino80 family of chromatin remodeling ATPases, which includes 2 yeast proteins – yIno80 and ySwr1. yINO80 and ySWR1 are both recruited to DSBs in yeast and loss of either component leads to significant defects in both checkpoint activation and DSB repair (Downs et al., 2004; Papamichos-Chronakis et al., 2006; van Attikum et al., 2007). Members of the Ino80 family, including the mammalian p400 ATPase, can exchange histone H2A for the H2A variant H2A.Z (Fuchs et al., 2001; Gevry et al., 2007; Kusch et al., 2004), suggesting that Ino80 family members may regulate H2A.Z exchange during DSB repair. Indeed, in yeast, loss of H2A.Z leads to increased sensitivity to DNA damaging agents (Morillo-Huesca et al., 2010; Papamichos-Chronakis et al., 2011) and defective repair of DSBs (Kalocsay et al., 2009). Although a transient increase in H2A.Z deposition at DSBs in yeast has been reported (Kalocsay et al., 2009), other studies suggest that Ino80 and Swr1 may function antagonistically to regulate or maintain H2A.Z at DSBs (Papamichos-Chronakis et al., 2006; van Attikum et al., 2007), with no overall increase in H2A.Z exchange at DSBs in yeast (van Attikum et al., 2007). However, in mammalian cells, the hNuA4 complex promotes not only H4 acetylation by the Tip60 subunit but also the rapid exchange of H2A for H2A.Z at DSBs (figure 2) (Xu et al., 2012). H2A.Z exchange requires the ATPase activity of the p400 motor protein and creates chromatin domains containing H2A.Z-nucleosomes that extend away from the DSB. Surprisingly, H2A.Z preceeds, and is required for, both the acetylation of histone H4 by Tip60 and the creation of open chromatin domains at DSBs (Downs et al., 2004; Murr et al., 2007; Xu et al., 2010). The exchange of H2A.Z onto nucleosomes at DSBs leads to an increase in the salt solubility of the histones (Xu et al., 2012), indicating the formation of open chromatin at the site of damage. This is consistent with published work demonstrating that H2A.Z-nucleosomes are less stable than H2A nucleosomes and are more sensitive to extraction at low salt concentrations (Henikoff et al., 2009; Jin and Felsenfeld, 2007; Weber et al., 2010; Zhang et al., 2005). However, other studies have shown that H2A.Z stabilizes nucleosomes (Fan et al., 2004; Park et al., 2004). These opposing effects of H2A.Z on nucleosome structure have been extensively reviewed by others (Billon and Cote, 2012; Zlatanova and Thakar, 2008). However, it has been noted that the ability of H2A.Z to reduce nucleosome stability is dependent on both histone modifications and the presence of additional histone variants, including histone H3.3, on the nucleosome (Henikoff et al., 2009; Jin and Felsenfeld, 2007; Jin et al., 2009). The ability of H2A.Z to destabilize nucleosomes at DSBs may therefore depend on both the presence of other histone variants (such as H3.3) and histone post-translational modifications, including acetylation of H2A.Z, on nucleosomes. Consistent with this, the ability of H2A.Z to create open chromatin structures at DSBs requires both the presence of H2A.Z and acetylation of histone H4 tails by the Tip60 acetyltransferase (Xu et al., 2012) (figure 2). That is, H2A.Z appears to only be capable of destabilizing nucleosomes at DSBs in the context of an acetylated H4 tail.
Figure 2. H2A.Z exchange drives H4 acetylation.
Exchange of H2A for H2A.Z alters interaction between the N-terminal tail of histone H4, exposing it to acetylation by Tip60. The combination of H2A.Z exchange and H4 acetylation (Ac) functions to shift chromatin into the open, relaxed conformation required for DSB repair.
How the presence of H2A.Z promotes the acetylation of the n-terminal of H4 by Tip60 is less clear. Nucleosomes containing H2A.Z exhibit only subtle differences in structure from H2A-nucleosomes (Suto et al., 2000). The N-terminal tail of histone H4 interacts with an acidic patch on the surface of the nucleosome and promotes packing into 30nm fibers (Robinson et al., 2008; Shogren-Knaak et al., 2006). In H2A.Z, this acidic patch is extended in length, and it has been proposed that this extended acidic region stabilizes the interaction between H2A.Z and H4, promoting packing of nucleosome fibers (Fan et al., 2004). This would tend to restrict the ability of Tip60 to acetylate the N-terminus of H4. However, as discussed above, the ability of H2A.Z to impact chromatin organization can be modulated by the presence of histone H3.3 or by additional histone modifications within the nucleosome (Jin and Felsenfeld, 2007; Jin et al., 2009; Zlatanova and Thakar, 2008) (figure 2). H2A.Z exchange may therefore be only part of the equation, with the potential for exchange of H3.3, specific acetylation of H2A.Z or additional remodeling motor ATPases contributing to acetylation of histone H4 in response to DSBs. Unraveling these early events will provide new insight into H2A.Z-mediated shifts in chromatin structure at DSB.
Rapid Chromatin Remodeling Promotes Ordered Chromatin Modification
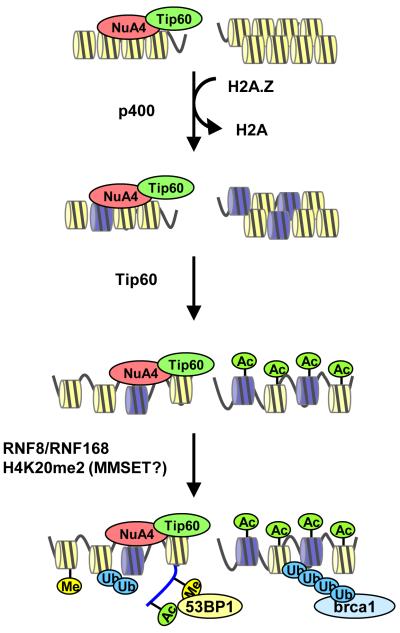
The NuA4 driven changes in chromatin organization (figure 2) have a significant impact on the mechanism of DSB repair. In particular, the formation of open chromatin domains through H2A.Z exchange and H4 acetylation facilitates further DNA damage-dependent modification of the chromatin by both ubiquitination and methylation of histone H4 (figure 3). Inactivation of components of hNuA4, including p400, Tip60 or Trrap, blocks the ubiquitination of histone H2A/H2AX by RNF8/RNF168 and inhibits the subsequent loading of several effector proteins including brca1, 53BP1 and rad51, onto chromatin (figure 3) (Courilleau et al., 2012; Murr et al., 2006; Xu et al., 2012; Xu et al., 2010). Brca1 recruitment requires interaction between the RAP80 subunit of the brca1 complex and ubiquitinated chromatin at DSBs (Sobhian et al., 2007). The NuA4-dependent shift in chromatin structure at DSBs may therefore reveal cryptic sites for H2A/H2AX ubiquitination by RNF8/RNF168 and drive loading of brca1. The recruitment of 53BP1, a DNA repair protein that regulates NHEJ (Bunting et al., 2010), is complex and can also be regulated by RNF8/RNF168-mediated chromatin ubiquitination (Doil et al., 2009; Huen et al., 2007). However, 53BP1 does not posses an identifiable ubiquitin binding motif. It has also been shown that 53BP1 recruitment to DSBs requires H4 acetylation (Murr et al., 2006; Xu et al., 2010) and H4K20 methylation (Botuyan et al., 2006). In fact, 53BP1’s tudor domain can bind to histone H4 dimethylated on lysine 20 (H4K20me2) (Botuyan et al., 2006). Because a significant fraction (>80%) of H4K20 is dimethylated in mammalian cells, the increased acetylation of histone H4 at DSBs may function to both unpack closely opposed chromatin fibers and reveal H4K20me2 for 53BP1 binding. Also, H2A/H2AX ubiquitination by RNF8 and RNF168 may further promote 53BP1 loading by altering the accessibility of 53BP1 to H4K20me2 (Figure 3). Interestingly, mice lacking both of the Suv4-20h H4K20me2 methyltransferases have almost no H4K20me2 and display increased genomic instability, yet maintain normal recruitment of 53BP1 to DSBs (Schotta et al., 2008). While this may suggest that H4K20me2 is dispensable for 53BP1 recruitment to DSBs, it has recently been reported that the methyltransferase MMSET is recruited to DSBs and promotes the formation of H4K20me2 (Pei et al., 2011). Recruitment of MMSET may provide the mechanism for methylation of the small fraction of H4K20 that is not constitutively methylated and may partially compensate for loss of constitutive H4K20me2 in the Suv4-20h1/suv4-20h2 double knockout mice. In fact, 53BP1 has been reported to promote long range interactions between DNA ends (Difilippantonio et al., 2008) suggesting that 53BP1 binding may itself play a role in regulating or stabilizing chromatin structure after DNA damage (Noon et al., 2010). Thus the initial change in nucleosome function imposed by H2A.Z exchange promotes an ordered series of histone modifications, including acetylation of histone H4 and ubiquitination of the chromatin (figure 3). This may then unmask H4K20me2 buried within the nucleosome structure, or promote H4K20 methylation by MMSET, and facilitate loading of both 53BP1 and brca1 complexes onto the chromatin. The early remodeling events therefore play a critical role in directing the ordered recruitment of DSB repair proteins to the site of damage.
Figure 3. H2A.Z exchange drives chromatin changes that direct chromatin modification at DSBs.
At double strand breaks, H2A.Z exchange promotes H4 acetylation (Ac) by Tip60, which in turn directs ubiquitination (Ub) of the chromatin by the RNF8/RNF168 ubiquitin ligases and exposure and/or methylation (Me) of H4K20me2 by MMSET. Association of NuA4-Tip60 with mdc1 is omitted for clarity. Abbreviation: P, phosphorylation.
Impact of H2A.Z on DSB repair
Cells lacking H2A.Z or components of NuA4 exhibit are hypersensitive to ionizing radiation and have defects in both NHEJ and HR directed repair (Downs et al., 2004; Ikura et al., 2000; Murr et al., 2006; Xu et al., 2012; Xu et al., 2010). This wide range of defects reflects the early and critical role of hNuA4 in promoting access to sites of damage, and reflects both the failure to create open chromatin structures and the lack of recruitment of brca1, which is essential for HR-mediated DSB repair. Intriguingly, when H2A.Z exchange at DSBs is inhibited, cells undergo unrestricted end resection, leading to accumulation of excess ssDNA and the loss of Ku70/80 binding (Xu et al., 2012). Further, this defect can be reversed by depletion of CtIP, suggesting that H2A.Z exchange functions to restrain or restrict the ability of the CtIP-MRN nuclease complex to initiate end resection of the DSB. In yeast, loss of the ySwr1 ATPase also leads to defects in Ku70 recruitment and defects in error-free NHEJ (van Attikum et al., 2007), although this is not directly linked to H2A.Z exchange. Recent work on the role of H2A.Z at transcriptional start sites (TSS) provides some potential insight into how H2A.Z may restrict end resection. The TSS of many genes is flanked by H2A.Z-nucleosomes (Jin et al., 2009; Zhang et al., 2005), which may function to fix the position of nucleosomes on either side of the TSS and thereby maintain nucleosome free DNA for transcription factor binding. Nucleosomes are also lost at DSBs, creating nucleosome free regions (Tsukuda et al., 2005). The placement of H2A.Z nucleosomes either side of nucleosome-free regions at the DSB therefore creates a structure similar to that reported at the TSS of genes. Positioning of H2A.Z on either side of the DSB may therefore define the limits of the nucleosome-free region and create a chromatin template which restricts or limits end resection by the CtIP-MRN complex. The early remodeling of the chromatin at DSBs through H2A.Z exchange and H4 acetylation are therefore critical for setting the scene for further processing and eventual repair of the DSB through either NHEJ or HR pathways
Accessing DSBs in Heterochromatin
How cells access and repair DSBs within the higher order chromatin environment of heterochromatin has been the subject of recent studies. Heterochromatin is classically described as condensed, densely-staining regions of DNA that contain few active genes but are enriched for repetitive sequences. Mammalian heterochromatin is characterized by high levels of the histone modifications H3K9me3 and H3K27me3 and low levels of histone acetylation. Heterochromatin is maintained by a dense array of specific chromatin binding proteins, including members of the HP1 family (which bind to methylated H3K9), kap-1, histone deacetylases, and histone methyltransferases. From the perspective of DSB repair, it is important to determine if the dense packing and unique array of heterochromatin binding proteins present a specific barrier to the DSB repair machinery. Further, the presence of repetitive DNA within heterochromatin may provide a significant challenge for HR-mediated repair, requiring more stringent control of HR to prevent inappropriate recombination events.
Kap-1 is a repressor protein that interacts with HP1, histone deacetylases (HDACs), and histone methyltransferases and functions to maintain heterochromatin (Iyengar and Farnham, 2011). In response to DSBs, kap-1 is phosphorylated by ATM (Goodarzi et al., 2008; Ziv et al., 2006), promoting a general relaxation of the chromatin structure. Repair of DSBs (as measured by loss of γH2AX foci) is significantly slower within heterochromatin regions, and is dependent on phosphorylation of kap-1 by ATM. Further, kap-1 phosphorylation promotes release of the CHD3 remodeling ATPase from heterochromatin (Goodarzi et al., 2011), a process required for efficient repair. It is currently unclear how loss of CHD3 or phosphorylation of kap-1 (which remains associated with the DSB regions) impacts overall chromatin structure at DSBs. In addition to kap-1 phosphorylation, HP1 proteins (including HP1α, β and γ) can repress heterochromatin repair. Depletion of HP1 proteins (or depletion of the H3K9 methyltransferases) can decondense heterochromatin and promote repair of DSBs even in the absence of ATM kinase activity (Chiolo et al., 2011; Goodarzi et al., 2011; Goodarzi et al., 2008). Further, there is some evidence to suggest that HP1 proteins are actively ejected from the chromatin during DNA repair (Ayoub et al., 2008). These observations are consistent with the idea that the dense packing of nucleosomes and the presence of specific heterochromatin binding complexes is a significant barrier to repair of heterochromatic DSBs. Further, these results indicate a critical role for phosphorylation of kap-1 by the ATM kinase in promoting the unpacking of heterochromatin and thereby facilitating repair of heterochromatic DSBs. Currently it is unclear if, for example, the NuA4-Tip60 complex acetylates histones at heterochromatic DSBs or whether the phosphorylation of kap-1 within heterochromatin is sufficient to create the required open chromatin structure. Further, given that H2A.Z is found at heterochromatin boundaries, it will be interesting to determine if this variant is important for heterochromatic DSB repair as well.
Spacing of H2AX Nucleosomes and Heterochromatin
Studies on DSB repair in heterochromatin utilize microscopy to monitor the appearance of γH2AX foci and either DAPI (to detect dense chromatin domains) or antibodies to locate regions of heterochromatin (Chiolo et al., 2011; Goodarzi et al., 2008; Noon et al., 2010). Several studies indicate that γH2AX foci preferentially assemble in euchromatin or are predominantly located at the boundary of the heterochromatin (Goodarzi et al., 2008; Kim et al., 2007; Noon et al., 2010). However, studies using enzymatically-generated DSBs coupled with chromatin immunoprecipitation indicate that γH2AX does not spread uniformly along the chromosome (Iacovoni et al., 2010; Meier et al., 2007; Savic et al., 2009) and that the size of the γH2AX domain varies between different chromatin locations (Xu et al., 2012). Further, in yeast, γH2AX does not spread through heterochromatin regions (Kim et al., 2007). H2AX is unique compared to other DSB repair proteins because it is prepositioned on nucleosomes rather than recruited to DSBs. To function as a DSB detector, and to allow for γH2AX propagation along the chromatin, it would be expected that H2AX should be evenly deposited along the chromatin. However, the amount of H2AX in cells can vary from 2% to 20% of the total H2A (Rogakou et al., 1998). That is, in some cells, 1 in 2.5 nucleosomes contain H2AX, whereas in other cells as few as 1 in 25 nucleosomes may contain H2AX. In fact, high-resolution microscopy indicates that H2AX is concentrated in specific domains (Bewersdorf et al., 2006), and ChIP-Seq analysis indicates that H2AX is concentrated within gene-rich regions (Iacovoni et al., 2010). This raises the possibility that H2AX density or distribution within heterochromatin is significantly lower than in other domains. The failure to detect γH2AX foci in heterochromatin using microscopy may therefore reflect altered H2AX distribution in heterochromatin and a reduced need for H2AX function in heterochromatin.
In addition to differential H2AX distribution in heterochromatin, recent work in Drosophila has provided an alternative explanation for why γH2AX foci are only detected at the periphery of the heterochromatin. This work demonstrates that phosphorylation of H2AX and initial recruitment of DSB repair proteins to the break occurs normally within the heterochromatin. However, these heterochromatic DSBs rapidly migrated out of the heterochromatin; hence the actual DSB repair is carried out within euchromatin (Chiolo et al., 2011; Jakob et al., 2011). Further, this relocation of the DSB is only partly dependent on ATM, indicating that phosphorylation of kap-1 by ATM does not contribute to this process. Moving the DSB out of the heterochromatin may limit recombination with repetitive sequences and allow increased mobility and easier access to the DSB. However, it should be noted that experiments in mammalian cells have indicated only limited mobility for DSBs, so it will be important to explore DSB mobility in the heterochromatin of mammalian cells (Krawczyk et al., 2012; Soutoglou et al., 2007). Finally, it is interesting to note that, in yeast, exchange of H2A.Z into the chromatin is required for relocalization of persistent DSBs to the nuclear periphery (Kalocsay et al., 2009). The NuA4-mediated exchange of H2A.Z at heterochromatin DSBs (figure 2) may potentially promote relocation of DSBs out of the heterochromatin. Clearly, our understanding of the mechanism of DSB repair within heterochromatin is limited. Developing new approaches, such as coupling synthetic nucleases to create DSBs in heterochromatin with ChIP-Seq approaches, may provide a more directed approach to understanding DSB repair within specific chromatin domains.
Early Recruitment Events: HP1
It is now clear that additional chromatin based events occur prior to the NuA4-mediated chromatin relaxation. In particular, 2 heterochromatin-associated proteins, HP1 and kap-1, participate in the early response to DSBs in euchromatin. HP1α and kap-1 are rapidly recruited to DSBs within seconds to minutes after damage induction ((Baldeyron et al., 2011; Luijsterburg et al., 2009) reviewed in (Soria et al., 2012)). The recruitment of HP1α and kap-1 is essential for loading 53BP1 and brca1 and for HR directed repair. Kap-1 and HP1 proteins may be recruited to DSBs as a single complex, although HP1α loading requires the histone chaperone ASF1 (Baldeyron et al., 2011). Importantly, HP1 and kap-1 recruitment to euchromatin is transient, with both proteins dissociating from the break a few minutes after damage induction (Baldeyron et al., 2011). It is currently unclear if HP1 and kap-1 have distinct roles in heterochromatin and euchromatin during DSB repair, and why transient recruitment and release of HP1 is important remains to be investigated. One potential explanation is that kap-1 exists as a complex with repressive factors including HDACs and H3K9 methyltransferases (Iyengar and Farnham, 2011). Recruitment of repressive kap-1 complexes may rapidly “heterochromatinize” the DSB region, preventing transcription and stabilizing the chromatin structure. Further, since the Tip60 sub-unit of NuA4 requires interaction with H3K9me3 for stimulation of its acetyltransferase activity (Sun et al., 2009), recruitment of kap-1/HP1 complexes may provide a mechanism for the rapid methylation of H3K9 and therefore facilitate the activity of both Tip60 and the NuA4 complex. The transient accumulation of kap-1 and HP1 complexes may rewrite local histone modification signatures, thereby increasing available H3K9me3 and promoting the activity of the Tip60 sub-unit of NuA4 and other factors.
Early Recruitment of NuRD and ALC1 complexes through PARylation
Similarly to recruitment of kap-1/HP1, there is also a rapid and transient accumulation of the NuRD (Chou et al., 2010; Larsen et al., 2010; Polo et al., 2010; Smeenk et al., 2010) and ALC1 (Ahel et al., 2009) remodeling complexes at DSBs (figure 4). NuRD complexes contain either the CHD3 or CHD4 ATPase, HDAC1 or HDAC2, and associated regulatory subunits (Clapier and Cairns, 2009). NuRD is a repressive complex that maintains higher order chromatin structure. Inactivation of NuRD or ALC1 leads to defects in DSB repair and increased sensitivity to DNA damage (Ahel et al., 2009; Chou et al., 2010; Polo et al., 2010; Smeenk et al., 2010). NuRD regulates the acetylation of p53 and thereby controls the extent of G1-S arrest following DNA damage (Larsen et al., 2010; Polo et al., 2010). Second, NuRD, like NuA4, is required for chromatin ubiquitination by RNF8/RNF168 and for loading of brca1 (Larsen et al., 2010; Smeenk et al., 2010). The recruitment of NuRD complexes to DSBs requires PARylation of the chromatin by PARP1 (Chou et al., 2010; Polo et al., 2010). PARP1 belongs to a family of polyADP ribose polymerases that play a central role in both transcription and DNA repair (Gibson and Kraus, 2012). Chromatin at DSBs is rapidly and transiently PARylated (figure 4) and it is this modification, rather than γH2AX or ATM signaling, that localizes NuRD at the DSB (Chou et al., 2010; Polo et al., 2010).
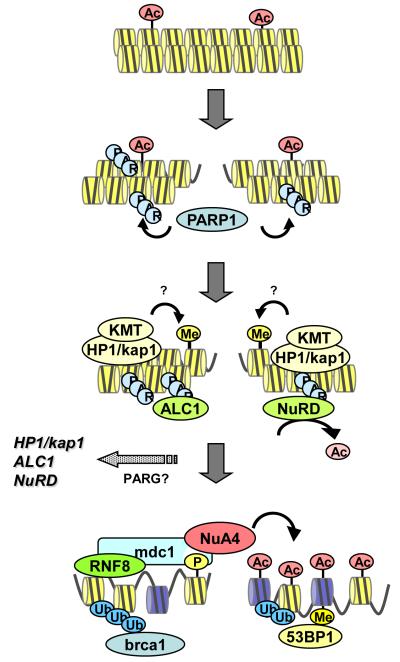
Figure 4. Creating access to Double strand Breaks.
Chronological sequence of steps in remodeling of a double strand break (DSB). Initial polyADP-ribosylation (PARylation) by PARP1 leads to rapid recruitment of NuRD and ALC1 (through interaction with PAR) and Kap-1/HP1 complexes (possibly through interaction with PAR). Deacetylation (by NurD) and proposed H3K9 methylation by lysine methyltransferases (KMTs), including suv39h1 and G9a, creates a temporary repressive chromatin structure. Subsequent phosphorylation (P) of γH2AX recruits NuA4-Tip60, promoting the ordered remodeling of the chromatin through H2A.Z exchange, histone acetylation (Ac) and chromatin ubiquitination (Ub). This creates a common chromatin template for DSB repair by either nonhomologous end joining (NHEJ) -or homologous recombination (HR)-mediated repair._ KMT, lysine methyltransferases.
Similarly, ALC1, a remodeling ATPase that functions to reposition nucleosomes on the chromatin, is also rapidly recruited to DSBs through direct interaction with PAR chains on the chromatin (Ahel et al., 2009; Gottschalk et al., 2009). ALC1 loading is also rapid and transient after DNA damage, and may favor the formation of open chromatin (Ahel et al., 2009). Thus at least 3 remodeling complexes, HP1/kap-1, NuRD, and ALC1 are rapidly, but transiently, recruited to DSBs (figure 4). Because PARylation of the chromatin is transient, yet independent of γH2AX formation, the recruitment of HP1/kap-1, NuRD, and ALC1 likely precedes the recruitment and loading of the NuA4-Tip60 complex (figure 4). However, whether these complexes work sequentially or in parallel is not yet known. For example, whether the recruitment of NuA4-Tip60 or H2A.Z exchange requires prior processing of the chromatin by either ALC1 or NuRD, or is dependent on chromatin PARylation, is not known. Further, it remains to be seen if the HP1/kap-1 complex is recruited to DSBs through PARylation or some other mechanism. Finally, the rapid release of ALC1, NuRD and HP1/kap-1 complexes may be brought about by dePARylation of the chromatin by polyADP-ribose glycohydrolases (PARGs) (figure 4). Understanding the regulation of PARGs may provide new insight into some of the earliest events occurring during DSB repair.
The HP1-kap-1, ALC1 and NuRD complexes deploy a wide range of chromatin remodeling activities, including HDACs (NuRD), methyltransferases (HP1/kap-1), and remodeling ATPase activities (NuRD and ALC1) at the DSB. Because these complexes are only retained at the DSBs for a short time period (minutes), they must play a critical role in the initial detection and processing of the chromatin at the DSB. This role could include the rapid termination of local transcription by promoting histone deacetylation (NuRD) and/or the formation of repressive chromatin through histone methylation and loading of kap-1/HP1 complexes. By erasing previous histone acetylation marks, NuRD and the other these complexes may prime the chromatin for uniform acetylation by the NuA4-Tip60 complex. Further, ALC1 may function to reposition nucleosomes at the DSB and to stabilize the chromatin and facilitate further processing and repair. These events may rapidly and transiently stabilize the local chromatin structure by creating a temporary, compacted, repressive chromatin environment at the DSB. Subsequently, DSB signaling, including γH2AX formation and ATM activation, leads to the ordered recruitment of DSB repair proteins to the chromatin at DSBs. The transient creation of PAR chains at DSBs by PARP1, which allows the rapid recruitment of NuRD, ALC1, and potentially kap-1/HP1, is therefore a critical early event in the DNA damage response.
Conclusions and Future Directions
A eukaryotic cell must integrate classical DSB repair signaling and repair by NHEJ and HR pathways with the complexity of the local chromatin architecture. Functional chromatin domains, such as replication forks, genes, or heterochromatin, differ significantly in the patterns of histone modifications, the types of chromatin binding proteins, and the degree of nucleosome packing. Each of these domains may therefore require unique chromatin remodeling complexes to alter the local chromatin architecture at individual DSBs. Identifying the protein remodeling complexes that are essential for repair in specific chromatin structures is therefore of key importance. Such processes may be critical for reshaping the local chromatin structure and for creating a common DNA template that can be presented to the DSB repair machinery. It is clear that some of the earliest events in DSB repair occurring in the first few minutes after damage can have a profound impact on processing of the damaged chromatin template. However, in addition to these early events, there are many additional steps in DSB repair that require chromatin remolding, such as homology searching during HR-directed repair or regulation of end-resection during repair. In addition, resetting the chromatin structure and restoring the original epigenetic code to the repaired chromatin are vital to ensure that normal functionality is restored to the damaged chromatin.
Acknowledgements
This work was supported by NIH grants CA64585 and CA93602 to BDP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- Baldeyron C, Soria G, Roche D, Cook AJ, Almouzni G. HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol. 2011;193:81–95. doi: 10.1083/jcb.201101030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewersdorf J, Bennett BT, Knight KL. H2AX chromatin structures and their response to DNA damage revealed by 4Pi microscopy. Proc Natl Acad Sci U S A. 2006;103:18137–18142. doi: 10.1073/pnas.0608709103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Cote J. Precise deposition of histone H2A.Z in chromatin for genome expression and maintenance. Biochimica et biophysica acta. 2012;1819:290–302. doi: 10.1016/j.bbagrm.2011.10.004. [DOI] [PubMed] [Google Scholar]
- Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8:957–967. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothmer A, Robbiani DF, Feldhahn N, Gazumyan A, Nussenzweig A, Nussenzweig MC. 53BP1 regulates DNA resection and the choice between classical and alternative end joining during class switch recombination. The Journal of experimental medicine. 2010;207:855–865. doi: 10.1084/jem.20100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr Opin Genet Dev. 2005;15:185–190. doi: 10.1016/j.gde.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO reports. 2008;9:795–801. doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen Z, Chen H, Su Z, Yang J, Lin F, Shi S, He X. Nucleosomes suppress spontaneous mutations base-specifically in eukaryotes. Science. 2012;335:1235–1238. doi: 10.1126/science.1217580. [DOI] [PubMed] [Google Scholar]
- Chiolo I, Minoda A, Colmenares SU, Polyzos A, Costes SV, Karpen GH. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Adamson B, Dephoure NE, Tan X, Nottke AC, Hurov KE, Gygi SP, Colaiacovo MP, Elledge SJ. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci U S A. 2010;107:18475–18480. doi: 10.1073/pnas.1012946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Courilleau C, Chailleux C, Jauneau A, Grimal F, Briois S, Boutet-Robinet E, Boudsocq F, Trouche D, Canitrot Y. The chromatin remodeler p400 ATPase facilitates Rad51-mediated repair of DNA double-strand breaks. The Journal of cell biology. 2012;199:1067–1081. doi: 10.1083/jcb.201205059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, van Steensel B. Chromatin domains in higher eukaryotes: insights from genome-wide mapping studies. Chromosoma. 2009;118:25–36. doi: 10.1007/s00412-008-0186-0. [DOI] [PubMed] [Google Scholar]
- Difilippantonio S, Gapud E, Wong N, Huang CY, Mahowald G, Chen HT, Kruhlak MJ, Callen E, Livak F, Nussenzweig MC, et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature. 2008;456:529–533. doi: 10.1038/nature07476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doil C, Mailand N, Bekker-Jensen S, Menard P, Larsen DH, Pepperkok R, Ellenberg J, Panier S, Durocher D, Bartek J, et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 2009;136:435–446. doi: 10.1016/j.cell.2008.12.041. [DOI] [PubMed] [Google Scholar]
- Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Binding of Chromatin-Modifying Activities to Phosphorylated Histone H2A at DNA Damage Sites. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–154. doi: 10.1016/j.gde.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Molecular cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. The p400 complex is an essential E1A transformation target. Cell. 2001;106:297–307. doi: 10.1016/s0092-8674(01)00450-0. [DOI] [PubMed] [Google Scholar]
- Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nature reviews Molecular cell biology. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Kurka T, Jeggo PA. KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nature structural & molecular biology. 2011;18:831–839. doi: 10.1038/nsmb.2077. [DOI] [PubMed] [Google Scholar]
- Goodarzi AA, Noon AT, Deckbar D, Ziv Y, Shiloh Y, Lobrich M, Jeggo PA. ATM signaling facilitates repair of DNA double-strand breaks associated with heterochromatin. Mol Cell. 2008;31:167–177. doi: 10.1016/j.molcel.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Gorrini C, Squatrito M, Luise C, Syed N, Perna D, Wark L, Martinato F, Sardella D, Verrecchia A, Bennett S, et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature. 2007;448:1063–U1012. doi: 10.1038/nature06055. [DOI] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, et al. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome research. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nature structural & molecular biology. 2010;17:11–16. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. Embo J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol Cell Biol. 2007;27:7028–7040. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. The Journal of biological chemistry. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob B, Splinter J, Conrad S, Voss KO, Zink D, Durante M, Lobrich M, Taucher-Scholz G. DNA double-strand breaks in heterochromatin elicit fast repair protein recruitment, histone H2AX phosphorylation and relocation to euchromatin. Nucleic acids research. 2011;39:6489–6499. doi: 10.1093/nar/gkr230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Shibata E, Dutta A. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Molecular and cellular biology. 2008;28:2690–2700. doi: 10.1128/MCB.01983-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–1529. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Kennedy RD, D’Andrea AD. DNA repair pathways in clinical practice: lessons from pediatric cancer susceptibility syndromes. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:3799–3808. doi: 10.1200/JCO.2005.05.4171. [DOI] [PubMed] [Google Scholar]
- Kim JA, Kruhlak M, Dotiwala F, Nussenzweig A, Haber JE. Heterochromatin is refractory to gamma-H2AX modification in yeast and mammals. J Cell Biol. 2007;178:209–218. doi: 10.1083/jcb.200612031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318:1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczyk PM, Borovski T, Stap J, Cijsouw T, ten Cate R, Medema JP, Kanaar R, Franken NA, Aten JA. Chromatin mobility is increased at sites of DNA double-strand breaks. Journal of cell science. 2012;125:2127–2133. doi: 10.1242/jcs.089847. [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Celeste A, Dellaire G, Fernandez-Capetillo O, Muller WG, McNally JG, Bazett-Jones DP, Nussenzweig A. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol. 2006;172:823–834. doi: 10.1083/jcb.200510015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. The Journal of cell biology. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin MF. Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat Rev Mol Cell Biol. 2008;9:759–769. doi: 10.1038/nrm2514. [DOI] [PubMed] [Google Scholar]
- Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21:187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nature reviews Molecular cell biology. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Dinant C, Lans H, Stap J, Wiernasz E, Lagerwerf S, Warmerdam DO, Lindh M, Brink MC, Dobrucki JW, et al. Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol. 2009;185:577–586. doi: 10.1083/jcb.200810035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, 3rd, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- Meier A, Fiegler H, Munoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, Jackson SP. Spreading of mammalian DNA-damage response factors studied by ChIP-chip at damaged telomeres. Embo J. 2007;26:2707–2718. doi: 10.1038/sj.emboj.7601719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillo-Huesca M, Clemente-Ruiz M, Andujar E, Prado F. The SWR1 histone replacement complex causes genetic instability and genome-wide transcription misregulation in the absence of H2A.Z. PloS one. 2010;5:e12143. doi: 10.1371/journal.pone.0012143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O. Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol. 2007;178:1101–1108. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Murr R, Vaissiere T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- Noon AT, Shibata A, Rief N, Lobrich M, Stewart GS, Jeggo PA, Goodarzi AA. 53BP1-dependent robust localized KAP-1 phosphorylation is essential for heterochromatic DNA double-strand break repair. Nat Cell Biol. 2010;12:177–184. doi: 10.1038/ncb2017. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. The Journal of biological chemistry. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- Pei H, Zhang L, Luo K, Qin Y, Chesi M, Fei F, Bergsagel PL, Wang L, You Z, Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr Opin Genet Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, An W, Routh A, Martino F, Chapman L, Roeder RG, Rhodes D. 30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction. J Mol Biol. 2008;381:816–825. doi: 10.1016/j.jmb.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S, Mello CC, Shimada A, Nakatani Y, Hashimoto S, Ogawa M, Matsushima K, Gu SG, Kasahara M, Ahsan B, et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic V, Yin B, Maas NL, Bredemeyer AL, Carpenter AC, Helmink BA, Yang-Iott KS, Sleckman BP, Bassing CH. Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol Cell. 2009;34:298–310. doi: 10.1016/j.molcel.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotta G, Sengupta R, Kubicek S, Malin S, Kauer M, Callen E, Celeste A, Pagani M, Opravil S, De La Rosa-Velazquez IA, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes & development. 2008;22:2048–2061. doi: 10.1101/gad.476008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smerdon MJ. DNA repair and the role of chromatin structure. Curr Opin Cell Biol. 1991;3:422–428. doi: 10.1016/0955-0674(91)90069-b. [DOI] [PubMed] [Google Scholar]
- Smerdon MJ, Tlsty TD, Lieberman MW. Distribution of ultraviolet-induced DNA repair synthesis in nuclease sensitive and resistant regions of human chromatin. Biochemistry. 1978;17:2377–2386. doi: 10.1021/bi00605a020. [DOI] [PubMed] [Google Scholar]
- Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G, Polo SE, Almouzni G. Prime, repair, restore: the active role of chromatin in the DNA damage response. Molecular cell. 2012;46:722–734. doi: 10.1016/j.molcel.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Dorn JF, Sengupta K, Jasin M, Nussenzweig A, Ried T, Danuser G, Misteli T. Positional stability of single double-strand breaks in mammalian cells. Nat Cell Biol. 2007;9:675–682. doi: 10.1038/ncb1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A. 2005;102:13182–13187. doi: 10.1073/pnas.0504211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Price BD. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle. 2010;9:930–936. doi: 10.4161/cc.9.5.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–1382. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A. Z. Nat Struct Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- Sykes SM, Mellert HS, Holbert MA, Li K, Marmorstein R, Lane WS, McMahon SB. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell. 2006;24:841–851. doi: 10.1016/j.molcel.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annual review of genetics. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Tolstorukov MY, Volfovsky N, Stephens RM, Park PJ. Impact of chromatin structure on sequence variability in the human genome. Nature structural & molecular biology. 2011;18:510–515. doi: 10.1038/nsmb.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H, Fritsch O, Gasser SM. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. Embo J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber CM, Henikoff JG, Henikoff S. H2A.Z nucleosomes enriched over active genes are homotypic. Nature structural & molecular biology. 2010;17:1500–1507. doi: 10.1038/nsmb.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z Controls a Critical Chromatin Remodeling Step Required for DNA Double-Strand Break Repair. Molecular cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–267. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J Cell Biol. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]