Abstract
The replacement histone variant H2AX senses DNA double-strand breaks (DSBs) and recruits characteristic sets of proteins at its phosphorylated (γ-H2AX) foci for concurrent DNA repair. We reasoned that the H2AX interaction network, or interactome formed in the tumor-associated DNA DSB environment such as in hepatocellular carcinoma (HCC) cells, where pre-neoplastic lesions frequently occur, is indicative of HCC pathogenic status. By using an in vivo dual-tagging quantitative proteomic method, we identified 102 H2AX-specific interacting partners in HCC cells that stably expressed FLAG-tagged H2AX at close to the endogenous level. Using bioinformatics tools for data-dependent network analysis, we further found binary relationships among these interactors in defined pathway modules, implicating H2AX in a multi-functional role of coordinating a variety of biological pathways involved in DNA damage recognition and DNA repair, apoptosis, nucleic acid metabolism, Ca2+-binding signaling, cell cycle, etc. Furthermore our observations suggest that these pathways interconnect through key pathway components or H2AX interactors. The physiological accuracy of our quantitative proteomic approach in determining H2AX-specific interactors was evaluated by both co-immunoprecipitation/ immunoblotting and confocal co-localization experiments performed on HCC cells. Due to their involvement in diverse functions, the H2AX interactors involved in different pathway modules, such as Poly(ADP-ribose) polymerase1, 14-3-3ζ, coflin1, and peflin1, were examined for their relative H2AX binding affinities in paired hepatocytes and HCC cells. Treatment with the DSB-inducing agent bleomycin enhanced binding of these proteins to H2AX, suggesting an active role of H2AX in coordinating the functional pathways of each protein in DNA damage recognition and repair.
Keywords: H2AX, DSBs, DNA repair, protein-protein interactions, in vivo dual-tagging quantitative proteomic method, hepatocellular carcinoma pathogenesis
Introduction
DNA double-strand breaks (DSBs) induced by various stresses1–5 represent a common genomic damage/lesion that may lead to genomic instability and ultimately, to cancer development2, 6. The replacement histone variant H2AX7 8 plays a central role in both cellular responses to DNA damage and in repairing damaged DNA2, 3. In the early cellular response to DNA DSBs, H2AX is phosphorylated at serine 139, probably by ataxia telangiectasia mutated (ATM), triggering various signal transduction cascades for DNA damage recognition and DNA repair9, 10. The phosphorylated form of H2AX or γ-H2AX can recruit particular sets of proteins such as MRE11, RAD50, and NBS1 (MRN) in a complex with Brca1, 53BP1, and NFBD1/DNA damage checkpoint 1(MDC1) to form γ-H2AX foci at DNA DSB sites11–13. For example, MDC1 recognizes γ-H2AX through its BRCT domain, and facilitates recruitment of additional MRN and ATM proteins that subsequently leads to the phosphorylation of additional H2AX and MDC1 molecules14, 15. The MRN complex can interact directly with MDC1, then MDC1 binds ATM through its FHA domain and NBS1 through its Ser-Asp-Thr repeat14, 15. Other evidence suggests that the ubiquitin-interacting-motif-containing protein RAP80 binds K63-linked ubiquitin chains of ubiquitinated proteins and, therefore, is able to recognize ubiquitinated H2A/H2AX, resulting in formation of a larger BRCA1/BARD1/CCDC98/RAP80 protein complex targeted to DNA damage foci14, 16, 17. In addition to these H2AX interacting proteins, many pivotal DNA repair-related proteins such as 53BP118 have also been found to interact with H2AX through different recruiting mechanisms.
H2AX and its interacting proteins play synergistic roles in tumourigenesis19, 20. H2AX knockout mice were found with increased genomic instability and a higher risk of developing cancers11, 21. Mutations or deletions in the H2AX gene are frequently found associated with various human cancers including acute myeloid leukemia, acute lymphoid leukemia, head and neck squamous carcinoma, etc8, 22–26. In tumor cells from clinical specimens and cell culture, emerging evidences also showed increased levels of DNA DSBs and mutations in H2AX-interacting partner genes27, 28. Defects in DNA damage response pathways, such as the MRN complex or the kinases ATM and RAD3-related (ATR), are associated with cancer predisposition syndromes in humans, and disruption of DNA repair systems in mouse models leads to an increased risk of cancer29, 30. H2AX and its interacting proteins such as ATM, Chk2, p53 were found activated in bladder cancer31. Immunohistochemistry studies have shown DNA damage signal activation in precancerous bladder lesions, which is lost on progression, suggesting that the damage signaling acts as a brake to further tumorigenesis32. In addition, expression of RAD51, one of the critical H2AX interactors, is higher in breast cancer than in normal breast tissue33. We therefore reasoned that different compositions of H2AX complexes may be specifically correlated to different human cancers, wherein the H2AX-associated pathways for DNA damage recognition and repair could be dys-regulated leading to un-repairable DNA DSBs.
Emerging mass spectrometry(MS)-based proteomic approaches with improved sensitivity, signal specificity, and throughput have been extended to efficiently and systematically analyze protein complexes and decipher protein interaction networks34–36. Metabolic labeling strategies such as SILAC/AACT26 have improved the signal specificity in distinguishing genuine protein-protein interactions in protein complexes37, 38. In our previous studies of ionizing radiation-induced dynamic changes in the H2AX interactome using a dual-tagging quantitative proteomic approach38, we found several novel binding partners of H2AX in human embryonic kidney (HEK) 293T cells under defined conditions, and demonstrated that the H2AX protein complex underwent dynamic changes upon induction of DNA damage and during DNA repair. Further analysis revealed a critical role for Ca2+/calmodulin in ionizing radiation-induced cell cycle arrest in HEK cells. Despite growing evidence of correlations between an abnormal H2AX interactome, and carcinogenesis, little is known how a characteristic H2AX interactome is structured in cancer cells and how it is dysregulated in respond to DNA DSBs and repair. Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related death worldwide39. This high mortality is mainly due to unknown mechanisms of HCC pathogenesis and lack of precise HCC-characteristic markers for early diagnosis and therapeutic intervention39, 40. One of the conspicuous features of HCC development is the frequent occurrence of chromosomal abnormalities39. Microarray analyses revealed that expression of many DNA repair-related proteins was up-regulated in HCC cells40. Also HCC tissues accumulate higher endogenous levels of γ-H2AX foci41. Some evidence suggests that the regulatory level of γ-H2AX may be tissue-specific42, 43. Also, variable expression patterns of an H2AX partner, NBS1, in testis, thymus, spleen telencephalon, diencephalon, liver, lung, kidney, and gut are observed in mice44. Organ-specific factors may have an impact on PARP-1, another H2AX interactor, to protect against genotoxic damage in mice45. We therefore hypothesize that H2AX may recruit particular sets of protein interactors in HCC cells whereby H2AX coordinates complex cellular signals indicative of the pathological environment in HCC.
We describe here the use of DNA nuclease digestion of nuclei and a dual tagging (both epitope and SILAC tag) quantitative proteomics strategy37, 38, to profile proteins interacting with H2AX in HCC cells. Using bioinformatics tools, we performed data-dependent network analysis to first validate protein interactions with H2AX and second to sort out their binary interactions. As an immediate outcome, multiple pathway modules were mapped and their interconnected links were established. The proteomic dataset and the selected components of particular pathway modules were further validated by co-immunoprecipitation/immunoblotting. The endogenous interactions between H2AX and selected interacting partners representing the pathways involved in DNA damage recognition and repair were localized by confocal scanning. Using bleomycin (BLM), the most used agent to induce DNA DSBs46, 47 we then examined the possible role of certain H2AX interactors in DNA damage recognition and repair in HCC cells. We also compared the H2AX binding affinities of the proteins in paired hepatocytes and HCC cells48. All of these results provide insight into an HCC-characteristic H2AX interactome indicative of the pathological status of hepatocellular carcinoma.
Experimental Procedures
Materials
Deuterium-labeled leucine (5, 5, 5-d3) was purchased from Cambridge Isotope (Andover, MA). Trypsin was purchased from Promega (Madison, WI). Hoechst 33342 was purchased from Sigma (St. Louis, MO). All components of cell culture medium were purchased from Invitrogen Corporation (Carlsbad, CA) except for puromycin, which was obtained from Calbiochem (San Diego, CA) and fetal bovine serum (FBS), which was obtained from PAA Laboratories GmbH (Linz, Austria). Chemicals for IP, SDS-PAGE gel electrophoresis, visualization, peptide extraction, and sample preparation of LTQ-Orbitrap were purchased from Sigma (St. Louis, MO). All the chemicals were sequence- or HPLC -grade unless specifically mentioned.
The antibodies used in immunoblotting and immunofluorescence studies were purchased from different companies, such as H2AX(ab11175, ab22551), γ-H2AX(ab2893), β-actin (ab16039) and GAPDH(ab8245) from Abcam; PARP-1 (sc-1561), 14-3-3ζ(sc-1019) from Santa Cruz Biotechnology; CALR(10292-A-AP), NONO(11058-1-AP), CFL1(10960-1-AP), PEF1(10151-1-AP) and NCL(10556-1-AP) from Protein Tech Group; anti-FLAG(F1860) from Sigma; FITC-conjugated goat anti-rabbit secondary antibody (75230) and TRITC-conjugated goat anti-mouse secondary antibody (61551) from Jackson ImmunoResearch Laboratories, Inc.
The cell line QSG-7701 and QGY-7703 were purchased from Shanghai Institute of Biochemistry and Cell Biology (SIBCB, Shanghai, China). QSG-7701 and QGY-7703 were originally established in QiDong, the highest morbidity area of HCC in China. The hepatoma cell line QGY-7703 was derived from primary carcinoma of a 35 years old female, and its normal liver counterpart QSG-7701 was from peripheral non-tumor tissue of the same surgery specimen.
Plasmids, Stable Cell Lines, and Dual-tagging (FLAG tag and isotope tag) Quantitative Proteomics Approach
Expression plasmids for H2AX were described previously37, 38. The construct containing the FLAG tag alone or FLAG-H2AX was transfected into human HCC cells (QGY-7703) using the Lipofectemin™ 2000 reagent according to the manufacturer, and the cells were selected in RPMI-1640 supplemented with 10% dialyzed FBS, 100U/ml penicillin and streptomycin, and 1.0 μg/ml puromycin. Whereas the cells containing the tag alone (control cells) were cultured in regular unlabeled RPMI-1640 medium, the cells containing FLAG-tagged H2AX (FLAG-H2AX cells) were maintained in RPMI-1640 containing Leu-d3 to isotopically label the proteome. A detailed procedure has been described previously37, 38.
Histone Isolation
Histone extraction was performed as described previously15. Briefly, cells were lysed in 0.5% NP40, 100 mM NaCl, 50 mM Tris, 1 mM EDTA. After centrifugation, histones were extracted by resuspending pellets with 0.2 N HCl. The HCl extracts were centrifuged, and the supernatants (histone extracts) were then neutralized with 1 N NaOH and blotted with anti-H2AX or anti-γ-H2AX antibodies.
Protein Extraction and Purification of H2AX Protein Interaction Partners
Protein extraction and purification were performed as described previously with minor modifications38. Briefly, approximately 8×108 FLAG-H2AX cells and control cells were harvested, then equal numbers of each cell line were mixed. The mixed cells were lysed. After nuclear isolation and nuclease digestion, the nuclear extract was incubated with 200 μl of M2 anti-FLAG beads at 4°C for 6 h with 20% glycerol and 150 mM NaCl. The beads were then washed four times with TBS. The bound protein complexes were then solubilized in sample buffer, and separated by 12% SDS-PAGE. After staining with Coomassie Brilliant Blue, visible bands were cut for in-gel digestion. The extracted peptides were lyophilized for LC-MS/MS.
LC-MS/MS Analysis
LC-MS/MS experiments were performed on a LTQ-Orbitrap hybrid mass spectrometer (Thermo Finnigan, Bremen, Germany) equipped with a Finnigan Dynamic nano-spray source. Sample was injected via an SIL-20 AC auto-sampler (Shimadzu Corporation, Tokyo, Japan) and loaded onto a CAPTRAP column (0.5×2 mm, Michrom Bioresources Inc., Auburn, CA) for 5 min at a flow rate of 60μL/min. The peptide mixtures was subsequently separated by a PICOFRIT C18 reverse-phase column (0.075×100 mm, New Objective Inc., Woburn, MA) at a flow rate of 300nL/min with a 110 min-gradient. Buffer A was 5% ACN, 0.1% formic acid; buffer B was 95% ACN, 0.1% formic acid. Samples were loaded in solvent A, and peptides were eluted by 5% solvent B for 5 min followed by a linear gradient to 45% solvent B in the next 90 min, ramping to 95% solvent B in 4 min and dropping to 90% solvent B for 4 min before re-equilibrating the system to 10% solvent B for 7 min. The LTQ-Orbitrap mass spectrometry was operated in the data-dependent mode using the TOP3 strategy. In brief, a scan cycle was initiated with a full scan of mass range (m/z 400–2000) in the Orbitrap under the target mass resolution of 60,000, which was followed by MS/MS scans in the linear ion trap on the 3 most abundant precursor ions. Single charged ions were excluded from MS/MS analysis.
Database Search and Protein Identification
Applying the TurboSequest V.27 (rev.12) search engine, we searched all MS/MS spectra against human reference database (IPI 3.27). Searches were performed with the following parameters: Trypsin enzyme specificity with allowing one missed cleavage; a precursor tolerance of 10 ppm and fragment tolerance of 1 Da; dynamic modifications of d3-labeled Leu (+3.018). Positive identifications were made according to a rigorous statistical model as Peptide Prophet and reverse database search49, 50. In detail, for SEQUEST engine, all peptides must have a ΔCn of at least 0.1 and cross correlation values (XCorr) were assigned with at least 1.90 (+1 charge), 3.23 (+2 charge) and 3.66(+3 charge) by reverse database search evaluation (p=0.05). Additionally, the computed probability through Peptide Prophet of trans-proteomic pipeline (TPP) tools must be over 0.99 for each peptide51. Protein assignments were made only if the protein had at least two different peptides passing the above criteria. In order to eliminate redundant protein identifications that matched the same set of peptides (protein group), a parsimonious interpretation method was applied to result in a minimized dataset. Reproducibility was assessed primarily according to the method of Blagoev et al52.
Data-dependent bioinformatics for network analysis
Functional classification of identified proteins was accomplished by using Entrez Gene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db_gene) and DAVID (http:/www.david.abcc.ncifcrf.gov). Proteins with multiple functions were assigned to those that are best known. Pathway module analysis of the functional clusters was performed by STRING (http://www.string.embl.de), a database of known and predicted protein-protein interactions.
Immunoblotting Analysis
Cells (7×107 cells/assay) were harvested for nuclei isolation/digestion, immunoprecipitation, and immunoblotting. The procedures for nuclei isolation/digestion and immunoprecipitation were essentially the same as described as above, except that buffer volumes in each step were reduced ~10-fold. The immunoprecipitated proteins were eluted by boiling the beads and separated on SDS-PAGE. The proteins were then transferred to a PVDF membrane (Bio-Rad) and incubated with the specified primary antibody followed by incubation with a secondary antibody conjugated to horseradish peroxidase. The ECL substrate was then added and the blot was developed.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 15 min, washed in PBS, and blocked in 10% normal goat serum with 0.1% Triton X-100 for 1 h at room temperature. The fixed cells were incubated with anti-γ-H2AX (phosphorylated form) antibody overnight at 4°C, followed by incubation with TRITC-conjugated goat anti-mouse secondary antibody for 1 h at room temperature. Cells were washed in PBS. Slides were incubated sequentially with other primary antibodies overnight at 4°C and FITC-conjugated goat anti-rabbit secondary antibody for 1 h at room temperature. After washing, cells were counterstained with Hoechst 33342. Then slides were mounted in glycerol/PBS solution. All primary antibodies were used at dilutions of 1:200 to 1:800. The secondary antibodies included anti-rabbit FITC and anti- mouse TRITC and were used at dilutions of 1:200. Immunofluorescence images were captured using a laser scanning confocal microscope (Zeiss, LSM 510 system or Leica TCS-SP15, Germany).
Bleomycin Stimulation
HCC cells were seeded in 10 cm dishes. When they were 90–95% confluent, the cells were treated with BLM. γ- H2AX was detected with different concentrations of BLM and different times of treatment. The cells treated with optimized conditions (40mU/ml BLM and harvested after 3 hours) were selected for immunoblotting analysis.
Binding Comparative Assay
A stable cell line of hepatocytes (QSG-7701) was produced as described above. Hepatocytes and HCC cells were harvested for nuclei isolation digestion, immunoprecipitation, and immunoblotting analysis.
Results and Discussion
In vivo expression and subcellular location of epitope-tagged H2AX in hepatocellular carcinoma (HCC) cells
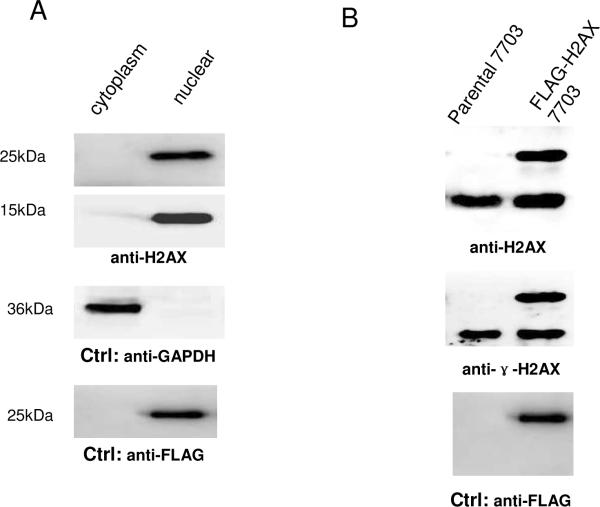
To identify H2AX-interacting proteins from HCC cells, we generated a HCC cell line stably expressing human FLAG tagged-H2AX at close to the natural level, using the retroviral gene transfer method which we reported previously37, 38. In the stable cell line, we first examined the subcellular location of the tagged-H2AX by using a sequential histone extraction method15. Each fraction of the extraction was examined by immunoblotting against H2AX antibody. As shown in Fig.1A, FLAG-tagged H2AX was detected only in the nuclear/histone components, suggesting that the FLAG-tagged H2AX was incorporated into chromatin similar to our previous observations38. Also expression of FLAG-tagged H2AX was found at a level similar to that of endogenous H2AX (Fig.1B, left). The phosphorylated form of FLAG-tagged-H2AX, γ-H2AX, was also detected at an abundance slightly higher than that of its endogenous counterpart (Fig. 1B, middle). Similarly, anti-FLAG was used to detect ectopically expressed ɣ-H2AX and H2AX (Fig. 1B, right).
Figure 1. Expression of FLAG-tagged H2AX in stable QGY-7703 cells.
(A). FLAG-tagged H2AX was detected only in the nuclear fraction, not in the cytoplasmic fraction. Equal amounts of protein (40μg) extracted from cytoplasm and nuclear fraction of cells stably expressing FLAG-tagged H2AX (FLAG-H2AX cells) were loaded onto a 12% SDS-polyacrylamide gel for Western blotting (WB) analysis. GAPDH and endogenous H2AX were detected and used as the control markers of the cytoplasmic and nuclear fractions, respectively. (B). FLAG-tagged H2AX and its phosphorylated form were expressed at a level comparable to that of its endogenous counterpart. Equal amounts of protein (15μg) extracted from the parental 7703 cells and the FLAG-H2AX cells were loaded onto a 15% SDS-polyacrylamide gel for Western blotting (WB) analysis using either anti-H2AX or anti-phospho-H2AX or anti-FLAG antibody. The bands appearing close to 15 or 25 kDa corresponded to the endogenous or FLAG-tagged H2AX, respectively.
Given the fact that H2AX senses DNA DSBs through site-specific phosphorylation53, we expected to observe a portion of γ-H2AX in HCC cells where many un-repairable DSBs may exist. In fact, after several passages the expression level of the epitope-tagged H2AX became even lower than the untagged endogenous form (Table S1). Furthermore, the stable HCC cells expressing the FLAG-tagged H2AX showed a growth rate and morphology similar to the parental cells, indicating that expression of the FLAG-tagged H2AX had no effect on the phenotype of the stable cells. Collectively the above results suggested that the FLAG-tagged H2AX, like its endogenous counterpart, was correctly packaged into nucleosomes and functioned in a physiologically relevant condition in chromatin.
Identification of H2AX-interacting components formed in HCC cells using a dual-tagging (FLAG tag and isotope tag) quantitative proteomics approach
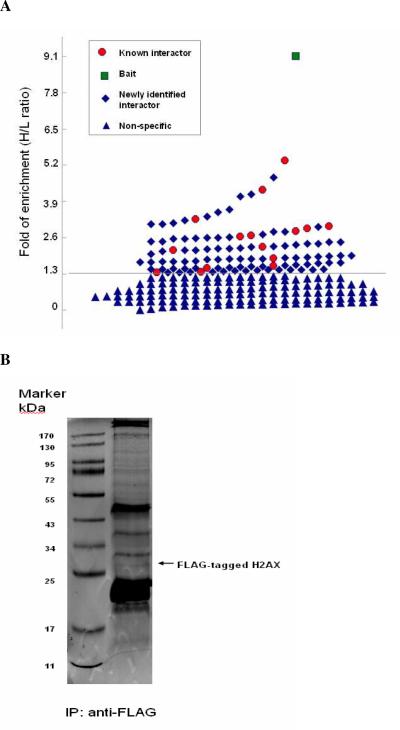
The strategy for affinity purification and identification of H2AX-interacting proteins is, with a few modifications, similar to that described previously38. Briefly, the HCC cell line stably expressing FLAG-H2AX was maintained in a “heavy” medium containing leucine-d3, whereas the control cells were cultured in the regular (or “light”) medium. Equal amounts of cells from each culture were combined. After nuclei isolation/digestion, extraction and IP, the immunoprecipitated proteins were eluted and separated on SDS-PAGE (Fig 2B). All gel bands were digested with trypsin followed by LC-MS/MS for peptide separation and protein identification. In our MS analysis, all of the leucine-containing peptides appeared as pairs in the MS spectrum with one set of peaks from the stable isotope-labeled cells and the other set of peaks from the unlabeled cells. The intensity ratios of the paired peaks reflect the binding profiles of the protein to the bait protein H2AX. For those H2AX-interacting proteins, the heavy isotope signals should be more intense than their lighter counterparts, suggesting enrichment of these proteins around the H2AX bait. Because background binding occurs randomly in both IP purifications, the ratio of the “heavy” (H) versus “light” (L) peptide intensities (H/L) for bait-nonspecific binding proteins should be 1:1.
Figure 2. AACT/SILAC-based MS analysis of H2AX complexes.
(A) The distribution of H/L ratio of the identified proteins in the H2AX complex formed in HCC cells. The threshold for distinguishing specific interactors from non-specific contaminants was an H/L of 1.34. The sign of diamond, circle and square indicate those proteins with H/L over 1.34, suggesting specificity in associating with H2AX, while a triangle represents a protein with H/L less than 1.34 or a non-specific protein in the complex. The green square represents the bait protein. The red circles indicate proteins known previously to interact with H2AX in other types of cells. The blue diamonds suggest newly identified H2AX-interacting partners. (B) SDS-PAGE analysis of FLAG-immunoprecipitates as visualized by Coomassie Brilliant staining. The band of FLAG-tagged H2AX is indicated by arrow.
The procedure for protein identification and quantitative measurements was carried out similarly to that described previously37, 54. As shown in Figure 2A, for the distribution of H/L for all proteins identified, the proteins previously known to interact with H2AX such as Calreticulin (CALR) were found with H/L ratios > 1.34. Empirically, we selected this value as the threshold for distinguishing specific interactors from non-specific contaminants as all proteins with H/L less than 1.34 were considered as non-specific background. We then searched the available information about the known functional category and the possible H2AX-interacting nature of our identified interactors by using public interaction databases such as oPHID, IntAct, BioGRID, and HPRD. As summarized in Table 1, among 102 proteins distinguished as the H2AX interactors 15 of them are known H2AX interacting partners.
Table 1.
Identified H2AX-interacting Proteins in HCC QGY7703 Cells.
| IPI no. | Gene symbol | Protein namea | No.of peptide-matched | Enrichment-foldb | S.D. |
|---|---|---|---|---|---|
| IPI00219037.5 | H2AX | Histone H2AX (bait) | 4 | 9.11 | |
| Apoptosis | |||||
| IPI00012011.6 | CFL1 | Cofilin-1 | 2 | 1.90 | 0.014 |
| IPI00291262.3 | CLU | Clusterin precursor | 2 | 2.26 | 0.56 |
| IPI00027230.3 | HSP90B1 | Endoplasmin precursor | 8 | 1.60 | 0.07 |
| IPI00304925.4 | HSPA1A | Heat shock 70 kDa protein 1 | 3 | 1.50 | 0.16 |
| IPI00555692.2 | ANXA4 | ANXA4 protein | 2 | 1.49 | 0.72 |
| IPI00329801.12 | ANXA5 | Annexin A5 | 6 | 1.36 | 0.11 |
| IPI00025277.5 | PDCD6 | Programmed cell death protein 6 | 3 | 1.745 | 0.19 |
| IPI00171438.2 | TXNDC | Thioredoxin domain-containing protein 5 precursor | 2 | 1.62 | 0.08 |
| IPI00291467.7 | SLC25A6 | ADP/ATP translocase 3 | 2 | 1.72 | 0.057 |
| Response to protein stimulus, oxidative stress and hypoxia | |||||
| IPI00023137.1 | TOR1B | Torsin B precursor | 2 | 2.71 | 0.70 |
| IPI00382470.3 | HSP90AA1 | Heat shock protein HSP 90-alpha 2 | 3 | 2.57 | 0.19 |
| IPI00301277.1 | HSPA1L | Heat shock 70 kDa protein 1L | 3 | 1.45 | 0.13 |
| IPI00030275.5 | TRAP1 | Heat shock protein 75 kDa mitochondrial precursor | 9 | 2.45 | 0.24 |
| IPI00220301.5 | PRDX6 | Peroxiredoxin-6 | 3 | 1.73 | 0.38 |
| IPI00027192.5 | PLOD1 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase precursor | 2 | 2.46 | 0.03 |
| IPI00022314.1 | SOD2 | Superoxide dismutase90, | 2 | 2.56 | 0.22 |
| DNA repair (14) | |||||
| IPI00000690.1 | AIFM1 | Isoform 1 of Apoptosis-inducing factor 1, mitochondrial precursor | 6 | 2.93 | 0.37 |
| IPI00304596.3 | NONO | Non-POU domain-containing octamer-binding protein | 2 | 1.93 | 0.02 |
| IPI00449049.5 | PARP1 | Poly [ADP-ribose] polymerase 1 | 2 | 1.81 | 0.07 |
| IPI00020599.1 | CALR | Calreticulin precursor | 7 | 1.34 | 0.20 |
| IPI00021263.3 | YWHAZ | 14-3-3 protein zeta/delta | 3 | 1.37 | 0.11 |
| IPI00000816.1 | YWHAE | 14-3-3 protein epsilon | 3 | 1.71 | 0.10 |
| IPI00413293.5 | TOR1A | Torsin A precursor | 2 | 1.81 | 0.56 |
| IPI00000877.1 | HYOU1 | 150 kDa oxygen-regulated protein precursor | 4 | 2.68 | 0.28 |
| IPI00029744.1 | SSBP1 | Single-stranded DNA-binding protein, mitochondrial precursor | 2 | 1.74 | 1.12 |
| IPI00169383.3 | PGK1 | Phosphoglycerate kinase 1 | 4 | 2.58 | 0.61 |
| IPI00219365.3 | MSN | Moesin | 2 | 1.96 | 0.76 |
| IPI00025491.1 | EIF4A1 | Eukaryotic initiation factor 4A-I | 2 | 1.41 | 0.04 |
| IPI00008438.1 | RPS10 | 40S ribosomal protein S10 | 2 | 2.14 | 0.03 |
| IPI00011062.1 | CPS1 | Isoform 1 of Carbamoyl-phosphate synthase [ammonia], mitochondrial precursor | 4 | 1.44 | 0.05 |
| DNA metabolism | |||||
| IPI00005198.2 | ILF2 | Interleukin enhancer-binding factor 2 | 3 | 1.71 | 0.12 |
| IPI00453473.6 | HIST2H4A | Histone H4 | 5 | 2.27 | 0.44 |
| Ca2+ binding signaling (6) | |||||
| IPI00418169.3 | ANXA2 | annexin A2 isoform 1 | 3 | 1.67 | 0.27 |
| IPI00334627.3 | ANXA2P2 | Similar to annexin A2 isoform 1 | 6 | 1.84 | 0.20 |
| IPI00075248.11 | CALM2 | Calmodulin | 6 | 5.35 | 0.97 |
| IPI00032313.1 | S100A4 | Protein S100-A4 | 2 | 1.87 | 0.01 |
| IPI00018235.3 | PEF1 | Peflin | 2 | 3.08 | 0.54 |
| IPI00002459.4 | ANXA6 | annexin VI isoform 2 | 8 | 2.19 | 0.22 |
| Cell cycle | |||||
| IPI00216318.5 | YWHAB | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | 2 | 1.48 | 0.04 |
| IPI00017297.1 | MATR3 | Matrin-3 | 2 | 2.44 | 0.02 |
| Alcohol metabolic process | |||||
| IPI00465439.5 | ALDOA | Fructose-bisphosphate aldolase A | 2 | 2.015 | 0.049 |
| IPI00220644.8 | PKM2 | pyruvate kinase 3 isoform 2 | 6 | 2.45 | 0.27 |
| IPI00479186.5 | PKM2 | pyruvate kinase 3 isoform 1 | 2 | 2.34 | 0.17 |
| IPI00465248.5 | ENO1 | enolase 1 | 2 | 1.945 | 0.16 |
| IPI00058192.1 | POFUT1 | Isoform 1 of GDP-fucose protein O-fucosyltransferase 1 precursor | 3 | 1.86 | 0.05 |
| IPI00219217.3 | LDHB | L-lactate dehydrogenase B chain | 2 | 2.15 | 0.19 |
| RNA metabolism | |||||
| IPI00301936.3 | ELAVL1 | ELAV-like protein 1 | 3 | 1.47 | 0.22 |
| IPI00028888.1 | HNRPD | Isoform 1 of Heterogeneous nuclear ribonucleoprotein D0 | 3 | 1.39 | 0.49 |
| IPI00011274.2 | HNRPDL | heterogeneous nuclear ribonucleo protein D-like | 2 | 1.84 | 0.10 |
| IPI00013877.2 | HNRPH3 | Isoform 1 of Heterogeneous nuclear ribonucleoprotein H3 | 2 | 3.12 | 0.36 |
| IPI00012074.3 | HNRPR | Heterogeneous nuclear ribonucleoprotein R | 5 | 3.26 | 0.31 |
| IPI00106509.2 | HNRPAB | Isoform 4 of Heterogeneous nuclear ribonucleoprotein A/B | 2 | 4.57 | 0.13 |
| IPI00012066.2 | PCBP2 | poly(rC)-binding protein 2 isoform b | 2 | 1.86 | 0.77 |
| IPI00008524.1 | PABPC1 | Isoform 1 of Polyadenylate-binding protein 1 | 2 | 4.18 | 0.54 |
| IPI00221035.3 | BTF3 | Isoform 1 of Transcription factor BTF3 | 2 | 2.63 | 0.05 |
| IPI00073713.3 | MSI2 | Isoform 1 of RNA-binding protein Musashi homolog 2 | 2 | 2.81 | 0.65 |
| IPI00328840.9 | THOC4 | THO complex subunit 4 | 3 | 4.76 | 0.70 |
| IPI00018140.3 | SYNCRIP | Isoform 1 of Heterogeneous nuclear ribonucleoprotein Q | 9 | 2.62 | 0.35 |
| IPI00215965.2 | HNRPA1 | heterogeneous nuclear ribonucleoprotein A1 isoform b | 2 | 1.42 | 0.20 |
| IPI00386854.5 | HNRPA2B1 | HNRPA2B1 protein | 4 | 2.59 | 0.17 |
| IPI00216592.2 | HNRPC | Isoform C1 of Heterogeneous nuclear ribonucleoproteinsC1/C2 | 3 | 1.84 | 0.04 |
| Establishment of cellular localization | |||||
| IPI00018931.6 | VPS35 | Vacuolar protein sorting-associated protein35 | 3 | 1.845 | 0.15 |
| IPI00024911.1 | ERP29 | Endoplasmic reticulum protein ERp29 precursor | 5 | 1.49 | 0.16 |
| IPI00156689.3 | VAT1 | Synaptic vesicle membrane protein VAT-1 homolog | 3 | 2.27 | 0.45 |
| IPI00021405.3 | LMNA | Isoform A of Lamin-A/C | 3 | 1.53 | 0.06 |
| IPI00045839.3 | LEPRE1 | Isoform 3 of Prolyl 3-hydroxylase 1 precursor | 3 | 1.35 | 0.15 |
| IPI00009904.1 | PDIA4 | Protein disulfide-isomerase A4 precursor | 11 | 1.58 | 0.08 |
| IPI00020436.4 | RAB11B | Ras-related protein Rab-11B | 3 | 1.47 | 0.60 |
| IPI00023748.3 | NACA | Nascent polypeptide-associated complex subunit alpha | 2 | 1.50 | 0.63 |
| Cellular macromolecule metabolism | |||||
| IPI00021728.3 | EIF2S2 | Eukaryotic translation initiation factor 2 subunit 2 | 2 | 1.36 | 0.13 |
| IPI00178440.3 | EEF1B2 | Elongation factor 1-beta | 2 | 1.52 | 0.58 |
| IPI00299571.5 | PDIA6 | Isoform 2 of Protein disulfide-isomerase A6 precursor | 9 | 1.60 | 0.05 |
| IPI00010720.1 | CCT5 | T-complex protein 1 subunit epsilon | 2 | 1.57 | 012 |
| IPI00011229.1 | CTSD | Cathepsin D precursor | 3 | 3.61 | 0.39 |
| IPI00386755.2 | ERO1L | ERO1-like protein alpha precursor | 2 | 1.56 | 0.11 |
| IPI00010796.1 | P4HB | Protein disulfide-isomerase precursor | 6 | 1.47 | 0.42 |
| IPI00011937.1 | PRDX4 | Peroxiredoxin-4 | 3 | 2.19 | 0.27 |
| IPI00419585.9 | PPIA | Peptidyl-prolyl cistrans isomerase A | 4 | 1.44 | 0.06 |
| IPI00028004.2 | PSMB3 | Proteasome subunit beta type 3 | 2 | 2.26 | 0.74 |
| IPI00008529.1 | RPLP2 | 60S acidic ribosomal protein P2 | 2 | 1.40 | 0.01 |
| IPI00555956.2 | PSMB4 | Proteasome subunit beta type 4 precursor | 2 | 1.76 | 0.43 |
| IPI00303300.3 | FKBP10 | FK506-binding protein 10 precursor | 3 | 2.39 | 0.96 |
| IPI00171412.1 | SUMF2 | Isoform 1 of Sulfatase-modifying factor 2 precursor | 2 | 2.38 | 0.02 |
| IPI00003128.1 | P4HA2 | Isoform IIb of Prolyl 4-hydroxylase subunit alpha-2 precursor | 3 | 2.3 | 0.14 |
| IPI00028635.4 | RPN2 | Dolichyl-diphosphooligosaccharide—protein glycosyltransferase 63 kDa subunit precursor | 7 | 2.20 | 0.02 |
| Cell redox homeostasis | |||||
| IPI00026328.3 | TXNDC12 | Thioredoxin domain-containing protein 12 precursor | 3 | 1.35 | 0.04 |
| IPI00303568.3 | PTGES2 | Prostaglandin E synthase 2 | 6 | 1.71 | 0.34 |
| Carboxylic acid metabolism | |||||
| IPI00017726.1 | HSD17B10 | hydroxysteroid (17-beta) dehydrogenase 10isoform 1 | 5 | 2.15 | 0.35 |
| IPI00643920.2 | TKT | Transketolase | 3 | 3.52 | 0.31 |
| Carbohydrate catabolism | |||||
| IPI00011454.1 | GANAB | Isoform 2 of Neutral alpha-glucosidase AB precursor | 3 | 1.44 | 0.42 |
| IPI00441344.1 | GLB1 | Beta-galactosidase precursor | 2 | 4.08 | 0.38 |
| IPI00383046.3 | CMBL | carboxymethylenebutenolidase-like | 7 | 4.15 | 0.29 |
| IPI00026154.2 | PRKCSH | Glucosidase 2 subunit beta precursor | 4 | 2.21 | 1.00 |
| IPI00023673.1 | LGALS3BP | Galectin-3-binding protein Precursor | 3 | 3.22 | 0.29 |
| IPI00027851.1 | HEXA | Beta-hexosaminidase alpha chain Precursor | 2 | 3.01 | 0.16 |
| Others | |||||
| IPI00020075.4 | ABHD10 | CDNA FLJ11342 fis, clone PLACE1010800 | 3 | 2.10 | 0.05 |
| IPI00168479.2 | APOA1BP | apolipoprotein A-I binding protein precursor | 3 | 3.36 | 0.22 |
| IPI00550363.3 | CCDC19 | Transgelin-2 | 3 | 1.44 | 0.67 |
| IPI00329696.1 | FAM82B | Protein FAM82B | 3 | 3.08 | 0.08 |
| IPI00007765.5 | HSPA9 | Stress-70 protein, mitochondrial precursor | 9 | 1.63 | 0.74 |
| IPI00024919.3 | PRDX3 | Thioredoxin-dependent peroxide reductase, mitochondrial precursor | 3 | 2.81 | 0.34 |
| IPI00377161.7 | HIBCH | 3-hydroxyisobutyryl-Coenzyme A hydrolase isoform 2 | 2 | 4.32 | 0.63 |
Only the proteins with two or more peptides matched are listed.
Fold of enrichment was calculated as the ratio of Leu-d3-labeled peptide to unlabeled peptide.
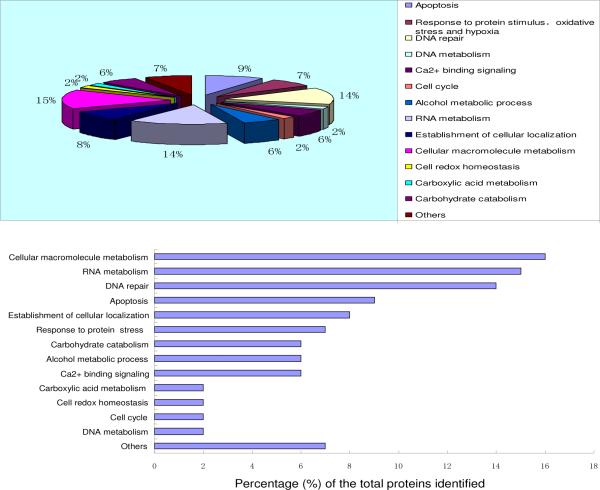
Functional categories of the identified H2AX-interacting proteins in HCC cells
By searching Entrez Gene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db_gene) and DAVID (http://www.david.abcc.ncifcrf.gov), we found that most of the H2AX interacting proteins identified by our proteomic method were clustered in diverse functional categories. As shown in Table 1 and Fig. 3, these functional categories include DNA damage recognition/repair, cell cycle, apoptosis, cellular localization, response to stimulus/oxidative stress/hypoxia, alcohol metabolic processes, DNA and RNA metabolism, cellular macromolecule metabolism, redox homeostasis, Ca2+-binding signaling, etc.
Figure 3. The distribution of HCC cell H2AX interacting proteins in different functional categories.
The identified proteins were analyzed by bioinformatics tools using Entrez Gene and KEGG databases. A total of 102 proteins identified as H2AX-interacting partners were found to be associated with fourteen functional categories or biological processes. Proteins with multiple functions were assigned to the best known function.
The distribution of the identified H2AX interactors in these functional categories is given in Figure 3. Using a similar dual-tagging strategy, those proteins including Calmodulin, CALR, PARP-1, histone H4, etc, were also found to interact with H2AX in the human embryonic kidney (HEK) 293T cells38. The identification of these H2AX-interacting partners in both HEK and hepacellular carcinoma cell types suggested there are core components in the H2AX complex regardless of the difference in cell types. Because of their functional relevance to Ca2+ signaling-related cell cycle, apoptosis, and DNA damage recognition and repair, etc, the core role of H2AX-interacting complexes in coordinating these biological processes was clearly suggested. In addition we have identified many more H2AX-intercating partners in HCC cells in which some may be HCC-specific and others may be due to the higher sensitivity of LTQ-orbitrap compared to the ABI Qstar/Qtof used previously. The largest portion of the H2AX interactome in HCC cells was represented by proteins involved in macromolecule metabolism including both nucleic acid and protein metabolism, and carbohydrate catabolism. This observation suggested that H2AX may recruit these proteins for readily activating multiple functional pathways such as energy generation, protein synthesis, RNA splicing and transport, translational control, gene regulation, etc24, 55–57. Interestingly, in our dataset (Table 1) many H2AX interactors such as CFL1, HSP90B1 and ANXA5 possess anti-apoptotic functions58 59. A possible explanation is that H2AX may recruit these proteins in a complex to protect the damaged or un-repairable cells from apoptosis in hepacellular carcinoma. In addition, one group of interactors has known functions in alcohol metabolic processes and oxidative stress. Alcohol has long been recognized as a major risk factor for HCC development60, 61 62, and acetaldehyde, the main metabolite of alcohol, was reported to cause hepatocellular injury and is an important factor in causing increased oxidative stress, which damages DNA62. Examples of our identified H2AX interactors in this category are enolase 1 (ENO1), Fructose-bisphosphate aldolase A (ALDOA), and Isoform 1 of GDP-fucose protein O-fucosyltransferase 1 precursor (POFUT1), which may take part in alcohol metabolism in liver physiology and pathology.
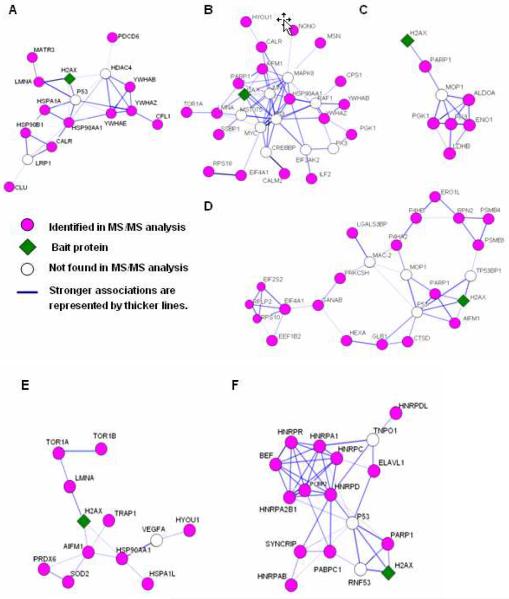
Data-dependent network analysis of the H2AX-interacting proteins identified in HCC cells reveals that H2AX coordinates cross-talk among diverse pathway modules
To elucidate how H2AX coordinates multiple functional pathways in the tumor environment and the possible functional role of the H2AX interactome in HCC, we explored the inter-connected relationship among the H2AX interactors and further identified each of different pathway modules. By using the STRING mapping tool, the sub-networks operating each pathway module could be mapped and specifically defined with the associated function. As shown in Figure 4, H2AX was found to be the critical network node in a variety of the identified pathway modules associated with apoptosis and cell cycle control (Fig. 4A), DNA repair (Fig. 4B), alcohol metabolism (Fig. 4C), carbohydrate catabolism and cellular macromolecule metabolism (Fig. 4D), stress response (Fig. 4E), and RNA processing (Fig. 4F). Further, given their previously known multi-functional nature, certain H2AX interactors, for instance, YWHAB and YWHAE (also named as 14-3-3 isomers), were found in multiple pathway modules related to apoptosis, cell cycle, and DNA repair (Fig. 4A&4B). EIF4A1 and RPS10 were shown in all pathway modules associating with carbohydrate catabolism, macromolecule metabolism, and DNA repair (Fig. 4B–4D). TOR1A, LMNA, and HYOU1 were shared by both pathway modules related to stress response and DNA repair (Fig.4B&4E). Notably, we have detected some vital proteins such as PARP-1, HSP90AA1, etc, previously known as components of multiple signaling pathways related to DNA damage recognition and repair. Although our network analysis suggested that p53, a key tumor suppressor protein,63, 64 could be a critical node in a number of pathway modules (Fig.4), it was not identified in our mass spectrometry experiments. In response to DNA damage, p53 accumulates in differentiated cells, and activates particular target genes that initiate cell cycle arrest or apoptosis63. The p53-regulated tumor suppressor pathway is frequently inactivated in cancer65. A possible explanation of our results was that most of the pathway modules identified in our networks analysis could be more p53-independent than p53-dependent in the HCC cells, or both mechanisms may co-exist. PARP-1 is multi-functional in many pathways including those related to DNA damage/repair3, 66, 67. HSP90AA1 is a molecular chaperone involved in both formation and maintenance of proper conformation of proteins and promoting cell survival in response to stress26, 68.
Figure 4. The H2AX-associating pathway modules found in the HCC-specific H2AX interactome.
The regulatory/signaling pathway modules of identified proteins were analyzed by DAVID and STRING 8.1 (http://www.string.embl.de/). (A) apoptosis and cell cycle, (B) DNA repair, (C) alcohol metabolism, (D) carbohydrate catabolism and cellular macromolecule metabolism, (E) stress response, (F) RNA processing and protein synthesis.
This data-dependent network analysis not only provided further support for the accuracy of our determination of the H2AX-interacting partners but also the binary relationships among these interactors, as most of them could be identified in the corresponding pathway module(s). More importantly, H2AX was shown to promote cross-talk in these interconnected pathway modules. Clearly, these pathway modules coordinated by H2AX may sense intrinsic stress such as un-repaired DNA DSBs in HCC cells, transduce the signal in recognizing DNA damage, and selectively activate biological processes such as apoptosis, cell cycle arrest, or DNA repair.
Validation of the HCC-specific H2AX interactome dataset using immunoblotting
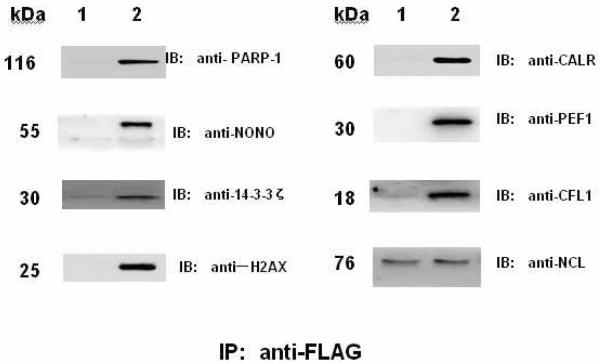
We used co-immunoprecipitation (co-IP) and concurrent immunoblotting to evaluate the physiologically relevant accuracy of our proteomics dataset of the HCC cell H2AX-interacting complex. In our selected pool of H2AX interactors, proteins such as PARP-1, CALR, non-pou domain-containing octamer-binding protein (NONO), 14-3-3ζ, CFL1, and PEF1 were known to be involved in individual or multiple pathway modules associated with DNA damage recognition/repair, apoptosis, and cell cycle control respectively and could represent each of individual pathway modules. The known H2AX interactors such as PARP-1, CALR and NONO were found to interact with H2AX in HCC cells while other proteins previously unknown to interact with H2AX such as 14-3-3ζ, CFL1 and PEF1 were also identified as H2AX-inetracting partners in HCC cells.
Poly(ADP-ribose) polymerases, or PARPs, which showed 1.8 fold enrichment in the H2AX complex (Table 1) are a class of cell signaling enzymes functioning in poly(ADP-ribosylation) of DNA-binding proteins69. PARP-1 is best known to be involved in the cellular response to DNA damage69, 70 in addition to its associations with various pathways including DNA replication, DNA repair, recombination, gene transcription, cell proliferation and death71. PARP-1 participates in DNA DSB repair through either nonhomologous end-joining (NHEJ) or homologous recombination (HR)3, and is believed to be a `sensor' in detecting DNA damage and initiating DNA repair72. Earlier reports suggested the increased level of PARP-1 in cancer cells might promote their survival72, 73. PARP-1 inhibitors have been used in cancer chemotherapy in combination with anti-tumor drugs having DNA-binding properties74, 75. A previous study also indicated that PARP-1 expression was significantly increased in human HCC compared to its surrounding liver tissue76, 77. Also PARP-1 was found to contribute to HBV-related hepatocarcinogenesis78. Our study here has provided direct evidence suggesting that the role of PARP-1 in HCC might be due to its interaction with H2AX in the HCC cells.
14-3-3ζ is one of the isoforms in the 14-3-3 family of regulators of diverse cellular responses in both physiological and pathological conditions79. 14-3-3ζ is an important regulatory protein in intracellular signaling pathways and is known to interact with more than 100 cellular proteins, including many oncogene and proto-oncogene products80. 14-3-3ζ blocks apoptosis by inhibiting activation of p38 mitogen-activated protein kinase (MAPK) and could be an anti-apoptotic factor in cells81. 14-3-3ζ was also reported to interact with β-catenin, and to enhance or to inhibit β-catenin-dependent transcription. It also facilitates activation of β-catenin through Akt, and is possibly involved in stem cell development82. Furthermore, 14-3-3ζ is up-regulated in a number of cancer cell types83, 84. Evidence indicates that 14-3-3ζ plays a role in keratin filament organization in hepatocytes, and regulates mitotic progression66, 85. In our analysis (Table 1), 14-3-3ζ was identified as an intrinsic interactor of H2AX in HCC cells.
Cofilin-1 (CFL1) is a small ubiquitous protein that binds both monomeric and filamentous actin, and, through its ability to sever actin filaments, is an essential regulator of actin dynamics at the plasma membrane during cell migration86. Aberrant regulation of cell migration drives the progression of many diseases, including cancer invasion and metastasis87. CFL1 may be involved in cancer cell migration, invasion, and metastasis88–90. In the highly metastatic human HCC cells such as MHCC97-H91, a decreased level of CFL1 was found. Furthermore, some studies suggest CFL1 may relate to radio-sensitivity by altering DNA repair capacity87 since key components for repair of DNA DSBs, including RAD51, RAD52, and Ku70/Ku80, were down-regulated in CFL1 over-expressing cells following ionizing radiation87. Interestingly, CFL1 was identified with 1.9 fold increase in its binding to H2AX in HCC cells.
One of the penta-EF-hand (PEF) proteins, PEF1, is a Ca2+-binding protein92 functioning in Ca2+-mediated signaling irrespective of cell type93. PEF1 dimerizes with apoptosis-linked gene 2 (ALG-2) and modulates the function of ALG-2 in Ca2+ signaling92. Our previous study in HEK 293 cells showed that the H2AX interactome plays a important role in ionizing radiation-induced cell cycle arrest through Ca2+ signaling pathways38.
Similar to the design of validation experiments38, the immunoblotting assay using corresponding protein antibodies was performed on the same amount of the immunoprecipitates derived from FLAG-alone HCC cells (parental) and FLAG-H2AX stable HCC cells, respectively. H2AX-specific interactions for the selected proteins found in HCC cells are shown in Fig. 5, and are consistent with our quantitative proteomic dataset.
Figure 5. Immunochemical analyses of selected proteins identified as H2AX-interacting partners in HCC cells.
The immunoprecipitated proteins from FLAG-QGY-7703 cells and FLAG-H2AX-QGY-7703 cells were analyzed by immunoblotting (IB). Anti-FLAG antibody was used to detect H2AX with nuleolin (NCL) as a loading control. Lane 1: the QGY-7703 cells expressing FLAG alone; lane 2: the QGY-7703 cells stably expressing FLAG-H2AX.
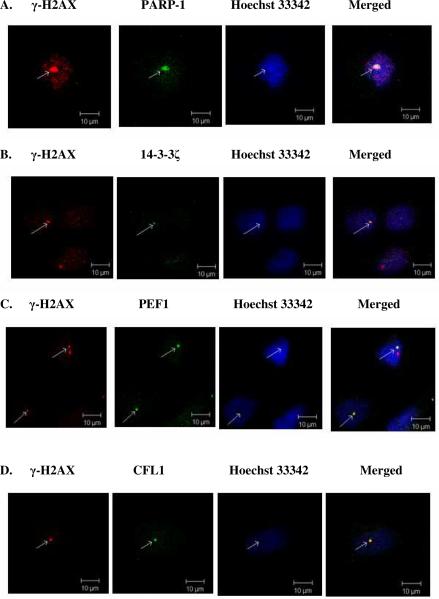
In situ localization of endogenous H2AX-specific interactions using confocal laser scanning
With the positive control of the known H2AX-PARP1 interaction, by using confocal scanning, we further examined the endogenous interactions of H2AX with its newly identified partners such as 14-3-3ζ, CFL1, and PEF1 in `resting' HCC cells. As shown in Figure 6, endogenous phosphorylated (γ)-H2AX stained with red fluorescence was observed in the nucleus. Stained with green fluorescence, either 14-3-3ζ, CFL1, PEF1, or PARP-1 were also found in the nucleus (in blue). Although in a small population, γ-H2AX was found to co-localize with either PARP-1, or 14-3-3ζ, or CFL1 or PEF1 in the nucleus of HCC cells as seen by the bright merged spots (arrowhead in Fig 6). As a result, for the first time, the interactions of H2AX with 14-3-3ζ, CFL1, PEF1 and PARP-1 were observed at the endogenous level in HCC cells based on both proteomic and immunoassay/confocal results.
Figure 6. Confocal laser scanning analysis of co-localized endogenous interactions between γ-H2AX and either 14-3-3ζ, or PEF1, or PARP-1 or CFL1 respectively using.
The parental QGY7703 cells were stained with TRITC for γ-H2AX (red), and with FITC for either PARP-1, or 14-3-3ζ, or PEF1 or CFL1 respectively (green). The nucleus was stained with Hoechst 33342 (blue). With a scale bar of 10 μm the merged images are indicated by white foci. All foci are indicated by white arrows. The confocal images of co-localized (A). γ-H2AX and PARP-1, (B). γ-H2AX and 14-3-3 ζ, (C). γ-H2AX and PEF1, and (D). γ-H2AX and CFL1.
The correspondence between methods indicates the accuracy and reliability of our dual-tagging quantitative method for profiling bait-specific interacting partners. The results also suggest that these H2AX interactors function in DNA damage recognition/repair, apoptosis, and cell cycle control through their interactions with H2AX or mediated by H2AX.
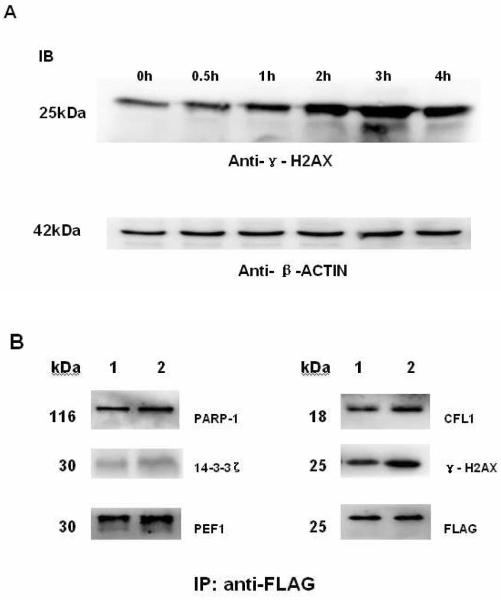
H2AX recruits particular interacting proteins in response to bleomycin (BLM)-induced DNA damage in HCC cells
To understand the functional role of the HCC-specific H2AX-interacting complexes in diverse functions of DNA damage recognition and repair, apoptosis, and cell cycle control in response to DNA DSBs, we next studied the changes in H2AX binding by the interactors representing each functional category. Bleomycin (BLM) was used to trigger the response to instant DNA DSBs47, 94. Because H2AX can sense the extent of BLM-induced DNA DSBs, we first examined both dose- and time course-dependent BLM-induced phosphorylation at H2AX to determine the condition of maximum effect of BLM. As shown in Fig.7A, the abundance of phosphorylated H2AX or γ-H2AX increased along with BLM treatment at a defined dose of 40 mg/ml for an extended period of time until a maximum level was reached after approximately 3 hours. Under this condition, the H2AX-interacting complex was first pulled down through immunoprecipitation using anti-FLAG beads. Using the same approach, the immunoprecipitate was obtained from non-stimulated HCC cells stably expressing FLAG-tagged H2AX. Using the antibodies against each of the H2AX interactors such as PARP-1, 14-3-3ζ, CFL1 and PEF1, immunoblotting experiments were performed on equal amounts of the immunoprecipitates derived from BLM-stimulated versus non-stimulated HCC cells. In these co-immunoprecipitation and immunoblotting experiments, the amount of FLAG tag detected was used as the control. As shown in Figure 7B, the abundance of PARP-1, CFL1, PEF1, or 14-3-3 ζ in the H2AX-interacting complex increased proportionally along with the BLM-induced increase of γ-H2AX (Table S2), suggesting that these proteins were recruited through interaction with H2AX to the complex that participates in the processes of recognizing BLM-induced DNA DSBs or DNA damage.
Figure 7. Immunoblotting analysis of bleomycin (BLM)-induced changes in the H2AX binding to its interacting partners.
(A) Time course-dependent BLM-induced phosphorylation of H2AX in HCC cells, and (B) Immunoprecipitates isolated from either untreated or BLM-stimulated cells (40 U/ml for 3 hours), respectively, were analyzed by immunoblotting (IB). The FLAG tag was detected and used as the loading control. Lane 1: from the untreated FLAG-H2AX-QGY-7703 cells; lane 2: from the BLM-stimulated FLAG-H2AX-QGY-7703 cells.
In addition to its known role in DNA repair, with still largely unknown mechanism γ-H2AX may participate in regulation of a variety of biological processes such as cell cycle, chromatin remodeling, etc9, 95. For example, two distinct γ-H2AX foci were found in non-irradiated or `resting' cells96. One predominant population of small foci did not co-localize with DSB repairing proteins, while another small population of large foci co-localized with many repair proteins and resembled IR-induced foci96. It has also been shown that H2AX can be phosphorylated independently of DNA DSBs, and that this phosphorylation could be involved in regulating the cell cycle independently of sensing DNA damage or promoting repair96, 97. Also H2AX may function to promote chromatin remodeling98.
Our results for BLM-inducible γ-H2AX interactions in HCC cells revealed that upon BLM-induced DSBs, γ-H2AX could interact with those proteins representing diverse functional categories of DNA DSB recognition such as PARP-1, the cytoskeleton such as CFL1, Ca2+ signaling such as PEF1, etc. The BLM-induced composition of γ-H2AX foci provides an insight into the molecular mechanisms underlying how γ-H2AX turnover keeps cell cycle checkpoints active until any DNA damage is repaired.
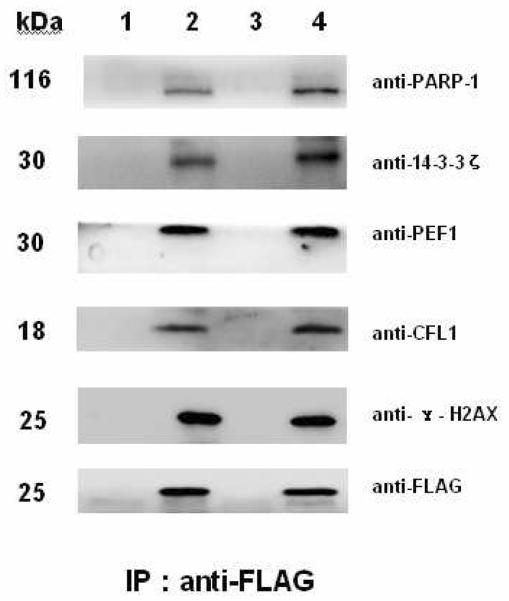
Comparative analysis of the H2AX interacting partners in paired hepatocytesversus HCC cells
On the basis of the profiling dataset of the H2AX interacting partners, we selected certain core components identified in the complex to compare their relative binding affinities to H2AX in the paired hepatocytes and HCC cells. These comparative studies were performed on two cell lines derived from healthy liver and carcinoma tissue of the same donor with the same genetic background, i.e., QSG-7701 for normal and QGY-7703 for HCC respectively48. We also generated the QSG-7701 or QGY-7703 cell lines expressing only FLAG (parental) and FLAG-tagged H2AX respectively. Co-immunoprecipitation and immunoblotting experiments were performed on the pair of QSG-7701 and QGY-7703 both stably expressing FLAG-tagged H2AX in contrast to their corresponding parental cells. As shown in Figure 8, with respect to the abundance of FLAG tag in the immunoprecipitates as the loading control, the expression of γ-H2AX did not show a significant difference between these cell lines. Measured by Fujifilm Las-3000 Luminescent image analyzer, the H2AX interacting partners such as PARP-1, 14-3-3ζ, and CFL1 displayed enhanced binding to H2AX in QGY-7703 HCC cells compared to that in normal QSG-7701 hepatocytes (Table S3). PARP-1 was among those interactors showing a relatively stronger enhancement for its binding to H2AX in HCC cells.
Figure 8. Comparative analysis of the H2AX binding with selected protein partners in paired hepatocytes (QSG-7701)versusHCC (QGY-7703) cells.
Using the antibodies as indicated in the Figure, immunoprecipitates were obtained from QSG-7701 cells expressing FLAG alone, QSG-7701 cells expressing FLAG-H2AX, the QGY-7703 cells expressing FLAG alone and QGY-7703 cells expressing FLAG-H2AX respectively. In immunoblotting (IB) analysis, the FLAG tag was detected and used as the loading control. Lane1: from the FLAG-QSG-7701 cells; lane2: from the FLAG-H2AX-QSG-7701 cells; lane3: from the FLAG-QGY-7703 cells; lane4: from the FLAG-H2AX-QGY-7703 cells.
Defects in the DNA damage recognition and repair pathways contribute to genome instability and promote tumorigenesis3. It has been documented that the processes of DNA damage response and repair remain active during tumourigenesis31, 99, probably due to stress-inducing DNA damage in the rapidly dividing pre-neoplastic lesions31, 99 100, 101. Improper DNA damage recognition and DNA repair could contribute to the survival and apoptotic resistance of cancer cells. Clearly, our observations support these notions at the molecular level with a systems view, suggesting that not only the phosphorylated H2AX but its interactors with characteristic binding strength are responsive to intrinsic DSBs/DNA damage in tumor cells.
Conclusions
Using a dual-tagging quantitative proteomic approach we have dissected a cell type-specific H2AX interactome. The HCC profile of the H2AX-interacting partners identified by our approach first suggested a multi-functional role of H2AX in mediating many biological pathways/processes such as DNA damage recognition, DNA repair, apoptosis, cell cycle, protein metabolism, cellular localization, etc. Network analysis further indicated the cross-talk among functional clusters. The physiologically relevant accuracy of these interactions was validated by immunoassys and the cellular localization of these interactions at the endogenous level was determined by confocal scanning microscopy. The stimulation of HCC by a DNA DSB-inducing agent, BLM, allowed us to determine the role of those interactions in the cellular response to DNA damage. The differential strengths of H2AX in recruiting its interactors were studied in paired hepatocyte and HCC cells, suggesting that H2AX mediates the differential cellular response in carcinogenesis through interacting with different strengths with its partners. Our results of dissecting HCC-characteristic H2AX interactome have provided insight into possible markers indicative of the pathological status of HCC.
Supplementary Material
Acknowledgments
We thank Mr. J. Yao for his assistance in mass spectrometry analysis. This work was supported by grants from Shanghai Science and Technology Development Program (Grants 03DZ14024 and 07ZR14010) and the 863 High Technology Foundation of China (Grant 2006AA02A310). This work was also supported by US NIH 1R01AI064806-01A2, and 5R21DK082706. We also thank Dr. Howard Fried for his proof-reading of the manuscript.
Abbreviations
- DSBs
double-strand breaks
- AACT
amino acid-coded tagging
- SILAC
stable isotope labeling with amino acid in cell culture
- 14-3-3ζ
14-3-3zeta
- CFL1
coflin1
- PEF1
peflin1
- PARP-1
Poly(ADP-ribose) polymerase1
- HCC
hepatocellular carcinoma
- γ-H2AX
phosphorylation H2AX
- MS
mass spectrometry
- BLM
bleomycin
- CALR
Calreticulin
- NONO
non-pou domain-containing octamer-binding protein
- NCL
nucleolin
- MRN
MRE11-NBS1-RAD50
- UIM
ubiquitin-interaction motif
- ATM
ataxia telangiectasia mutated
- MDC1
DNA damage checkpoint 1
- SDT
Ser-Asp-Thr
- Co-IP
co-immunoprecipitation
- IB
immunoblotting
- WB
Western blotting
- FITC
fluorescein isothiocyannate
- ECL
electrochemiluminescence
- HPLC
high performance liquid chromatography
- MS/MS
tandem mass spectrometry
- LTQ-Orbitrap
linear trap quadrupole-Orbitrap
Reference
- 1.Franco S, Alt FW, Manis JP. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst) 2006;5(9–10):1030–41. doi: 10.1016/j.dnarep.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 2.Phillips ER, McKinnon PJ. DNA double-strand break repair and development. Oncogene. 2007;26(56):7799–808. doi: 10.1038/sj.onc.1210877. [DOI] [PubMed] [Google Scholar]
- 3.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18(1):134–47. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 4.Pandita TK, Richardson C. Chromatin remodeling finds its place in the DNA double-strand break response. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkn1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009;417(3):639–50. doi: 10.1042/BJ20080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeggo PA, Lobrich M. DNA double-strand breaks: their cellular and clinical impact? Oncogene. 2007;26(56):7717–9. doi: 10.1038/sj.onc.1210868. [DOI] [PubMed] [Google Scholar]
- 7.Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W. Histone H2A variants H2AX and H2AZ. Curr Opin Genet Dev. 2002;12(2):162–9. doi: 10.1016/s0959-437x(02)00282-4. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava N, Gochhait S, de Boer P, Bamezai RN. Role of H2AX in DNA damage response and human cancers. Mutat Res. 2009;681(2–3):180–8. doi: 10.1016/j.mrrev.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36(17):5678–94. doi: 10.1093/nar/gkn550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Riches LC, Lynch AM, Gooderham NJ. Early events in the mammalian response to DNA double-strand breaks. Mutagenesis. 2008;23(5):331–9. doi: 10.1093/mutage/gen039. [DOI] [PubMed] [Google Scholar]
- 11.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. Genomic instability in mice lacking histone H2AX. Science. 2002;296(5569):922–7. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature. 2003;421(6926):961–6. doi: 10.1038/nature01446. [DOI] [PubMed] [Google Scholar]
- 13.Peng A, Chen PL. NFBD1/Mdc1 mediates ATR-dependent DNA damage response. Cancer Res. 2005;65(4):1158–63. doi: 10.1158/0008-5472.CAN-04-2508. [DOI] [PubMed] [Google Scholar]
- 14.Yan J, Jetten AM. RAP80 and RNF8, key players in the recruitment of repair proteins to DNA damage sites. Cancer Lett. 2008;271(2):179–90. doi: 10.1016/j.canlet.2008.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lou Z, Minter-Dykhouse K, Franco S, Gostissa M, Rivera MA, Celeste A, Manis JP, van Deursen J, Nussenzweig A, Paull TT, Alt FW, Chen J. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol Cell. 2006;21(2):187–200. doi: 10.1016/j.molcel.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316(5828):1194–8. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316(5828):1198–202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie A, Hartlerode A, Stucki M, Odate S, Puget N, Kwok A, Nagaraju G, Yan C, Alt FW, Chen J, Jackson SP, Scully R. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell. 2007;28(6):1045–57. doi: 10.1016/j.molcel.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teoh NC, Dan YY, Swisshelm K, Lehman S, Wright JH, Haque J, Gu Y, Fausto N. Defective DNA strand break repair causes chromosomal instability and accelerates liver carcinogenesis in mice. Hepatology. 2008;47(6):2078–88. doi: 10.1002/hep.22194. [DOI] [PubMed] [Google Scholar]
- 20.Downs JA. Chromatin structure and DNA double-strand break responses in cancer progression and therapy. Oncogene. 2007;26(56):7765–72. doi: 10.1038/sj.onc.1210874. [DOI] [PubMed] [Google Scholar]
- 21.Levitt PS, Zhu M, Cassano A, Yazinski SA, Liu H, Darfler J, Peters RM, Weiss RS. Genome maintenance defects in cultured cells and mice following partial inactivation of the essential cell cycle checkpoint gene Hus1. Mol Cell Biol. 2007;27(6):2189–201. doi: 10.1128/MCB.01763-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parikh RA, White JS, Huang X, Schoppy DW, Baysal BE, Baskaran R, Bakkenist CJ, Saunders WS, Hsu LC, Romkes M, Gollin SM. Loss of distal 11q is associated with DNA repair deficiency and reduced sensitivity to ionizing radiation in head and neck squamous cell carcinoma. Genes Chromosomes Cancer. 2007;46(8):761–75. doi: 10.1002/gcc.20462. [DOI] [PubMed] [Google Scholar]
- 23.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. GammaH2AX and cancer. Nat Rev Cancer. 2008;8(12):957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Srivastava N, Gochhait S, Gupta P, Bamezai RN. Copy number alterations of the H2AFX gene in sporadic breast cancer patients. Cancer Genet Cytogenet. 2008;180(2):121–8. doi: 10.1016/j.cancergencyto.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 25.Walsh SH, Rosenquist R. Absence of H2AX gene mutations in B-cell leukemias and lymphomas. Leukemia. 2005;19(3):464. doi: 10.1038/sj.leu.2403651. [DOI] [PubMed] [Google Scholar]
- 26.Chen B, Piel WH, Gui L, Bruford E, Monteiro A. The HSP90 family of genes in the human genome: insights into their divergence and evolution. Genomics. 2005;86(6):627–37. doi: 10.1016/j.ygeno.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 27.Yu T, MacPhail SH, Banath JP, Klokov D, Olive PL. Endogenous expression of phosphorylated histone H2AX in tumors in relation to DNA double-strand breaks and genomic instability. DNA Repair (Amst) 2006;5(8):935–46. doi: 10.1016/j.dnarep.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 28.Sedelnikova OA, Bonner WM. GammaH2AX in cancer cells: a potential biomarker for cancer diagnostics, prediction and recurrence. Cell Cycle. 2006;5(24):2909–13. doi: 10.4161/cc.5.24.3569. [DOI] [PubMed] [Google Scholar]
- 29.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85(4):509–20. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- 30.Lavin MF, Delia D, Chessa L. ATM and the DNA damage response. Workshop on ataxia-telangiectasia and related syndromes. EMBO Rep. 2006;7(2):154–60. doi: 10.1038/sj.embor.7400629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–70. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 32.Choudhury A, Elliott F, Iles MM, Churchman M, Bristow RG, Bishop DT, Kiltie AE. Analysis of variants in DNA damage signalling genes in bladder cancer. BMC Med Genet. 2008;9:69. doi: 10.1186/1471-2350-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishida T, Takizawa Y, Sakane I, Kurumizaka H. Altered DNA binding by the human Rad51-R150Q mutant found in breast cancer patients. Biol Pharm Bull. 2007;30(8):1374–8. doi: 10.1248/bpb.30.1374. [DOI] [PubMed] [Google Scholar]
- 34.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol. 2004;6(2):97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 35.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso G, St Onge P, Ghanny S, Lam MH, Butland G, Altaf-Ul AM, Kanaya S, Shilatifard A, O'Shea E, Weissman JS, Ingles CJ, Hughes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440(7084):637–43. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 36.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol Cell Proteomics. 2007;6(3):439–50. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Wang T, Gu S, Ronni T, Du YC, Chen X. In vivo dual-tagging proteomic approach in studying signaling pathways in immune response. J Proteome Res. 2005;4(3):941–9. doi: 10.1021/pr050031z. [DOI] [PubMed] [Google Scholar]
- 38.Du YC, Gu S, Zhou J, Wang T, Cai H, Macinnes MA, Bradbury EM, Chen X. The dynamic alterations of H2AX complex during DNA repair detected by a proteomic approach reveal the critical roles of Ca(2+)/calmodulin in the ionizing radiation-induced cell cycle arrest. Mol Cell Proteomics. 2006;5(6):1033–44. doi: 10.1074/mcp.M500327-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Wong CM, Ng IO. Molecular pathogenesis of hepatocellular carcinoma. Liver Int. 2008;28(2):160–74. doi: 10.1111/j.1478-3231.2007.01637.x. [DOI] [PubMed] [Google Scholar]
- 40.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, Bottinger E, Friedman S, Waxman S, Llovet JM. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–47. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 41.Nowsheen S, Wukovich RL, Aziz K, Kalogerinis PT, Richardson CC, Panayiotidis MI, Bonner WM, Sedelnikova OA, Georgakilas AG. Accumulation of oxidatively induced clustered DNA lesions in human tumor tissues. Mutat Res. 2008 doi: 10.1016/j.mrgentox.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koike M, Mashino M, Sugasawa J, Koike A. Histone H2AX phosphorylation independent of ATM after X-irradiation in mouse liver and kidney in situ. J Radiat Res (Tokyo) 2008;49(4):445–9. doi: 10.1269/jrr.08010. [DOI] [PubMed] [Google Scholar]
- 43.Koike M, Sugasawa J, Yasuda M, Koike A. Tissue-specific DNA-PK-dependent H2AX phosphorylation and gamma-H2AX elimination after X-irradiation in vivo. Biochem Biophys Res Commun. 2008;376(1):52–5. doi: 10.1016/j.bbrc.2008.08.095. [DOI] [PubMed] [Google Scholar]
- 44.Wilda M, Demuth I, Concannon P, Sperling K, Hameister H. Expression pattern of the Nijmegen breakage syndrome gene, Nbs1, during murine development. Hum Mol Genet. 2000;9(12):1739–44. doi: 10.1093/hmg/9.12.1739. [DOI] [PubMed] [Google Scholar]
- 45.Louro H, Pinheiro I, Costa P, Sousa C, Dias A, Boavida MG, Silva MJ. Mutagenic effects of poly (ADP-ribose) polymerase-1 deficiency in transgenic mice. Mutat Res. 2008;640(1–2):82–8. doi: 10.1016/j.mrfmmm.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Morel F, Renoux M, Lachaume P, Alziari S. Bleomycin-induced double-strand breaks in mitochondrial DNA of Drosophila cells are repaired. Mutat Res. 2008;637(1–2):111–7. doi: 10.1016/j.mrfmmm.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 47.Schlade-Bartusiak K, Stembalska-Kozlowska A, Bernady M, Kudyba M, Sasiadek M. Analysis of adaptive response to bleomycin and mitomycin C. Mutat Res. 2002;513(1–2):75–81. doi: 10.1016/s1383-5718(01)00288-1. [DOI] [PubMed] [Google Scholar]
- 48.Bao M, Song P, Qingping L, Liu Y, Yun D, Hexige S, Du R, Yang Z, Fan H, Yang P, Chen X. Quantitative proteomic analysis of a paired human liver healthy versus carcinoma cell lines with the same genetic background to identify potential hepatocellular carcinoma markers. PROTEOMICS - CLINICAL APPLICATIONS. 2009;3(6):705–719. doi: 10.1002/prca.200780128. [DOI] [PubMed] [Google Scholar]
- 49.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74(20):5383–92. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 50.Moore RE, Young MK, Lee TD. Qscore: an algorithm for evaluating SEQUEST database search results. J Am Soc Mass Spectrom. 2002;13(4):378–86. doi: 10.1016/S1044-0305(02)00352-5. [DOI] [PubMed] [Google Scholar]
- 51.Keller A, Eng J, Zhang N, Li XJ, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:0017. doi: 10.1038/msb4100024. 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21(3):315–8. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 53.Escargueil AE, Soares DG, Salvador M, Larsen AK, Henriques JA. What histone code for DNA repair? Mutat Res. 2008;658(3):259–70. doi: 10.1016/j.mrrev.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 54.Luk SC, Ngai SM, Tsui SK, Fung KP, Lee CY, Waye MM. In vivo and in vitro association of 14-3-3 epsilon isoform with calmodulin: implication for signal transduction and cell proliferation. J Cell Biochem. 1999;73(1):31–5. doi: 10.1002/(sici)1097-4644(19990401)73:1<31::aid-jcb4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka T, Halicka HD, Huang X, Traganos F, Darzynkiewicz Z. Constitutive histone H2AX phosphorylation and ATM activation, the reporters of DNA damage by endogenous oxidants. Cell Cycle. 2006;5(17):1940–5. doi: 10.4161/cc.5.17.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iwabuchi K, Matsui T, Hashimoto M, Matsumoto Y, Kurihara T, Date T. Characterization of a cancer cell line that expresses a splicing variant form of 53BP1: separation of checkpoint and repair functions in 53BP1. Biochem Biophys Res Commun. 2008;376(3):509–13. doi: 10.1016/j.bbrc.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 57.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol Cell Biol. 2007;27(16):5806–18. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bruneel A, Labas V, Mailloux A, Sharma S, Royer N, Vinh J, Pernet P, Vaubourdolle M, Baudin B. Proteomics of human umbilical vein endothelial cells applied to etoposide-induced apoptosis. Proteomics. 2005;5(15):3876–84. doi: 10.1002/pmic.200401239. [DOI] [PubMed] [Google Scholar]
- 59.Bando Y, Katayama T, Aleshin AN, Manabe T, Tohyama M. GRP94 reduces cell death in SH-SY5Y cells perturbated calcium homeostasis. Apoptosis. 2004;9(4):501–8. doi: 10.1023/B:APPT.0000031446.95532.ad. [DOI] [PubMed] [Google Scholar]
- 60.Hamaguchi T, Iizuka N, Tsunedomi R, Hamamoto Y, Miyamoto T, Iida M, Tokuhisa Y, Sakamoto K, Takashima M, Tamesa T, Oka M. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int J Oncol. 2008;33(4):725–31. [PubMed] [Google Scholar]
- 61.Wang Y, Lee GF, Kelley RF, Spellman MW. Identification of a GDP-L-fucose:polypeptide fucosyltransferase and enzymatic addition of O-linked fucose to EGF domains. Glycobiology. 1996;6(8):837–42. doi: 10.1093/glycob/6.8.837. [DOI] [PubMed] [Google Scholar]
- 62.Davis GL, Dempster J, Meler JD, Orr DW, Walberg MW, Brown B, Berger BD, O'Connor JK, Goldstein RM. Hepatocellular carcinoma: management of an increasingly common problem. Proc (Bayl Univ Med Cent) 2008;21(3):266–280. doi: 10.1080/08998280.2008.11928410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Solozobova V, Rolletschek A, Blattner C. Nuclear accumulation and activation of p53 in embryonic stem cells after DNA damage. BMC Cell Biol. 2009;10:46. doi: 10.1186/1471-2121-10-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grover R, Candeias MM, Fahraeus R, Das S. p53 and little brother p53/47: linking IRES activities with protein functions. Oncogene. 2009;28(30):2766–72. doi: 10.1038/onc.2009.138. [DOI] [PubMed] [Google Scholar]
- 65.Puigvert JC, Huveneers S, Fredriksson L, op het Veld M, van de Water B, Danen EH. Cross-talk between integrins and oncogenes modulates chemosensitivity. Mol Pharmacol. 2009;75(4):947–55. doi: 10.1124/mol.108.051649. [DOI] [PubMed] [Google Scholar]
- 66.Sugimura K, Takebayashi S, Taguchi H, Takeda S, Okumura K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J Cell Biol. 2008;183(7):1203–12. doi: 10.1083/jcb.200806068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cohen-Armon M. PARP-1 activation in the ERK signaling pathway. Trends Pharmacol Sci. 2007;28(11):556–60. doi: 10.1016/j.tips.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 68.Marcos-Carcavilla A, Calvo JH, Gonzalez C, Moazami-Goudarzi K, Laurent P, Bertaud M, Hayes H, Beattie AE, Serrano C, Lyahyai J, Martin-Burriel I, Serrano M. Structural and functional analysis of the HSP90AA1 gene: distribution of polymorphisms among sheep with different responses to scrapie. Cell Stress Chaperones. 2008;13(1):19–29. doi: 10.1007/s12192-007-0004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cepeda V, Fuertes MA, Castilla J, Alonso C, Quevedo C, Soto M, Perez JM. Poly(ADP-ribose) polymerase-1 (PARP-1) inhibitors in cancer chemotherapy. Recent Patents Anticancer Drug Discov. 2006;1(1):39–53. doi: 10.2174/157489206775246430. [DOI] [PubMed] [Google Scholar]
- 70.Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y. Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs. Embo J. 2008;27(2):421–32. doi: 10.1038/sj.emboj.7601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piskunova TS, Iurova MN, Zabezhinskii MA, Anisimov VN. [Poly(ADP-ribosa)polymerase--the relationships with life span and carcinogenesis] Adv Gerontol. 2007;20(2):82–90. [PubMed] [Google Scholar]
- 72.Lewis C, Low JA. Clinical poly(ADP-ribose) polymerase inhibitors for the treatment of cancer. Curr Opin Investig Drugs. 2007;8(12):1051–6. [PubMed] [Google Scholar]
- 73.Elser M, Borsig L, Hassa PO, Erener S, Messner S, Valovka T, Keller S, Gassmann M, Hottiger MO. Poly(ADP-ribose) polymerase 1 promotes tumor cell survival by coactivating hypoxia-inducible factor-1-dependent gene expression. Mol Cancer Res. 2008;6(2):282–90. doi: 10.1158/1541-7786.MCR-07-0377. [DOI] [PubMed] [Google Scholar]
- 74.Zaremba T, Curtin NJ. PARP inhibitor development for systemic cancer targeting. Anticancer Agents Med Chem. 2007;7(5):515–23. doi: 10.2174/187152007781668715. [DOI] [PubMed] [Google Scholar]
- 75.Genovese T, Cuzzocrea S. Role of free radicals and poly(ADP-ribose)polymerase-1 in the development of spinal cord injury: new potential therapeutic targets. Curr Med Chem. 2008;15(5):477–87. doi: 10.2174/092986708783503177. [DOI] [PubMed] [Google Scholar]
- 76.Shimizu S, Nomura F, Tomonaga T, Sunaga M, Noda M, Ebara M, Saisho H. Expression of poly(ADP-ribose) polymerase in human hepatocellular carcinoma and analysis of biopsy specimens obtained under sonographic guidance. Oncol Rep. 2004;12(4):821–5. [PubMed] [Google Scholar]
- 77.Huang SH, Xiong M, Chen XP, Xiao ZY, Zhao YF, Huang ZY. PJ34, an inhibitor of PARP-1, suppresses cell growth and enhances the suppressive effects of cisplatin in liver cancer cells. Oncol Rep. 2008;20(3):567–72. [PubMed] [Google Scholar]
- 78.Dandri M, Burda MR, Burkle A, Zuckerman DM, Will H, Rogler CE, Greten H, Petersen J. Increase in de novo HBV DNA integrations in response to oxidative DNA damage or inhibition of poly(ADP-ribosyl)ation. Hepatology. 2002;35(1):217–23. doi: 10.1053/jhep.2002.30203. [DOI] [PubMed] [Google Scholar]
- 79.Niemantsverdriet M, Wagner K, Visser M, Backendorf C. Cellular functions of 14-3-3 zeta in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene. 2008;27(9):1315–9. doi: 10.1038/sj.onc.1210742. [DOI] [PubMed] [Google Scholar]
- 80.Kobayashi R, Deavers M, Patenia R, Rice-Stitt T, Halbe J, Gallardo S, Freedman RS. 14-3-3 zeta protein secreted by tumor associated monocytes/macrophages from ascites of epithelial ovarian cancer patients. Cancer Immunol Immunother. 2008 doi: 10.1007/s00262-008-0549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xing H, Zhang S, Weinheimer C, Kovacs A, Muslin AJ. 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. Embo J. 2000;19(3):349–58. doi: 10.1093/emboj/19.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tian Q, Feetham MC, Tao WA, He XC, Li L, Aebersold R, Hood L. Proteomic analysis identifies that 14-3-3zeta interacts with beta-catenin and facilitates its activation by Akt. Proc Natl Acad Sci U S A. 2004;101(43):15370–5. doi: 10.1073/pnas.0406499101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Z, Zhao J, Du Y, Park HR, Sun SY, Bernal-Mizrachi L, Aitken A, Khuri FR, Fu H. Down-regulation of 14-3-3zeta suppresses anchorage-independent growth of lung cancer cells through anoikis activation. Proc Natl Acad Sci U S A. 2008;105(1):162–7. doi: 10.1073/pnas.0710905105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fan T, Li R, Todd NW, Qiu Q, Fang HB, Wang H, Shen J, Zhao RY, Caraway NP, Katz RL, Stass SA, Jiang F. Up-regulation of 14-3-3zeta in lung cancer and its implication as prognostic and therapeutic target. Cancer Res. 2007;67(16):7901–6. doi: 10.1158/0008-5472.CAN-07-0090. [DOI] [PubMed] [Google Scholar]
- 85.Toivola DM, Nieminen MI, Hesse M, He T, Baribault H, Magin TM, Omary MB, Eriksson JE. Disturbances in hepatic cell-cycle regulation in mice with assembly-deficient keratins 8/18. Hepatology. 2001;34(6):1174–83. doi: 10.1053/jhep.2001.29374. [DOI] [PubMed] [Google Scholar]
- 86.Bamburg JR, Bernstein BW. ADF/Cofilin. Curr Biol. 2008;18(7):R273–5. doi: 10.1016/j.cub.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 87.Lee YJ, Sheu TJ, Keng PC. Enhancement of radiosensitivity in H1299 cancer cells by actin-associated protein cofilin. Biochem Biophys Res Commun. 2005;335(2):286–91. doi: 10.1016/j.bbrc.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 88.Tojima T, Ito E. Signal transduction cascades underlying de novo protein synthesis required for neuronal morphogenesis in differentiating neurons. Prog Neurobiol. 2004;72(3):183–93. doi: 10.1016/j.pneurobio.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 89.Ding Y, Milosavljevic T, Alahari SK. Nischarin inhibits LIM Kinase to regulate Cofilin phosphorylation and Cell Invasion. Mol Cell Biol. 2008 doi: 10.1128/MCB.01832-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sidani M, Wessels D, Mouneimne G, Ghosh M, Goswami S, Sarmiento C, Wang W, Kuhl S, El-Sibai M, Backer JM, Eddy R, Soll D, Condeelis J. Cofilin determines the migration behavior and turning frequency of metastatic cancer cells. J Cell Biol. 2007;179(4):777–91. doi: 10.1083/jcb.200707009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ding SJ, Li Y, Shao XX, Zhou H, Zeng R, Tang ZY, Xia QC. Proteome analysis of hepatocellular carcinoma cell strains, MHCC97-H and MHCC97-L, with different metastasis potentials. Proteomics. 2004;4(4):982–94. doi: 10.1002/pmic.200300653. [DOI] [PubMed] [Google Scholar]
- 92.Kitaura Y, Matsumoto S, Satoh H, Hitomi K, Maki M. Peflin and ALG-2, members of the penta-EF-hand protein family, form a heterodimer that dissociates in a Ca2+-dependent manner. J Biol Chem. 2001;276(17):14053–8. doi: 10.1074/jbc.M008649200. [DOI] [PubMed] [Google Scholar]
- 93.Kitaura Y, Watanabe M, Satoh H, Kawai T, Hitomi K, Maki M. Peflin, a novel member of the five-EF-hand-protein family, is similar to the apoptosis-linked gene 2 (ALG-2) protein but possesses nonapeptide repeats in the N-terminal hydrophobic region. Biochem Biophys Res Commun. 1999;263(1):68–75. doi: 10.1006/bbrc.1999.1189. [DOI] [PubMed] [Google Scholar]
- 94.Lieberman HB. DNA damage repair and response proteins as targets for cancer therapy. Curr Med Chem. 2008;15(4):360–7. doi: 10.2174/092986708783497328. [DOI] [PubMed] [Google Scholar]
- 95.Lowndes NF, Toh GW. DNA repair: the importance of phosphorylating histone H2AX. Curr Biol. 2005;15(3):R99–R102. doi: 10.1016/j.cub.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 96.McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell. 2005;16(10):5013–25. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ichijima Y, Sakasai R, Okita N, Asahina K, Mizutani S, Teraoka H. Phosphorylation of histone H2AX at M phase in human cells without DNA damage response. Biochem Biophys Res Commun. 2005;336(3):807–12. doi: 10.1016/j.bbrc.2005.08.164. [DOI] [PubMed] [Google Scholar]
- 98.Song X, Gjoneska E, Ren Q, Taverna SD, Allis CD, Gorovsky MA. Phosphorylation of the SQ H2A.X motif is required for proper meiosis and mitosis in Tetrahymena thermophila. Mol Cell Biol. 2007;27(7):2648–60. doi: 10.1128/MCB.01910-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr., Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 100.Nuciforo PG, Luise C, Capra M, Pelosi G, d'Adda di Fagagna F. Complex engagement of DNA damage response pathways in human cancer and in lung tumor progression. Carcinogenesis. 2007;28(10):2082–8. doi: 10.1093/carcin/bgm108. [DOI] [PubMed] [Google Scholar]
- 101.Knock E, Deng L, Wu Q, Lawrance AK, Wang XL, Rozen R. Strain differences in mice highlight the role of DNA damage in neoplasia induced by low dietary folate. J Nutr. 2008;138(4):653–8. doi: 10.1093/jn/138.4.653. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.