Abstract
Organisms have sophisticated subcellular compartments containing enzymes that function in tandem. These confined compartments ensure effective chemical transformation and transport of molecules, and the elimination of toxic metabolic wastes1,2. Creating functional enzyme complexes that are confined in a similar way remains challenging. Here we show that two or more enzymes with complementary functions can be assembled and encapsulated within a thin polymer shell to form enzyme nanocomplexes. These nanocomplexes exhibit improved catalytic efficiency and enhanced stability when compared with free enzymes. Furthermore, the co-localized enzymes display complementary functions, whereby toxic intermediates generated by one enzyme can be promptly eliminated by another enzyme. We show that nanocomplexes containing alcohol oxidase and catalase could reduce blood alcohol levels in intoxicated mice, offering an alternative antidote and prophylactic for alcohol intoxication.
Enzymes are exquisite biocatalysts mediating every biological process in living organisms. In eukaryotic cells, most enzymes do not freely diffuse within the cytosols, but are spatially defined within subcellular organelles or closely co-localized as enzyme complexes together with other enzymes.1–3 In consecutive reactions catalysed by multiple enzymes, such close confinement minimizes the diffusion of intermediates among the enzymes, enhancing the overall reaction efficiency and specificity4–8. Meanwhile, toxic intermediates generated during a metabolic process are promptly eliminated by the proximate enzymes co-localized within the confined structures9,10. Peroxisome, as an example, harbours a variety of oxidases with important metabolic and catabolic functions11,12. Toxic intermediates such as hydrogen peroxide (H2O2) are also produced during the enzymatic reactions in peroxisome; nature circumvents this dilemma by incorporating catalase (Cat) within the peroxisomes. Catalase is highly active and specific in decomposing H2O2, preventing its escape from the peroxisomes and subsequent damage to other cellular components13.
Inspired by natural multi-enzyme architectures, researchers have long directed their attention to the construction of enzyme complexes with synergic and complementary functions, mainly focusing on co-entrapment, co-immobilization, template assembly or fusion-protein techniques1. The first two approaches enable co-entrapment or co-immobilization of multiple enzymes within liposomes or solid particles4,5,14,15. However, it is difficult to control the number, type and spatial arrangement of the enzymes within the liposomes and particles. The latter two approaches enable the formation of enzyme complexes with significantly improved compositional and spatial controls8,10, but still have limitations, including inadequate translational capability of the host cells and insufficient enzyme stability against proteolysis and non-physiological environments16.
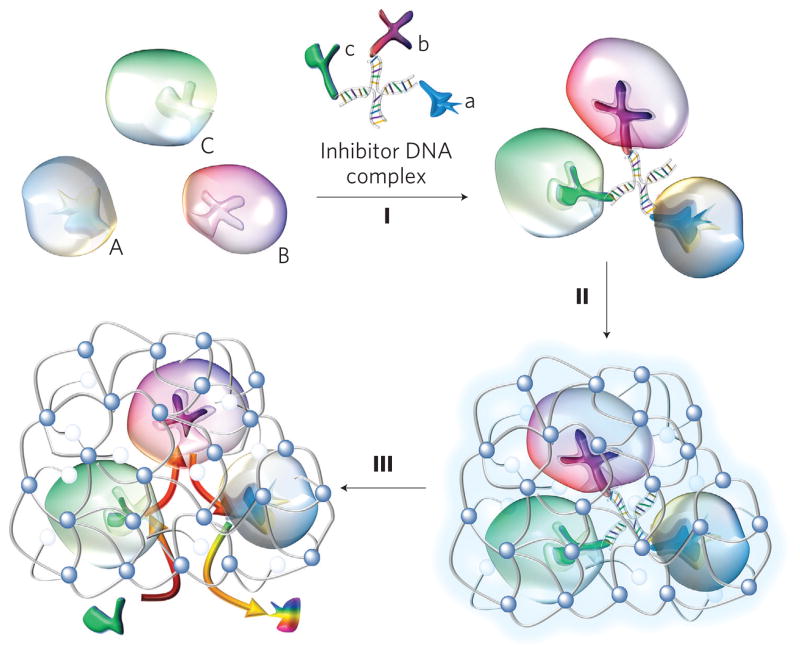
Here, we demonstrate a general design of robust enzyme nanocomplex with well-controlled enzyme composition and spatial arrangement. This is achieved by assembling or conjugating enzymes with synergic or complementary functions to form a nanocomplex, followed by encapsulation of the nanocomplex within a crosslinked polymer nanocapsule. Exemplified by the synthesis of a triple-enzyme nanocomplex (Fig. 1), inhibitors for each enzyme are respectively conjugated to a single-stranded DNA with a designed sequence. Complementary assembly of the DNA molecules forms a DNA-inhibitor scaffold linked with the three inhibitors, and specific binding of the inhibitors and the enzymes enables the construction of a triple-enzyme nanocomplex (step I, Fig. 1). Subsequent in situ polymerization leads to the growth of a thin layer of polymer network around each nanocomplex, and the formation of nanocapsules containing a triple-enzyme core and a permeable shell (step II). Finally, removal of the DNA-inhibitor scaffolds creates a highly robust enzyme nanocomplex denoted n(Enzymes), where ‘Enzymes’ within parentheses refers to the enzymes within the core of the nanocapsules (step III). It is important to point out that, without significant compromise of enzyme activity, encapsulating the enzymes within the nanocapsules effectively stabilizes them in a non-physiological environment and protects them against protease attack. Furthermore, the nanocomplexes can be readily functionalized to acquire both desired surface properties and targeting capability17.
Figure 1. Synthesis of enzyme nanocomplexes.
Schematic illustration of the synthesis of a model triple-enzyme nanocomplex by DNA-directed assembly and nano-encapsulation. Spontaneous assembly of invertase (Inv, A), glucose oxidase (GOx, B) and horseradish peroxidase (HRP, C) with an inhibitor-DNA scaffold containing their respective competitive inhibitors—lactobionic acid (a), glucosamine (b) and 4-dimethylaminoantipyrine (c)—leading to the formation of a triple-enzyme architecture (I). Confinement and stabilization of the triple-enzyme architecture by in situ growth of a thin network polymer around the enzyme nanocomplex (II). Removal of the DNA scaffold leading to the formation of triple-enzyme nanocomplexes with significantly enhanced stability and close-proximity definition. Such a close-proximity architecture enables active transport of their reaction intermediates among the enzymes, leading to significantly enhanced reaction efficiency and complementary function, such as the capability to eliminate toxic intermediates (III).
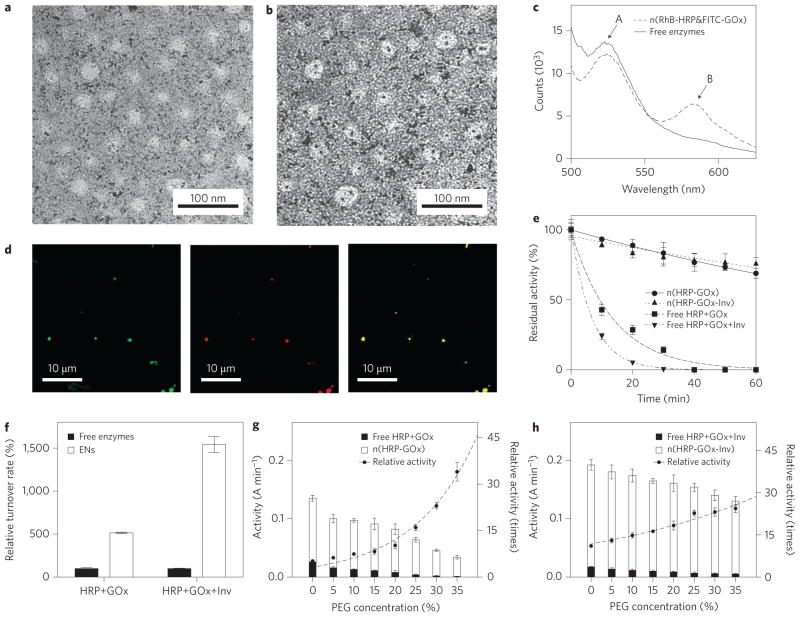
Successful construction of the enzyme nanocomplexes was demonstrated using horseradish peroxidase (HRP) and glucose oxidase (GOx) as model enzymes. Figure 2a presents a transmission electron microscope (TEM) image of n(HRP–GOx), which has an average diameter of 30±7 nm. To confirm the double-enzyme architecture, each HRP and GOx molecule was labelled with a single 1.4 nm gold nanoparticle. As shown in Fig. 2b, most gold-labelled enzyme nanocomplexes contain two gold nanoparticles, indicating that each nanocomplex indeed contains two enzyme molecules. By considering the specific bindings between these enzymes and their inhibitors (linked to the DNA scaffolds), it is reasonable to conclude that such a double-enzyme architecture includes one HRP molecule and one GOx molecule.
Figure 2. Structure and enhanced activity and stability of enzyme nanocomplexes.
a,b, Transmission electron micrograph showing the uniform size of n(HRP–GOx) and n(HRP–GOx) (a), prepared by labelling HRP and GOx with single gold nanoparticles (b). c, Fluorescence spectra of n(HRP–GOx) and a mixture of n(HRP) and n(GOx) with the same protein content. GOx and HRP were pre-labelled with FITC and RhB, respectively. The spectra were recorded with excitation at 450 nm. d, Confocal microscope images of n(FITC-labelled GOx) (left, excitation = 488 nm, emission = 510–530 nm), n(RhB-labelled HRP) (middle, excitation = 532 nm, emission = 570–600 nm) and n(RhB labelled HRP-FITC labelled GOx) (right, excitation = 488 nm; emission = 570–600 nm). e, Change in activity of n(HRP–GOx), n(HRP–GOx–Inv) and their native enzyme mixture counterparts during incubation at 65 °C. f, Turnover rates for n(HRP–GOx) and n(HRP–GOx–Inv) and their corresponding free enzyme mixtures. g, Activity of n(HRP–GOx) and a mixture containing the same amount of free HRP and GOx in the presence of increasing concentrations of PEG in phosphate buffer (50 mM, pH 7.0). h, Activity of n(HRP–GOx–Inv) and a mixture containing the same amount of free HRP, GOx and Inv as the complex in the presence of increasing PEG concentrations. Relative activities were normalized by the activities of the free enzyme mixtures with the same enzyme content and PEG concentration. The activity unit (A min−1) represents the absorbance change (at 460 nm) of the reaction solution per minute. Data represent mean±standard error of the mean (s.e.m.) from three independent experiments.
Such a nanocomplex architecture spatially locates the constituent enzymes within close proximity. Using rhodamine-B-labelled HRP (HRP–RhB) and fluorescein-isothiocyanate-labelled GOx (GOx–FITC) as an example, Fig. 2c compares the fluorescence spectra of a mixture of the two constituents (equal molar ratio) as well as the corresponding dual-enzyme nanocomplexes. Because FITC and RhB have excitation maxima at 495 and 540 nm, respectively, the mixture of GOx–FITC and HRP–RhB exhibits FITC emission centred at 520 nm when under 450 nm excitation. However, the spectrum for the nanocomplex exhibits intense emissions from both FITC and RhB (centred at 520 nm and 585 nm, respectively). This observation indicates an effective Förster resonance energy transfer (FRET) from GOx–FITC to HRP–RhB, confirming that the HRP and GOx molecules are closely associated within a short distance of one another (<10 nm)18. Figure 2d presents fluorescence microscope images of the nanocomplex, showing co-localization of GOx–FITC and HRP–RhB. Upon excitation at 488 nm, FRET emission at 580 nm was also detected at the same position, further confirming their close association.
Such a close-proximity architecture endows the nanocomplexes with significantly enhanced catalytic efficiency, as demonstrated by consecutive reactions of sucrose and glucose mediated by invertase (Inv), GOx and HRP. When glucose was added or generated, it was oxidized, producing H2O2, which further oxidized o-dianisidine via an HRP-mediated reaction. Figure 2f compares the turnover rates (o-dianisidine oxidation rates) of n(HRP–GOx) and n(HRP– GOx–Inv) with those of their native enzyme mixtures with the same enzyme content. Compared with the native enzyme mixtures, n(HRP–GOx) and n(HRP–GOx–Inv) demonstrated elevated turnover rates, by factors of 5 and 15 in phosphate buffer, respectively. To simulate the viscous environment within the cell or blood-stream19, poly(ethylene glycol) (PEG) was added to the reaction medium. The relative turnover rates increased with increasing PEG concentration. When 35 wt% PEG was added, 34-fold and 24-fold increases in turnover rate were observed for n(HRP–GOx) and n(HRP–GOx–Inv), respectively (Fig. 2g,h). Moreover, both nanocomplexes retained over 70% of their activity after incubation at 65 °C for 60 min, whereas their mixture counterparts lost more than 98% of their activity, indicating that the nanocomplexes have significantly enhanced stability (Fig. 2e).
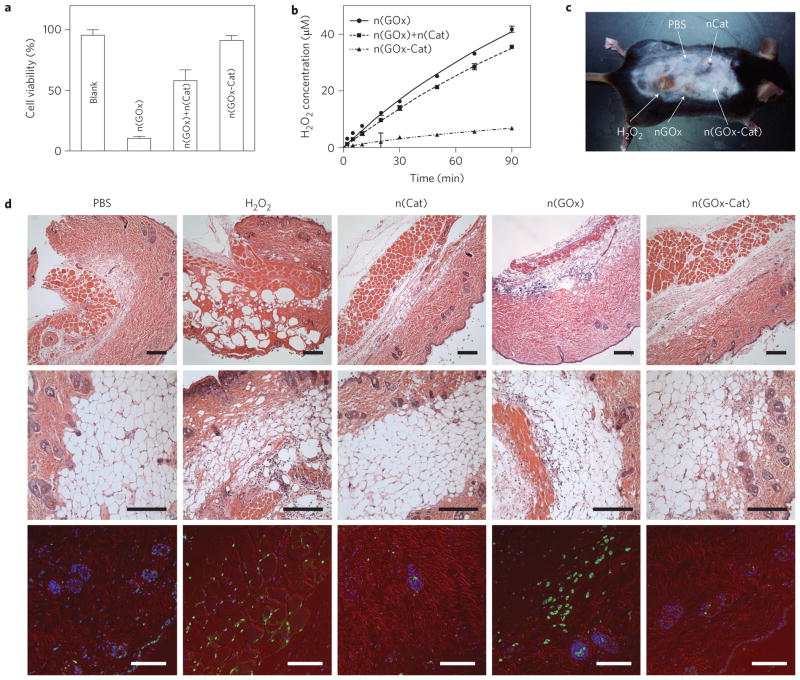
This unique architecture also affords the nanocomplexes with complementary functions. Oxidases, for example, are being used or proposed for various therapeutics20, but oxidase-mediated reactions produce toxic H2O2 (ref. 21). However, by constructing oxidase–catalase nanocomplexes, such toxic intermediates can be eliminated effectively. In an example using GOx, cells incubated with n(GOx) in glucose-containing media lose more than 90% of their viability. Adding n(Cat) into the media increases the viability to 42%. However, cells containing n(GOx–Cat) exhibit much higher viability (~91%), similar to that of the control, as a result of effective H2O2 elimination (Fig. 3a). The net rates of H2O2 production were quantified by adding the enzyme nanocapsules to glucose-containing buffer solutions, then measuring their temporal H2O2 concentration profiles (Fig. 3b). It was found that the net rate of H2O2 production for the n(GOx–Cat) system was consistently fivefold lower than the rates for the n(GOx)/n(Cat) mixture or n(GOx).
Figure 3. In vivo detoxifying capability of the catalase-containing enzyme nanocomplexes.
a, Cell viability assays after treated with n(GOx), mixture of n(GOx) and n(Cat), and n(GOx-Cat). Cell proliferation rates were normalized with those of the untreated cells. Cell viability assays were performed by incubating the treated cells with CellTiter Blue for 3 h at 37 °C. b, H2O2 concentration in glucose oxidation reaction catalysed by n(GOx), n(GOx)+n(Cat) or n(GOx-Cat). Enzyme reactions were performed in 1× phosphate buffer saline (PBS) and the initial concentration of glucose is 2 mg mL−1. The system with n(GOx), the mixture of n(GOx) and n(Cat), and n(GOx–Cat) show rates of 0.434±0.004 μMmin−1, 0.388± 0.005 μMmin−1, and 0.075± 0.007 μMmin−1, respectively. c, Photograph of a mouse cutaneously injected with PBS, n(Cat), H2O2, n(GOx) and n(GOx–Cat) at different sites. d, Micrographs of mouse skin tissue at the injection sites. Skin tissue was collected from the animal shown in c. Top and middle: H&E stained images. Bottom: Immunohistology stain with TUNEL assay (green), Cy3-conjugated monoclonal α-smooth muscle actin antibody (red) and DAPI (blue). Scale bars, 100 μm. Data represent mean±s.e.m. from three independent experiments.
The nanocapsules were then injected cutaneously into a mouse at different sites. Equal volumes of H2O2 solution (3% wt/vol) and phosphate buffer saline (PBS) were used as positive and negative controls, respectively (Fig. 3c). Skin lesions were observed in H2O2-treated and n(GOx)-treated sites 48 h after injection. In contrast, skin damage was not observed in the spots injected with PBS, n(Cat) or n(GOx–Cat). The skin tissue at the injection sites was sectioned and stained with haematoxylin and eosin (H&E) and a TUNEL staining kit. H2O2 administration caused tearing and ballooning in the dermis of the skin; n(GOx) administration resulted in similar tissue ballooning and neutrophil infiltration, albeit to a milder extent, indicating a pathophysiological response and injury due to the generated H2O2. No tissue damage was observed in the spots injected with PBS, n(Cat) or n(GOx–Cat) (Fig. 3d, top and middle). Consistently, cell apoptosis was evident in the skin tissue treated with either H2O2 or n(GOx), whereas the cell death in n(Cat)- or n(GOx–Cat)-treated skin tissues was minimal and comparable to that of the PBS-treated sample (Fig. 3d, bottom).
The capability to make robust enzyme nanocomplexes with enhanced and complementary functions offers a novel platform for various applications. For example, alcohol consumption is a millennium-old feature of human civilization, with unique social functions22. However, excessive consumption and abuse of alcohol is associated with a range of organ injuries and social problems23. Although an alcohol prophylactic does exist, the efficacy of this treatment has not yet been documented. In terms of an alcohol antidote, although various colloidal antidotes24,25 have been developed for drug overdoses, such antidotes may not be sufficient to treat alcohol intoxication. Recently, γ-aminobutyric-acid-based receptor antagonists were explored to resist the alcohol intoxication effect, but had side effects26. Here, based on nanocomplexes of alcohol oxidase (AOx) and catalase, we demonstrate designs for an alcohol prophylactic and antidote.
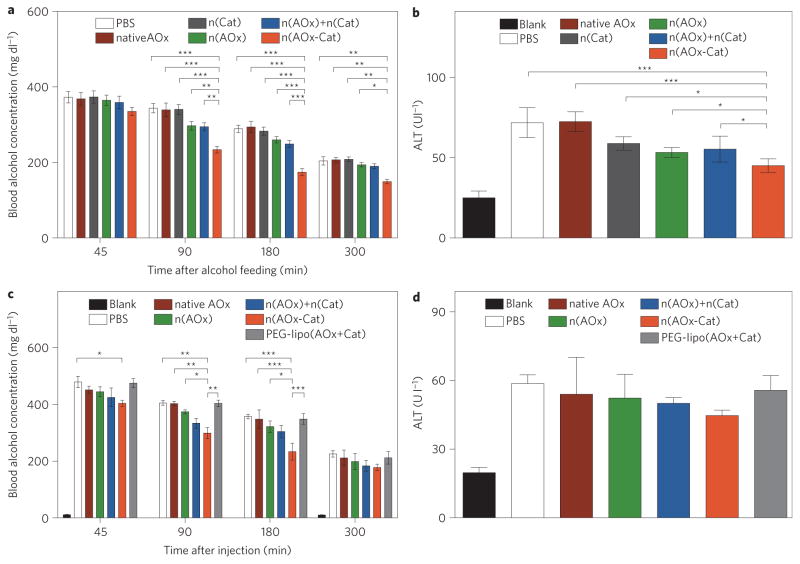
For the prophylactic studies, mice were gavaged with an alcoholic diet containing native AOx, n(AOx), n(Cat), a mixture of n(AOx) and n(Cat), or n(AOx–Cat). All mice were intoxicated and slept within 20 min of feeding. The group with n(AOx–Cat) woke up 1–2 h earlier than the other groups. The blood alcohol concentration (BAC) of the mice administered with n(AOx–Cat) was reduced by 10.1%, 31.8% and 36.8%, respectively, at 45 min, 90 min and 3 h of feeding (Fig. 4a). Significantly smaller BAC reductions were observed in those with n(AOx) (<8.5%) or with a mixture of n(AOx) and n(Cat) (<10.6%). Insignificant BAC reductions were observed for those with n(Cat) or native AOx. All alcohol-fed animals showed an increased plasma alanine aminotransferase (ALT) level; nevertheless, the mice treated with n(AOx–Cat) had a tendency for lower ALT levels (Fig. 4b). Most nanocomplexes were retained on the intestine surface, and no nanocomplexes could be detected in the liver or kidney (Supplementary Fig. S11).
Figure 4. Efficacy of n(AOx–Cat) as a prophylactic and antidote for alcohol intoxication.
Animals in the ‘Blank’ group were fed with a normal diet without alcohol and were used as the baseline group. There were four mice in each group. Data are presented as mean±standard error of the mean (s.e.m.) and the significance levels are *P<0.05, **P<0.01 and ***P<0.001. a,b, BAC (a) and ALT (b) in mice after gavage with an alcohol diet containing PBS, native AOx, n(Cat), n(AOx), a mixture of n(AOx) and n(Cat), or n(AOx–Cat), with equivalent amounts of enzymes. The amounts of AOx and Cat were fixed at 65 μg and 21μg, respectively, and the alcohol dosage was fixed at 6 mg ethanol per gram bodyweight. c,d, BAC (c) and ALT (d) of intoxicated mice after injection with PBS, native AOx, lipo(AOx+Cat) or n(AOx–Cat). Thirty minutes before injection, mice were gavaged with the alcohol diet at 6 mg ethanol per gram bodyweight. The volumes of PBS and enzyme solutions injected were maintained at 150 μl. The dose of enzyme injected was maintained at 65 μg AOx or 21 μg Cat per mouse. Three mice were used in each group.
For the antidote studies, PBS, native AOx, n(AOx), a mixture of n(Cat) and n(AOx), and n(AOx–Cat) were tail-vein-injected into intoxicated mice. The mice treated with n(AOx–Cat) presented with the lowest ALT level (Fig. 4d), and the most significant BAC reduction of 15.8%, 26.1%, 34.7% and 21.3% at 45 min, 90 min, 3 h and 5 h after the injection, respectively (Fig. 4c). Liposomes have been considered to be the most promising approach for protein delivery27. However, an insignificant BAC reduction was observed for the liposomes co-entrapping AOx and Cat. A dose dependence study suggests that BAC rapidly decreases with increasing n(AOx–Cat) dose from 30 to 60 μg AOx per animal, then remains almost constant with further increasing doses (Supplementary Fig. S10). Upon nanocomplex injection, the concentration of nanocomplexes in the blood decreases rapidly (Supplementary Fig. S12). Dynamic positron emission tomography–computed tomography (PET–CT) scanning suggests that the nanocomplexes accumulate in the liver shortly following the injection, whereas accumulation in the kidney increases with time, reaching a peak after ~40 min (Supplementary Figs S13 and S14).
Both the prophylactic and antidote studies suggest that n(AOx– Cat) is more effective than the mixture of n(AOx) and n(Cat) for BAC and ALT reduction. The unique n(AOx–Cat) architecture enables effective removal of toxic H2O2 (Supplementary Fig. S8) and prevents the AOx from being inactivated by the generated H2O2 (ref. 28). Moreover, the H2O2 elimination process regenerates molecular oxygen, ensuring fast alcohol oxidation kinetics and leading to more significant BAC reduction. The alcohol oxidation process also generates another toxic intermediate, acetaldehyde. More effective ALT reduction and complete liver protection will rely on the effective removal of acetaldehyde29; unfortunately, there is no available aldehyde dehydrogenase nor aldehyde oxidase (ADOx) that is sufficiently active. Better prophylactics and antidotes are anticipated when sufficiently active ADOx becomes available for the construction of n(AOx–ADOx–Cat) triple-enzyme nanocomplexes.
To conclude, we have demonstrated a design for a robust enzyme nanocomplex with improved efficiency and enhanced stability, as well as complementary and synergic functions, by precisely assembling and stabilizing multiple enzymes within a nanospace. Considering the vast library of enzymes that are currently or potentially available, novel classes of enzyme nanocomplexes could be built for a broad range of applications. Moreover, by judiciously choosing the constituent enzymes and harvesting their synergic and complementary effects, we foresee the creation of novel families of enzyme machineries with programmable function, possibly beyond those gifted by evolution.
Methods
Synthesis of enzyme nanocomplexes
The synthesis of DNA-inhibitor scaffolds was achieved by conjugating inhibitors to single-stranded DNA in designed sequences, followed by spontaneous assembly to form scaffolds. Enzymes were pre-conjugated with polymerizable acryloyl groups using N-hydroxysuccinimide ester, and complexed with the DNA scaffolds to form the enzyme complexes. The enzyme complexes were then encapsulated by in situ polymerization using acrylamide as the monomer, bis-methylacrylamide as the crosslinker, and ammonium persulphate/tetramethylethylenediamine as the initiator, at room temperature. The nanocomplexes were purified by dialysis against PBS and by size exclusion chromatography. The syntheses of single-enzyme nanocapsules—n(Cat), n(AOx) and n(GOx)—were achieved using a similar approach, in which polymer shells were grown directly around the pre-conjugated enzymes using the in situ polymerization technique. Detailed synthesis procedures and characterizations are provided in the Supplementary Information.
In vivo studies
All animals were treated in accordance with the Guide for Care and Use of Laboratory Animals, approved by local committee. Detailed procedures are listed as follows.
In alcohol antidote studies, male C57B6 mice (8–10 weeks old) from Jackson Laboratory were used. Eighteen mice were divided into six groups and fasted for 12 h before the studies. All mice were fed with an alcohol liquid diet at a dosage of 6 mg ethanol per gram bodyweight. The mice were intoxicated and fell asleep within 20 min of feeding. Thirty minutes after feeding, these intoxicated mice were tail-vein injected with PBS (control), native AOx, n(AOx), a mixture of n(AOx) and n(Cat), Lipo(AOx + Cat) and n(AOx-Cat), respectively. The dose of enzyme was maintained at ~65 μg AOx and ~21 μg Cat per mouse, and the volume of PBS or enzyme solutions was maintained at 150 μl per mouse. Blood samples (~10 μl) were taken from tails at 45 min, 90 min, 180 min and 300 min post-injection, and BACs were determined using an ethanol assay kit purchased from BioVision or Analox Instruments.
In alcohol prophylactic studies, 24 male mice (C57B6) were divided into six groups and gavaged with the alcohol diet at a dose of 6 mg ethanol per gram bodyweight. The doses of native AOx, n(Cat), n(AOx), a mixture of n(AOx) and n(Cat), or n(AOx–Cat) were maintained at 65 μg AOx and 21 μg Cat per animal. Another group of mice were fed with an isocaloric diet without ethanol and used as the control group. BACs were determined using the method described above. The levels of ALT were determined 6–8 h after alcohol gavage following methods described previously30.
In these in vivo studies, BACs were adjusted against the bodyweights of individual mice using the formula, BACadj = BACi × (BWTi/BWTave), where BACadj is the adjusted BAC, BACi and BWTi are the blood alcohol level and bodyweight of an individual mouse, and BWTave is the average weight of all mice in each set of experiments. Statistical analyses were performed using the Bonferroni post-test following a two-way ANOVA for BACs and the Newman–Keuls post-test following a one-way ANOVA for ALT data. P<0.05 or less was considered significant.
In assessing GOx-induced tissue damage in mice, male mice (C57B6) were anaesthetized using ketaime (85 mg per kg) combined with xylazine (10 mg per kg), intramuscularly injected into the posterior thighs. Hair on the backs of the animals was removed using an animal shaver. n(GOx), n(Cat) and n(GOx–Cat) were delivered into the skin via two cutaneous injections, at 0 and 24 h, with a dosage of 35 μg nanocapsules dispersed in 50 μl PBS buffer per group per injection. Equal volumes of H2O2 solution and PBS buffer were injected at different spots on the backs of the same animals, as positive and negative controls. The animals were anaesthetized 24 h following the second injection, and pieces of skin tissue were sampled, fixed and processed for H&E staining and fluorescent TUNEL staining using an in situ Cell Death Detection Kit from Roche. H&E images were captured with a Nikon Eclipse E600 microscope, and confocal fluorescence images were obtained with a Nikon PCM2000 confocal laser-scanning microscope using previous methods31,32.
Supplementary Material
Figure S1. Schematic illustration of the syntheses of ssDNA inhibitors for HRP (a), GOx (b), and Inv (c).
Figure S2. MALDI-TOF mass spectra of DNAs before (black) and after (red) 5′-modification with inhibitors. (a) ssDNA-1 and 4-Aminoantipyrine (4-AAP) modified ssDNA-1. (b) ssDNA-2 and glucosamine modified ssDNA-2. (c) ssDNA-I and glucosamine modified ssDNA-I. (d) ssDNA-II and 4-AAP modified ssDNA-II. (e) ssDNA-III and lactobionic acid modified ssDNA-III. (f) ssDNA-IV. As shown in the mass spectra (a–e), the molecular weights of ssDNAs increased after conjugating with enzyme inhibitors. Moreover, the increases between conjugated ssDNAs and their native counterparts are accordant with the molecular weights of corresponding enzyme inhibitors, respectively. (a. ΔMwssDNA-1 ≈ 200, Mw4-AAP = 203.4; b. ΔMwssDNA-2 ≈ 210, Mwgulcosamine = 180.1; c. ΔMwssDNA-I ≈ 200, Mwglucosamine = 180.1; d. ΔMwssDNA-II ≈ 250, Mw4-AAP = 203.4; e. ΔMwssDNA-III ≈ 400, Mwlactobionic acid = 358.3)
Figure S3. DNA-inhibitor scaffolds and DNA-enzymes complex for the syntheses of n(HRP-GOx) (a) and n(HRP-GOx-Inv) (b).
Figure S4. Elution curve of n(HRP-GOx)
Figure S5. Size distribution of PEG-lipo(AOx-Cat)
Figure S6. Size distribution of a. n(HRP-GOx) (~18 nm) (black) and n(HRP-GOx-Inv) (~37 nm)(red); b. n(GOx-Cat) (~17 nm); and c. n(AOx-Cat) (~24 nm).
Figure S7. Standard curve represents the relationship between absorbance and H2O2 concentration. Data represent average; errors come from three independent experiments performing triplicated.
Figure S8. Viability of the cells treated with EtOH in the presence of n(AOx-Cat), native AOx mixture of n(AOx) and n(Cat), or n(Cat).
Figure S9. Blood alcohol concentration (BAC) (a) and plasma alanine aminotransferase (ALT) (b) of mice gavaged with ethanol-containing diet followed by tail-vein injection of PBS (control), native AOx, n(AOx), n(Cat), mixture of n(AOx) and n(Cat), and n(AOx-Cat)). For mice treated with n(AOx-Cat), BAC decreases significantly before 3 hr (*P < 0.05; **P < 0.01, 4 mice used in each group), compared to the mice injected with PBS only. All the animals treated with alcohol show higher ALT levels; a tendency of lower ALT is also observed for the mice treated with n(AOx-Cat).
Figure S10. BAC of intoxicated mice injected with different doses of n(AOx-Cat) per mouse. Before the injection, mice were fasted for 12 hours and gavaged with the alcoholic liquid diet at 6 mg ethanol per gram of bodyweight. A series amount (30–240 μg AOx per mouse) of n(AOx-Cat) were then administrated to animals 30 min post alcohol feeding. The BAC was measured 75, 180, and 300 min after the injection of antidote. Due to a larger variation of animal weights in this experimental group of animals, the BAC was normalized with the body weight using the following equation: BACadj = BACi/[1 + ln2(BWTmax − BWTi)], where BWTmax is the highest bodyweight among this group of animals. All values are expressed as means ± s.e.m. unless otherwise indicated.
Figure S11. Confocal images of Kidney (a) Liver (b) and intestine (c) of animals treated with RhB-n(AOx) (red) and n(AOx) (no color).
Figure S12. Residual concentration of n(AOx) in the plasma of enzyme nanocomplex-treated mice.
Figure S13. CT, PET and PET/CT images of a mouse, injected with 18F-n(AOx-Cat). PET image were acquired with a 120-min dynamic PET scan started at 20 min post injection of 18F-ENs.
Figure S14. Residual concentration of 18F-n(AOx-Cat) in the organs of EN-treated mouse. Quantifications of 18F-n(AOx-Cat) in liver (red), kidney (blue), lung (black), and spleen (green) were achieved by monitoring the local radiation intensity with PET/CT.
Table S1. Sequences of DNA oligomers for the construction of DNA-enzyme complex
Table S2. Average number of acryloyl groups conjugated to the enzymes
Table S1. Kinetic parameters of HRP/GOx and HRP/GOx/Invertase free enzyme systems and their corresponding ENs System
Acknowledgments
This work was partially supported by the Defense Threat Reducing Agency (DTRA), the National Institutes of Health (NIH, grants R01AA018846 and R01AA018612), the National Natural Science Foundation of China (NSFC, grants 81025018, 91127045 and 50830103) and the National Basic Research Program of China (973 Program, no. 2011CB932500).
Footnotes
Supplementary information is available in the online version of the paper.
Reprints and permission information is available online at http://www.nature.com/reprints.
Author contributions
M.Y., L.S., W.C., O.Y., C.J. and Y. Lu conceived or designed the experiments. Y. Liu and J.D. performed the synthesis, characterization and data analysis. M. Lau, J.H., H.H. and J.L. performed the in vivo tests. S.L., W.W., X.Z. and H.W. performed the biodistribution and pharmacokinetic studies. Y. Liu, J.D., C.J. and Y. Lu co-wrote the paper. All authors discussed the results and commented on the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Schoffelen S, van Hest JCM. Multi-enzyme systems: bringing enzymes together in vitro. Soft Matter. 2012;8:1736–1746. [Google Scholar]
- 2.Conrado RJ, Varner JD, DeLisa MP. Engineering the spatial organization of metabolic enzymes: mimicking nature’s synergy. Curr Opin Biotechnol. 2008;19:492–499. doi: 10.1016/j.copbio.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Yan W, Aebersold R, Raines EW. Evolution of organelle-associated protein profiling. J Proteom. 2009;72:4–11. doi: 10.1016/j.jprot.2008.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumoto R, et al. A liposome-based energy conversion system for accelerating the multi-enzyme reactions. Phys Chem Chem Phys. 2010;12:13904–13906. doi: 10.1039/c0cp00556h. [DOI] [PubMed] [Google Scholar]
- 5.Van Dongen SFM, Nallani M, Cornelissen JJLM, Nolte RJM, van Hest JCM. A three-enzyme cascade reaction through positional assembly of enzymes in a polymersome nanoreactor. Chem Eur J. 2009;15:1107–1114. doi: 10.1002/chem.200802114. [DOI] [PubMed] [Google Scholar]
- 6.Wilner OI, et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nature Nanotech. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 7.Niemeyer CM, Koehler J, Wuerdemann C. DNA-directed assembly of bienzymic complexes from in vivo biotinylated NAD(P)H:FMN oxidoreductase and luciferase. ChemBioChem. 2002;3:242–245. doi: 10.1002/1439-7633(20020301)3:2/3<242::AID-CBIC242>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 8.Fierobe HP, et al. Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J Biol Chem. 2002;277:49621–49630. doi: 10.1074/jbc.M207672200. [DOI] [PubMed] [Google Scholar]
- 9.Kristensen C, et al. Metabolic engineering of dhurrin in transgenic Arabidopsis plants with marginal inadvertent effects on the metabolome and transcriptome. Proc Natl Acad Sci USA. 2005;102:1779–1784. doi: 10.1073/pnas.0409233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dueber JE, et al. Synthetic protein scaffolds provide modular control over metabolic flux. Nature Biotechnol. 2009;27:753–759. doi: 10.1038/nbt.1557. [DOI] [PubMed] [Google Scholar]
- 11.Wanders RJA, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 12.Schrader M, Fahimi HD. Mammalian peroxisomes and reactive oxygen species. Histochem Cell Biol. 2004;122:383–393. doi: 10.1007/s00418-004-0673-1. [DOI] [PubMed] [Google Scholar]
- 13.Sheikh FG, Pahan K, Khan M, Barbosa E, Singh I. Abnormality in catalase import into peroxisomes leads to severe neurological disorder. Proc Natl Acad Sci USA. 1998;95:2961–2966. doi: 10.1073/pnas.95.6.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scism RA, Bachmann BO. Five-component cascade synthesis of nucleotide analogues in an engineered self-immobilized enzyme aggregate. ChemBioChem. 2010;11:67–70. doi: 10.1002/cbic.200900620. [DOI] [PubMed] [Google Scholar]
- 15.Bäumler H, Georgieva R. Coupled enzyme reactions in multicompartment microparticles. Biomacromolecules. 2010;11:1480–1487. doi: 10.1021/bm1001125. [DOI] [PubMed] [Google Scholar]
- 16.Stempfer G, Höll-Neugebauer B, Kopetzki E, Rudolph R. A fusion protein designed for noncovalent immobilization: stability, enzymatic activity, and use in an enzyme reactor. Nature Biotechnol. 1996;14:481–484. doi: 10.1038/nbt0496-481. [DOI] [PubMed] [Google Scholar]
- 17.Yan M, et al. A novel intracellular protein delivery platform based on single-protein nanocapsules. Nature Nanotech. 2009;5:48–53. doi: 10.1038/nnano.2009.341. [DOI] [PubMed] [Google Scholar]
- 18.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nature Struct Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 19.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 20.Sherman MR, Saifer MGP, Perez-Ruiz F. PEG-uricase in the management of treatment-resistant gout and hyperuricemia. Adv Drug Deliv Rev. 2008;60:59–68. doi: 10.1016/j.addr.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Kehrer JP. Free radicals as mediators of tissue injury and disease. Crit Rev Toxicol. 1993;23:21–48. doi: 10.3109/10408449309104073. [DOI] [PubMed] [Google Scholar]
- 22.Cochrane J. Alcohol use in China. Alcohol Alcoholism. 2003;38:537–542. doi: 10.1093/alcalc/agg111. [DOI] [PubMed] [Google Scholar]
- 23.Lee K, Møller L, Hardt F, Haubek A, Jensen E. Alcohol-induced brain damage and liver damage in young males. Lancet. 1979;2:759–761. doi: 10.1016/s0140-6736(79)92113-5. [DOI] [PubMed] [Google Scholar]
- 24.Jamaty C, et al. Lipid emulsions in the treatment of acute poisoning: a systematic review of human and animal studies. Clin Toxicol. 2010;48:1–27. doi: 10.3109/15563650903544124. [DOI] [PubMed] [Google Scholar]
- 25.Bertrand N, Bouvet C, Moreau P, Leroux J-C. Transmembrane pH-gradient liposomes to treat cardiovascular drug intoxication. ACS Nano. 2010;4:7552–7558. doi: 10.1021/nn101924a. [DOI] [PubMed] [Google Scholar]
- 26.Shen Y, et al. Dihydromyricetin as a novel anti-alcohol intoxication medication. J Neurosci. 2012;32:390–401. doi: 10.1523/JNEUROSCI.4639-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martins S, Sarmento B, Ferreira DC, Souto EB. Lipid-based colloidal carriers for peptide and protein delivery—liposomes versus lipid nanoparticles. Int J Nanomed. 2007;2:595–607. [PMC free article] [PubMed] [Google Scholar]
- 28.Averbakh AZ, et al. Flavin-dependent alcohol oxidase from the yeast Pichia pinus. Spatial localization of the coenzyme FAD in the protein structure: hot-tritium bombardment and ESR experiments. Biochem J. 1995;310:601–604. doi: 10.1042/bj3100601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barry RE, McGivan JD. Acetaldehyde alone may initiate hepatocellular damage in acute alcoholic liver disease. Gut. 1985;26:1065–1069. doi: 10.1136/gut.26.10.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488–1499. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- 31.Ji C, Deng Q, Kaplowitz N. Role of TNF-α in ethanol-induced hyperhomocysteinemia and murine alcoholic liver injury. Hepatology. 2004;40:442–451. doi: 10.1002/hep.20309. [DOI] [PubMed] [Google Scholar]
- 32.Ji C, et al. Liver-specific loss of glucose-regulated protein 78 perturbs the unfolded protein response and exacerbates a spectrum of liver diseases in mice. Hepatology. 2011;54:229–239. doi: 10.1002/hep.24368. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Schematic illustration of the syntheses of ssDNA inhibitors for HRP (a), GOx (b), and Inv (c).
Figure S2. MALDI-TOF mass spectra of DNAs before (black) and after (red) 5′-modification with inhibitors. (a) ssDNA-1 and 4-Aminoantipyrine (4-AAP) modified ssDNA-1. (b) ssDNA-2 and glucosamine modified ssDNA-2. (c) ssDNA-I and glucosamine modified ssDNA-I. (d) ssDNA-II and 4-AAP modified ssDNA-II. (e) ssDNA-III and lactobionic acid modified ssDNA-III. (f) ssDNA-IV. As shown in the mass spectra (a–e), the molecular weights of ssDNAs increased after conjugating with enzyme inhibitors. Moreover, the increases between conjugated ssDNAs and their native counterparts are accordant with the molecular weights of corresponding enzyme inhibitors, respectively. (a. ΔMwssDNA-1 ≈ 200, Mw4-AAP = 203.4; b. ΔMwssDNA-2 ≈ 210, Mwgulcosamine = 180.1; c. ΔMwssDNA-I ≈ 200, Mwglucosamine = 180.1; d. ΔMwssDNA-II ≈ 250, Mw4-AAP = 203.4; e. ΔMwssDNA-III ≈ 400, Mwlactobionic acid = 358.3)
Figure S3. DNA-inhibitor scaffolds and DNA-enzymes complex for the syntheses of n(HRP-GOx) (a) and n(HRP-GOx-Inv) (b).
Figure S4. Elution curve of n(HRP-GOx)
Figure S5. Size distribution of PEG-lipo(AOx-Cat)
Figure S6. Size distribution of a. n(HRP-GOx) (~18 nm) (black) and n(HRP-GOx-Inv) (~37 nm)(red); b. n(GOx-Cat) (~17 nm); and c. n(AOx-Cat) (~24 nm).
Figure S7. Standard curve represents the relationship between absorbance and H2O2 concentration. Data represent average; errors come from three independent experiments performing triplicated.
Figure S8. Viability of the cells treated with EtOH in the presence of n(AOx-Cat), native AOx mixture of n(AOx) and n(Cat), or n(Cat).
Figure S9. Blood alcohol concentration (BAC) (a) and plasma alanine aminotransferase (ALT) (b) of mice gavaged with ethanol-containing diet followed by tail-vein injection of PBS (control), native AOx, n(AOx), n(Cat), mixture of n(AOx) and n(Cat), and n(AOx-Cat)). For mice treated with n(AOx-Cat), BAC decreases significantly before 3 hr (*P < 0.05; **P < 0.01, 4 mice used in each group), compared to the mice injected with PBS only. All the animals treated with alcohol show higher ALT levels; a tendency of lower ALT is also observed for the mice treated with n(AOx-Cat).
Figure S10. BAC of intoxicated mice injected with different doses of n(AOx-Cat) per mouse. Before the injection, mice were fasted for 12 hours and gavaged with the alcoholic liquid diet at 6 mg ethanol per gram of bodyweight. A series amount (30–240 μg AOx per mouse) of n(AOx-Cat) were then administrated to animals 30 min post alcohol feeding. The BAC was measured 75, 180, and 300 min after the injection of antidote. Due to a larger variation of animal weights in this experimental group of animals, the BAC was normalized with the body weight using the following equation: BACadj = BACi/[1 + ln2(BWTmax − BWTi)], where BWTmax is the highest bodyweight among this group of animals. All values are expressed as means ± s.e.m. unless otherwise indicated.
Figure S11. Confocal images of Kidney (a) Liver (b) and intestine (c) of animals treated with RhB-n(AOx) (red) and n(AOx) (no color).
Figure S12. Residual concentration of n(AOx) in the plasma of enzyme nanocomplex-treated mice.
Figure S13. CT, PET and PET/CT images of a mouse, injected with 18F-n(AOx-Cat). PET image were acquired with a 120-min dynamic PET scan started at 20 min post injection of 18F-ENs.
Figure S14. Residual concentration of 18F-n(AOx-Cat) in the organs of EN-treated mouse. Quantifications of 18F-n(AOx-Cat) in liver (red), kidney (blue), lung (black), and spleen (green) were achieved by monitoring the local radiation intensity with PET/CT.
Table S1. Sequences of DNA oligomers for the construction of DNA-enzyme complex
Table S2. Average number of acryloyl groups conjugated to the enzymes
Table S1. Kinetic parameters of HRP/GOx and HRP/GOx/Invertase free enzyme systems and their corresponding ENs System