Abstract
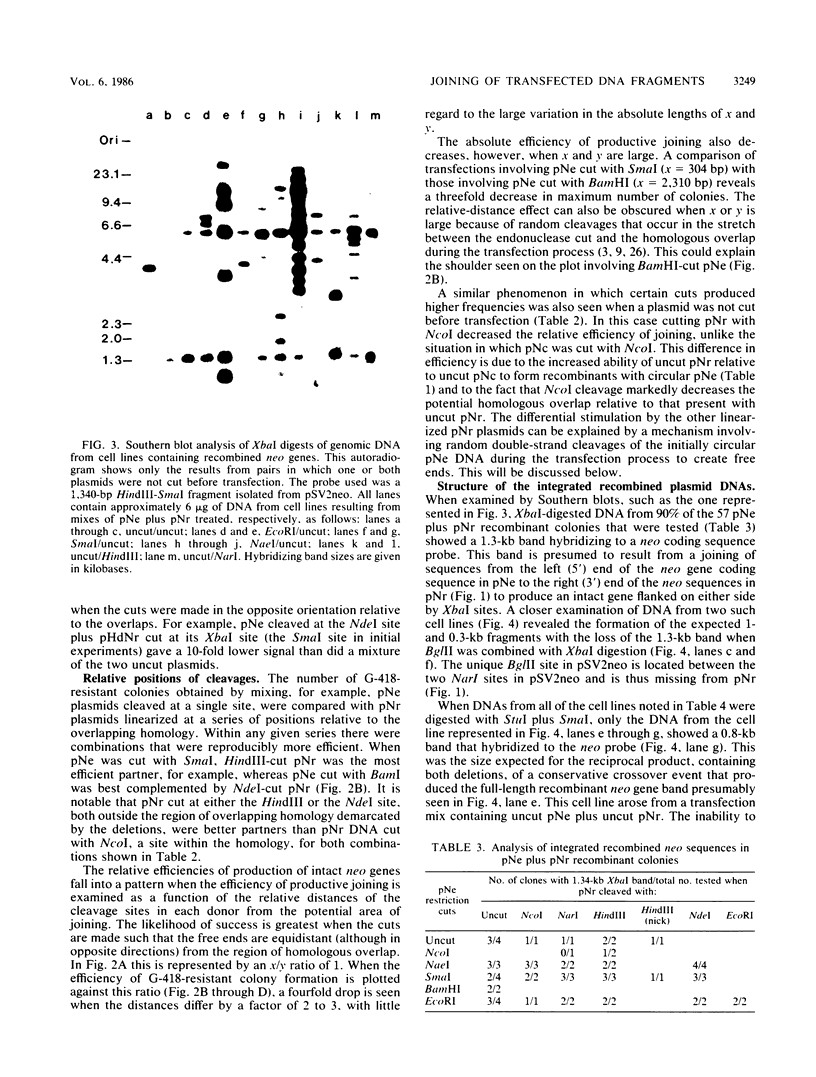
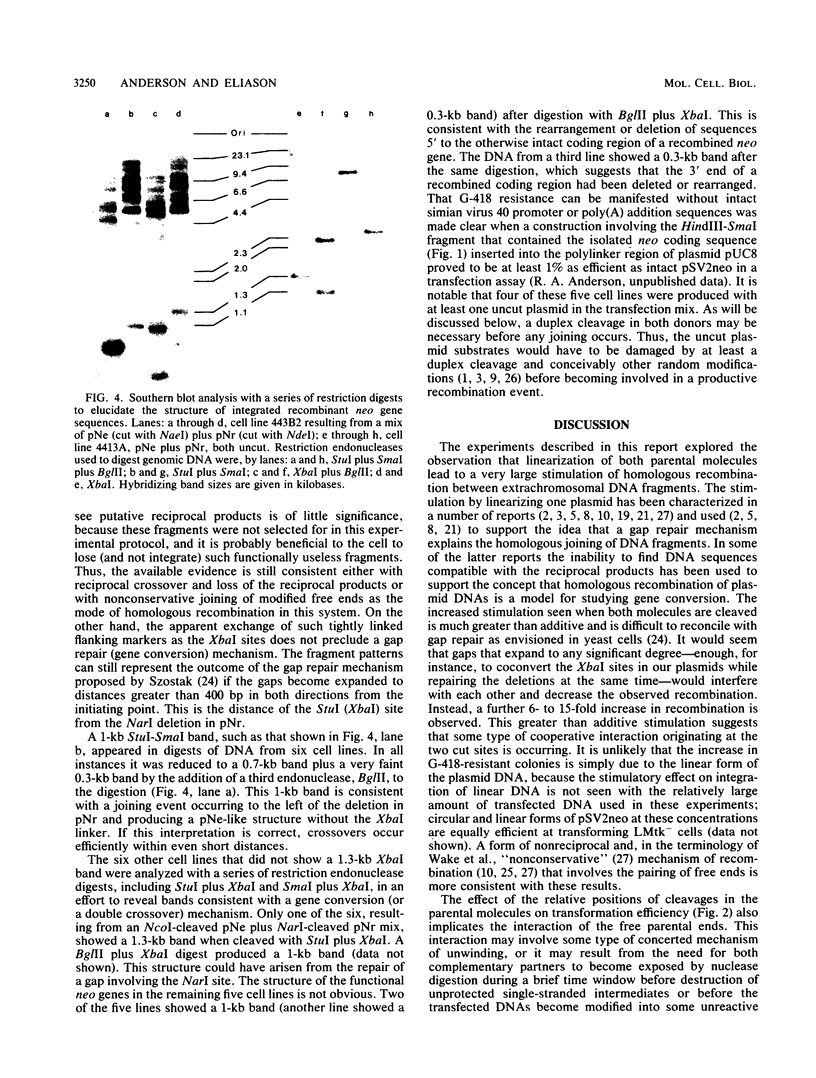
The mechanism by which double-strand cleavages stimulate the joining of plasmid DNA fragments introduced into cultured mammalian cells was investigated by cotransfecting pairs of plasmids encoding deletion mutations in a dominant selectable gene into LMtk- cells. Plasmid recombination substrates were produced by creating deletions of different sizes within the neo coding region of the pSV2neo plasmid. Complementing pairs of deleted plasmid DNAs were linearized at specific unique sites before cotransfection into mouse LMtk- cells by the calcium phosphate precipitation method. Cleaving one donor plasmid produced a 4- to 10-fold stimulation in the production of colonies able to survive in medium containing G-418. The linearization of the second plasmid further increased the efficiency by another factor of 6 to 15 when the cut was made on the opposite side of the homology, approximately equidistant from the center of the overlap. Fifty-seven individual G-418-resistant colonies representing the products of individual crosses were isolated, and the genomic DNAs containing the presumably integrated, functional recombinant neo genes were analyzed on Southern blots. A band consistent with the exchange of markers flanking the neo gene was present in 90% of the DNAs examined. In only one case was the pattern indicative of either a double crossover or a gene conversion event. These results support the idea that homologous extrachromosomal DNA fragments are joined through annealing of overlapping single-stranded ends. This DNA-joining phenomenon may represent the activity of cellular DNA repair enzymes; its relationship to genetic recombination occurring at the chromosomal level remains to be determined.
Full text
PDF
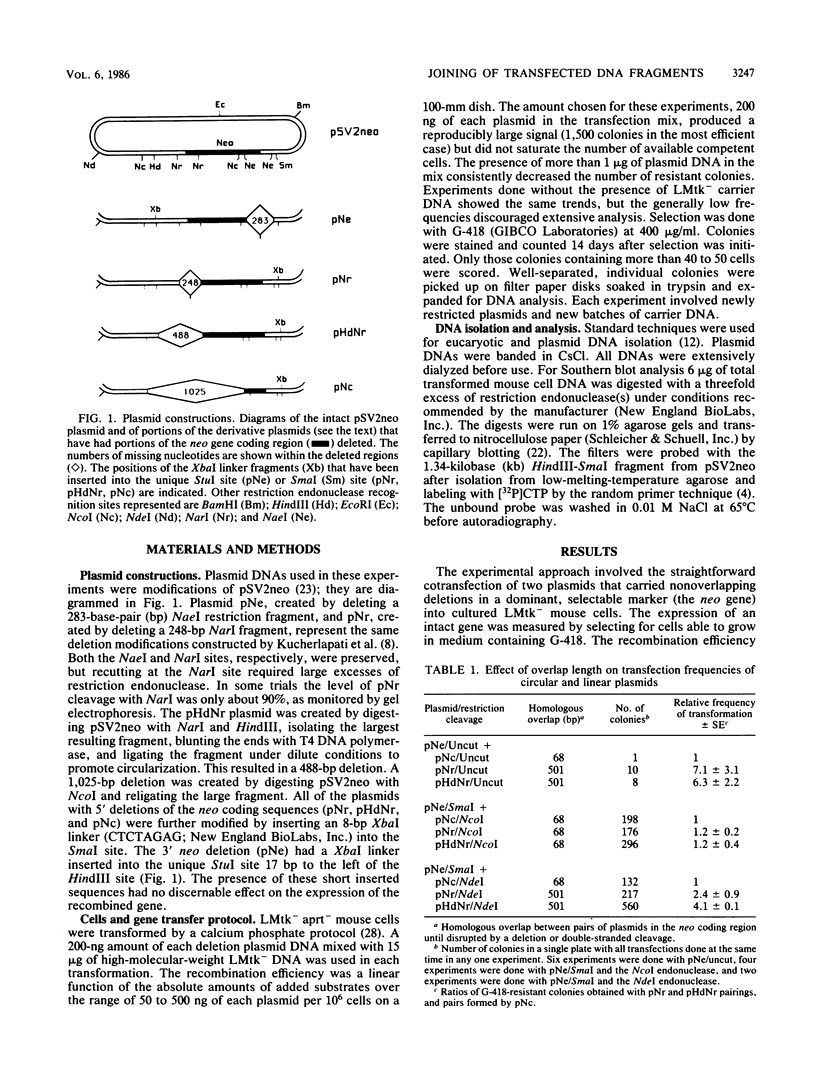
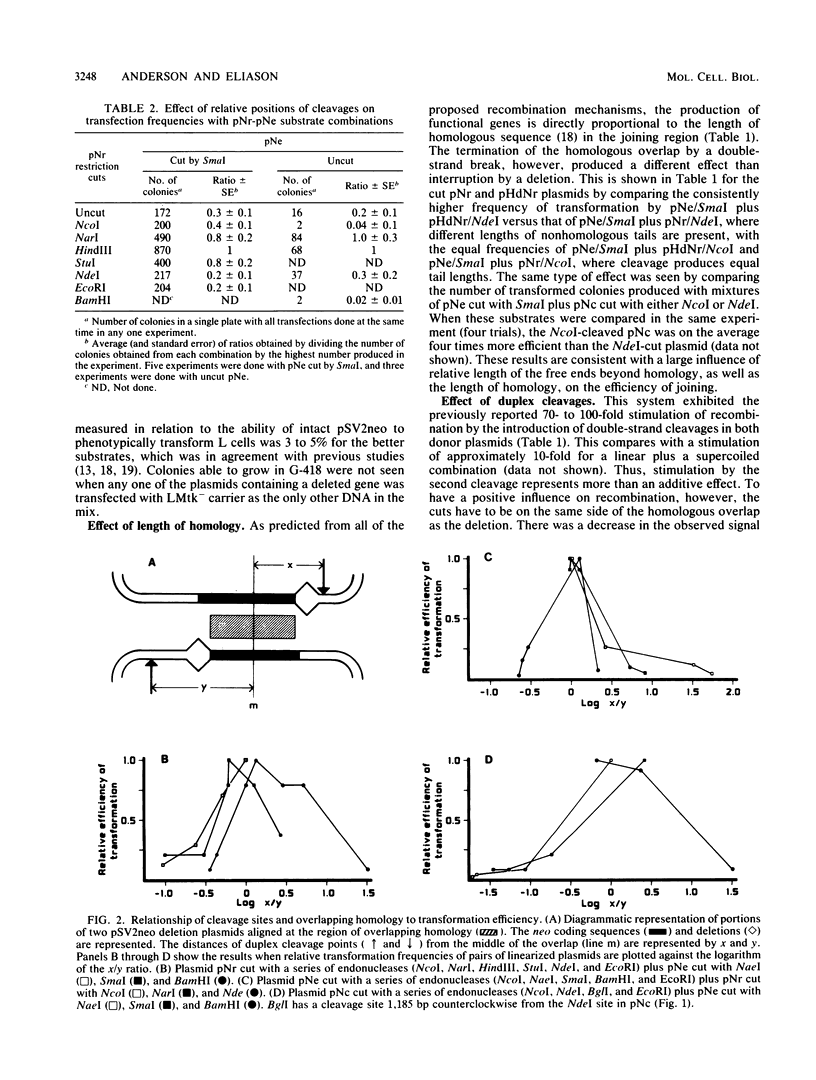


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brenner D. A., Kato S., Anderson R. A., Smigocki A. C., Camerini-Otero R. D. The recombination and integration of DNAs introduced into mouse L cells. Cold Spring Harb Symp Quant Biol. 1984;49:151–160. doi: 10.1101/sqb.1984.049.01.018. [DOI] [PubMed] [Google Scholar]
- Brenner D. A., Smigocki A. C., Camerini-Otero R. D. Effect of insertions, deletions, and double-strand breaks on homologous recombination in mouse L cells. Mol Cell Biol. 1985 Apr;5(4):684–691. doi: 10.1128/mcb.5.4.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Joffe S., Seidman M. M. Recombination and deletion of sequences in shuttle vector plasmids in mammalian cells. Mol Cell Biol. 1985 Sep;5(9):2265–2271. doi: 10.1128/mcb.5.9.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Folger K. R., Thomas K., Capecchi M. R. Nonreciprocal exchanges of information between DNA duplexes coinjected into mammalian cell nuclei. Mol Cell Biol. 1985 Jan;5(1):59–69. doi: 10.1128/mcb.5.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., de Villiers J., Weber F., Schaffner W. High frequency of homologous recombination in mammalian cells between endogenous and introduced SV40 genomes. Cell. 1985 Dec;43(3 Pt 2):695–703. doi: 10.1016/0092-8674(85)90242-9. [DOI] [PubMed] [Google Scholar]
- Kopchick J. J., Stacey D. W. Differences in intracellular DNA ligation after microinjection and transfection. Mol Cell Biol. 1984 Feb;4(2):240–246. doi: 10.1128/mcb.4.2.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucherlapati R. S., Eves E. M., Song K. Y., Morse B. S., Smithies O. Homologous recombination between plasmids in mammalian cells can be enhanced by treatment of input DNA. Proc Natl Acad Sci U S A. 1984 May;81(10):3153–3157. doi: 10.1073/pnas.81.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebkowski J. S., DuBridge R. B., Antell E. A., Greisen K. S., Calos M. P. Transfected DNA is mutated in monkey, mouse, and human cells. Mol Cell Biol. 1984 Oct;4(10):1951–1960. doi: 10.1128/mcb.4.10.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F. L., Sperle K., Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984 Jun;4(6):1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopata M. A., Cleveland D. W., Sollner-Webb B. High level transient expression of a chloramphenicol acetyl transferase gene by DEAE-dextran mediated DNA transfection coupled with a dimethyl sulfoxide or glycerol shock treatment. Nucleic Acids Res. 1984 Jul 25;12(14):5707–5717. doi: 10.1093/nar/12.14.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. K., Temin H. M. High-efficiency ligation and recombination of DNA fragments by vertebrate cells. Science. 1983 May 6;220(4597):606–609. doi: 10.1126/science.6301012. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Lebkowski J. S., Greisen K. S., Calos M. P. Specificity of mutations induced in transfected DNA by mammalian cells. EMBO J. 1984 Dec 20;3(13):3117–3121. doi: 10.1002/j.1460-2075.1984.tb02267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund M., Luthman H., Nilsson S. V., Magnusson G. Ethidium-bromide-inhibited restriction endonucleases cleave one strand of circular DNA. Gene. 1982 Nov;20(1):121–125. doi: 10.1016/0378-1119(82)90093-2. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Porter T. N., Wilson J. H. Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol. 1985 Oct;5(10):2599–2607. doi: 10.1128/mcb.5.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci U S A. 1985 May;82(10):3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. Rapid assay for extrachromosomal homologous recombination in monkey cells. Mol Cell Biol. 1985 Mar;5(3):529–537. doi: 10.1128/mcb.5.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz J., Subramani S. The minimum amount of homology required for homologous recombination in mammalian cells. Mol Cell Biol. 1984 Nov;4(11):2253–2258. doi: 10.1128/mcb.4.11.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J., Scangos G. Recombination during gene transfer into mouse cells can restore the function of deleted genes. Science. 1983 Jan 14;219(4581):174–176. doi: 10.1126/science.6294829. [DOI] [PubMed] [Google Scholar]
- Song K. Y., Chekuri L., Rauth S., Ehrlich S., Kucherlapati R. Effect of double-strand breaks on homologous recombination in mammalian cells and extracts. Mol Cell Biol. 1985 Dec;5(12):3331–3336. doi: 10.1128/mcb.5.12.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Southern P. J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J Mol Appl Genet. 1982;1(4):327–341. [PubMed] [Google Scholar]
- Szostak J. W., Orr-Weaver T. L., Rothstein R. J., Stahl F. W. The double-strand-break repair model for recombination. Cell. 1983 May;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Volkert F. C., Young C. S. The genetic analysis of recombination using adenovirus overlapping terminal DNA fragments. Virology. 1983 Feb;125(1):175–193. doi: 10.1016/0042-6822(83)90072-7. [DOI] [PubMed] [Google Scholar]
- Wake C. T., Gudewicz T., Porter T., White A., Wilson J. H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984 Mar;4(3):387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake C. T., Vernaleone F., Wilson J. H. Topological requirements for homologous recombination among DNA molecules transfected into mammalian cells. Mol Cell Biol. 1985 Aug;5(8):2080–2089. doi: 10.1128/mcb.5.8.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Pellicer A., Silverstein S., Axel R. Biochemical transfer of single-copy eucaryotic genes using total cellular DNA as donor. Cell. 1978 Jul;14(3):725–731. doi: 10.1016/0092-8674(78)90254-4. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Berget P. B., Pipas J. M. Somatic cells efficiently join unrelated DNA segments end-to-end. Mol Cell Biol. 1982 Oct;2(10):1258–1269. doi: 10.1128/mcb.2.10.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]