Highlights
► The reticulon family is involved in shaping and bending the endoplasmic reticulum. ► Reticulons interact with proteins of several positive-strand RNA viruses. ► Reticulons are involved in forming the replication compartments of brome mosaic virus. ► We hypothesize on the role of reticulons in forming the replication compartments of other RNA viruses.
Abstract
Positive-strand RNA [(+)RNA] viruses are responsible for numerous human, animal, and plant diseases. Because of the limiting coding capacity of (+)RNA viruses, their replication requires a complex orchestration of interactions between the viral genome, viral proteins and exploited host factors. To replicate their genomic RNAs, (+)RNA viruses induce membrane rearrangements that create membrane-linked RNA replication compartments. Along with substantial advances on the ultrastructure of the membrane-bound RNA replication compartments, recent results have shed light into the role that host factors play in rearranging these membranes. This review focuses on recent insights that have driven a new understanding of the role that the membrane-shaping host reticulon homology domain proteins (RHPs) play in facilitating the replication of various (+)RNA viruses.
Current Opinion in Microbiology 2012, 15:519–524
This review comes from a themed issue on Host–microbe interactions: Viruses
Edited by Marco Vignuzzi
For a complete overview see the Issue and the Editorial
Available online 21st May 2012
1369-5274/$ – see front matter, © 2012 Elsevier Ltd. All rights reserved.
Introduction
(+)RNA viruses reorganize intracellular membranes to assemble their RNA replication compartments, which are intricate factories or mini-organelles featuring the close association of both viral and host components [1]. Cellular lipid synthesis as well as appropriate lipid composition is required for viral replication, implying that the membrane is an essential, functional component of the RNA replication complex [2]. Pioneering ultrastructural studies using three-dimensional imaging by electron microscope tomography (EMT) have substantially enhanced the understanding of the RNA replication compartments induced by (+)RNA viruses [3, 4, 5, 6]. As noted below, various (+)RNA viruses induce membrane rearrangements with distinct morphologies. Nevertheless, in general, the membrane is thought to provide a surface to help localize and increase the local concentration of components required for viral replication [1], protect dsRNA replication intermediates and viral proteins from sensors of the innate immune system [7], and in some cases provide host factors that help target viral proteins or genomic RNAs to the appropriate membrane [8].
Host factors have been shown to be involved in every step of viral life cycles [9•, 10, 11, 12]. In keeping with this, recent results show that host factors also are key determinants in forming viral RNA replication compartments, including by generating appropriate lipid composition of the membranes [2, 13, 14•, 15••, 16, 17] and by structurally inducing/supporting relevant membrane curvature [17, 18, 19••]. Since genomic RNA replication is a highly conserved step in the viral life cycle, understanding the role that such host factors play in the formation and function of the viral-induced RNA replication compartments could provide a target to develop broad-spectrum antivirals.
Reticulon homology domain proteins
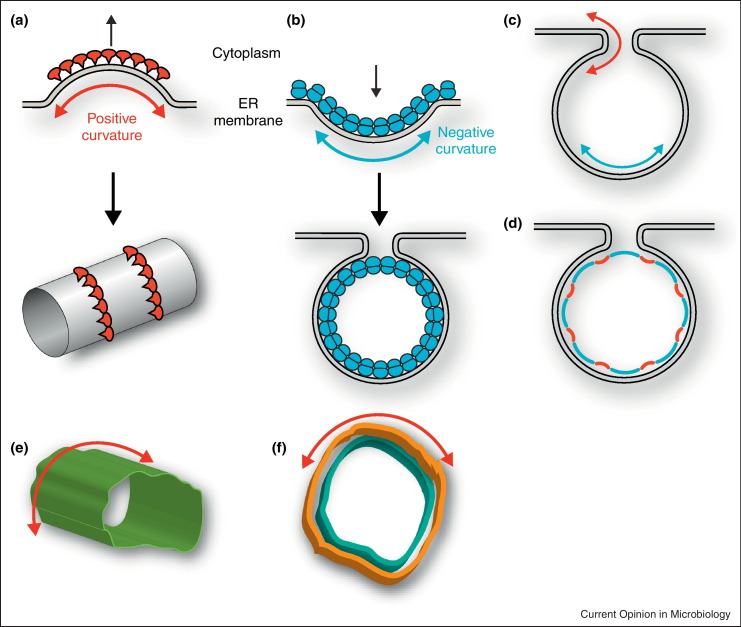
The reticulons are a group of morphogenic, ER membrane-shaping proteins that partition to and stabilize highly curved ER membrane tubules [20••], and as such are candidates for host factors that may affect the generation and/or maintenance of virus-induced membrane rearrangements. The diversity of the reticulon gene family varies among hosts, with four reticulon genes in mammals (RTN1-4), two genes in yeast (RTN1 and RTN2), and 21 and 17 genes, respectively, in the plants Arabidopsis thaliana and Oryza sativa [21, 22, 23, 24•]. A related protein family that is also necessary for ER tubule formation consists of six mammalian DP1/REEP proteins and yeast Yop1 [20••, 25]. The reticulon and DP1 families share little overall homology but do exhibit an important common structural feature—two elongated hydrophobic segments that appear to form partially membrane-spanning hairpin domains [20••, 26, 27]. For simplicity, for the rest of this review we will refer to the reticulon and DP1 families as the reticulon homology domain proteins (RHPs). The bulk of the hydrophobic portions of the RHPs is predicted to reside in the outer leaflet of the phospholipid bilayer, thus inducing curvature by hydrophobic wedging [20••]. The oligomerization of the RHPs is important both for their ability to localize to and to form tubular ER domains, suggesting that such scaffolding might also have a role in curvature induction and stabilization [28]. Consistent with their role in tubule formation, RHPs primarily partition to and stabilize peripheral ER membrane tubules while avoiding the low-curvature ER domains of the nuclear envelope and peripheral ER sheets [20••, 29]. Such ER tubules are said to have positive membrane curvature — that is, curved to protrude into the cytoplasm (Figure 1a).
Figure 1.
Positive and negative curvature found within viral induced replication compartments. (a) RHPs (red) induce curvature by protruding the membrane towards the cytoplasm to form tubules. (b) BMV replicase protein 1a (blue) forms spherules by invaginating the membrane away from the cytoplasm. (c) In viral induced vesicles that remain connected to the parental membrane, the area around the neck has positive curvature while the body of the vesicle has negative curvature. (d) Model for potential role of RHPs in formation and/or maintenance of BMV induced spherules. 1a (blue arc) might induce and stabilize negative curvature in the vesicle body whereas RHPs (red arcs) would partially cancel negative curvature at the body to allow expansion of the vesicle and stabilize the spherule neck. The poliovirus membranous replication complexes found in early (e) and late (f) stages of development both have positive curvature. For (f), cyan and orange indicate inner and outer membrane of double-membrane structures.
(d) was adapted from [16].
Role of RHPs in brome mosaic virus RNA replication complex formation
The replication cycle of brome mosaic virus (BMV), a small alphavirus-like virus, has been well characterized, making BMV an advantageous model to study some of the general features of (+)RNA virus RNA replication [30]. EM shows that for BMV, RNA synthesis localizes to 50–80 nm vesicular compartments, also known as spherules, in which the outer perinuclear ER membrane is invaginated away from the cytoplasm and into the ER lumen [31]. The interior of these vesicles is connected to the cytoplasm through a neck-like opening, providing a channel for ribonucleotide import and product RNA export. The only BMV protein necessary to induce spherule formation is replication factor 1a [31]. The self-interacting 1a protein is membrane associated, localizes to spherule interiors, and is present at high enough levels to imply that it must form a shell covering much of the spherule interior [31, 32, 33]. Such a rigid protein shell could contribute substantially to forming and maintaining the high-energy membrane deformation state of the spherules (Figure 1b). However, in addition to 1a, RHPs also were recently shown to play key roles in BMV RNA replication compartment formation and function [19••].
Using a loss-of-function experimental approach, RHPs were shown to be required for efficient BMV RNA replication in yeast [19••]. In keeping with the altered RNA replication phenotype, BMV 1a interacts with and incorporates RHPs into the spherule interior, and deleting all three yeast RHPs abolishes 1a-induced spherule formation [19••]. Topologically, the spherule body has negative curvature in that the membrane of the vesicle sunshine is curved away from the cytoplasm, although the neck has positive curvature and can be thought of as a tubule cut in half (Figure 1c). RHPs polymerize into short arcs to form perfect cylindrical tubes [34] and are involved in forming nuclear pores [35], which are topologically equivalent to spherule necks. Thus, RHPs are likely necessary for both spherule formation and maintenance of an open channel to the cytoplasm to facilitate import of RNA templates and nucleotides and export of progeny RNA (Figure 1d). In addition to stabilizing spherule necks, a subset of RHPs must also be incorporated within the vesicular portion of spherules as partial RHP depletion results in spherules with reduced diameter [19••]. In this instance, the positively curving RHPs might partially cancel the negative membrane curvature induced by 1a, allowing expansion of the spherule body (Figure 1d). Although RHPs are involved in shaping smooth ER [36], the site of many lipid synthesis steps [37], RHP deletion did not alter fatty acid levels or composition, suggesting that their main contribution to RNA synthesis is in RNA replication compartment formation. Further supporting this notion, RHP-GFP fusions retain 1a interaction but shift 1a-induced membrane rearrangements from spherules to double-membrane layers of ER surrounding the nucleus [19••].
Role of RHPs in Picornaviridae RNA replication
The importance of RHPs in (+)RNA virus replication was first demonstrated when human reticulon 3 (RTN3) was found to interact with the 2C proteins of poliovirus, coxsackievirus A16 (CA16) and enterovirus 71 (EV71) [38]. Moreover, immunoprecipitation showed that EV71 2C protein interacts with the reticulon homology domain of RTN1C, RTN2, and RTN4. siRNA knockdown of RTN3 resulted in inhibited synthesis of viral double-stranded RNA (dsRNA) and proteins in EV71-infected cells [38]. Ectopic expression of a form of RTN3 that was not degraded by siRNA in siRTN3 knockdowns rescued EV71 infectivity, suggesting that RTN3 plays a key role in EV71 replication. Although ultrastructural studies were not performed, immunofluorescence showed that endogenous RTN3 colocalized not only with EV71 2C but also with dsRNA, implying that RTN3 is likely associated with the viral replication compartment [38]. The 2C protein is one of the most highly conserved viral proteins among all picornaviruses and poliovirus 2C has been proposed to be involved in the formation of the vesicle-associated replication compartments [39].
Because of the limitations of conventional, two-dimensional electron microscopy, the RNA replication structures induced by poliovirus and other enteroviruses had been previously described as either single-membrane [40] or double-membrane vesicles (DMVs) [41]. However, EM tomograms of enterovirus-infected cells showed that, during the exponential phase of viral RNA synthesis, closed smooth single-membrane tubules constitute the predominant virus-induced membrane structure, whereas DMVs become increasingly abundant at the expense of these tubules as infection progresses [42•]. Similar observations have been made in poliovirus-infected cells. Early in infection, poliovirus induces the formation of single-membrane connecting and branching tubular compartments [43•]. As infection progresses, the tubular compartments gradually transform into double-membrane structures [43•]. Unlike the negatively curved spherular compartments induced by BMV, the enterovirus71-induced and poliovirus-induced tubule-like and vesicle-like structures are both positively curved (Figure 1e and f). Thus, RHPs may play a role in forming the single-membrane tubules, analogous to their normal role in shaping smooth ER [36], whereas later on they would stabilize the positively curved DMVs. The transformation from single-membrane tubules to DMVs would require both close apposition of the inner tubule membrane with a source for the outer membrane and the induction of curvature to wrap this outer membrane around the tubules, which is another role that RHPs might be involved in.
Potential RHP role in forming replication compartments of other (+)RNA viruses
A yeast-two hybrid system used to screen a liver cDNA library showed that RTN3 interacts with the NS4B protein of hepatitis C virus (HCV) [44]. When expressed individually or in the context of the entire HCV polyprotein, NS4B localizes to the ER [45]. Moreover, NS4B is a key organizer of the HCV RNA replication compartment by inducing the formation of ER-derived membranous vesicles, which accumulate in large cytosolic clusters, referred to as the membranous web [46]. Although the role that RTN3 plays in HCV RNA replication was not investigated further in these studies, given the particular functions of NS4B in inducing HCV RNA replication compartments, it is tempting to speculate that RTN3 might be involved in modulating the formation of the membranous web.
Dengue virus (DENV) infection induces several distinct ER-derived membrane structures, including vesicle packets (VPs) and convoluted membranes (CMs) [5, 47]. EM tomography shows that the inner vesicles of VPs are invaginations of the outer ER membrane [5], thus, the inner vesicle interiors are connected to the cytoplasm by neck-like pores similar to those in the spherules induced by BMV and many other (+)RNA viruses. Likewise, SARS coronavirus-infected cells contain a mixture of VP and CM rearrangements [4]. However, tomographic reconstructions failed to reveal any membrane openings between the SARS virus-induced DMV interiors and the cytoplasm, as was observed for BMV and DENV, suggesting that the DMVs might be more related to the DMVs formed by poliovirus. On the basis of such ultrastructural data, alpha-like and some flavi-like viruses appear to induce structures with negative membrane curvature along the dome of the vesicle [5], with areas of positive curvature along the neck, while picorna-like and coronavirus-like viruses induce structures with positive membrane curvature [4, 42•, 43•]. Despite the topological differences, one common theme might be the role of RHPs in forming the various RNA replication compartments, perhaps but not necessarily limited to those viruses that use membranes derived from the ER.
Concluding remarks
With RHPs now identified as playing crucial roles in the replication of multiple (+)RNA viruses, the stage is set to identify not only additional participating host factors, but also the detailed molecular mechanisms of how different viruses orchestrate the varied and often complex membrane rearrangements associated with their replication processes. Moreover, the role of RHPs in (+)RNA replication aside from genome amplification remains to be explored, as do the potential interactions of RHPs with other cellular pathways that contribute to virus replication, such as factors that regulate membrane synthesis/composition [2], trafficking, and membrane remodeling. Although mainly found in the ER, RHPs also localize to the Golgi and plasma membrane, implying a potential role in membrane trafficking [48]. Along these lines, poliovirus vesicle formation has been linked to COPI vesicle trafficking [49], tombusvirus (TBSV) recruitment of another set of membrane-shaping proteins ESCRT (endosomal sorting complexes required for transport) proteins apparently facilitates the assembly of TBSV-induced spherules [18] and enteroviruses and flaviviruses exploit host phospholipid-modifying enzyme PI4KIIIβ and replicate their respective viral RNA on phosphatidylinositol-4-phosphate lipid-enriched membranes [50••]. Cryo-EM tomography and super resolution imaging should help to resolve the three-dimensional organization at molecular resolution of viral and host proteins that are part of replication compartments and to enhance our understanding of the role that RHPs and other host factors play in forming the membrane-associated RNA replication compartments.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
Research related to these topics in the authors’ laboratory was supported by NIH Grant GM35072. P.A. is an HHMI investigator. A.D. was partially supported by NIH training grants T32 GM07215 and T32 AI078985.
References
- 1.den Boon J.A., Ahlquist P. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol. 2010;64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 2.Heaton N.S., Randall G. Multifaceted roles for lipids in viral infection. Trends Microbiol. 2011;19:368–375. doi: 10.1016/j.tim.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopek B.G., Perkins G., Miller D.J., Ellisman M.H., Ahlquist P. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 2007;5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fontana J., Lopez-Iglesias C., Tzeng W.P., Frey T.K., Fernandez J.J., Risco C. Three-dimensional structure of Rubella virus factories. Virology. 2010;405:579–591. doi: 10.1016/j.virol.2010.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen S., Thomsen A.R. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J Virol. 2012;86:2900–2910. doi: 10.1128/JVI.05738-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamanaka T., Ohta T., Takahashi M., Meshi T., Schmidt R., Dean C., Naito S., Ishikawa M. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc Natl Acad Sci U S A. 2000;97:10107–10112. doi: 10.1073/pnas.170295097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Nagy P.D., Pogany J. The dependence of viral RNA replication on co-opted host factors. Nat Rev Microbiol. 2011;10:137–149. doi: 10.1038/nrmicro2692. [DOI] [PMC free article] [PubMed] [Google Scholar]; Review highlighting the role that host factors play in the different steps of the cycle of positive-strand RNA viruses.
- 10.Coyne C.B., Bozym R., Morosky S.A., Hanna S.L., Mukherjee A., Tudor M., Kim K.S., Cherry S. Comparative RNAi screening reveals host factors involved in enterovirus infection of polarized endothelial monolayers. Cell Host Microbe. 2011;9:70–82. doi: 10.1016/j.chom.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sessions O.M., Barrows N.J., Souza-Neto J.A., Robinson T.J., Hershey C.L., Rodgers M.A., Ramirez J.L., Dimopoulos G., Yang P.L., Pearson J.L. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–1050. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tai A.W., Benita Y., Peng L.F., Kim S.S., Sakamoto N., Xavier R.J., Chung R.T. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Diaz A., Hao L., Gancarz B., den Boon J.A., Ahlquist P. Intersection of the multivesicular body pathway and lipid homeostasis in RNA replication by a positive-strand RNA virus. J Virol. 2011;85:5494–5503. doi: 10.1128/JVI.02031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Perera R., Riley C., Isaac G., Hopf-Jannasch A.S., Moore R.J., Weitz K.W., Pasa-Tolic L., Metz T.O., Adamec J., Kuhn R.J. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012;8:e1002584. doi: 10.1371/journal.ppat.1002584. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors used high resolution mass spectrometry to show that DENV causes significant changes to the membrane organization of infected cells and that there was a selective enrichment of lipids that influence membrane structure.
- 15••.Heaton N.S., Perera R., Berger K.L., Khadka S., Lacount D.J., Kuhn R.J., Randall G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc Natl Acad Sci U S A. 2010;107:17345–17350. doi: 10.1073/pnas.1010811107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Elegant study that combined RNAi analysis with pharmacological inhibitors to show that DENV co-opts the fatty acid biosynthetic pathway to establish its replication complexes, providing mechanistic insight into DENV membrane remodeling.
- 16.Reiss S., Rebhan I., Backes P., Romero-Brey I., Erfle H., Matula P., Kaderali L., Poenisch M., Blankenburg H., Hiet M.S. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J., Diaz A., Mao L., Ahlquist P., Wang X. Host acyl coenzyme a binding protein regulates replication complex assembly and activity of a positive-strand RNA virus. J Virol. 2012;86:5110–5121. doi: 10.1128/JVI.06701-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barajas D., Jiang Y., Nagy P.D. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 2009;5:e1000705. doi: 10.1371/journal.ppat.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Diaz A., Wang X., Ahlquist P. Membrane-shaping host reticulon proteins play crucial roles in viral RNA replication compartment formation and function. Proc Natl Acad Sci U S A. 2010;107:16291–16296. doi: 10.1073/pnas.1011105107. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study describes for the first time the role of the reticulons in shaping the membrane-bound replication compartments of a positive-strand RNA virus.
- 20••.Voeltz G.K., Prinz W.A., Shibata Y., Rist J.M., Rapoport T.A. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]; Comprehensive study demonstrating that the tubular ER is shaped by two families of curvature-stabilizing proteins, the reticulons and DP1/Yop1.
- 21.Nziengui H., Schoefs B. Functions of reticulons in plants: what we can learn from animals and yeasts. Cell Mol Life Sci. 2009;66:584–595. doi: 10.1007/s00018-008-8373-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y.S., Strittmatter S.M. The reticulons: a family of proteins with diverse functions. Genome Biol. 2007;8:234. doi: 10.1186/gb-2007-8-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 2010;22:1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24•.Di Sano F., Bernardoni P., Piacentini M. The reticulons: guardians of the structure and function of the endoplasmic reticulum. Exp Cell Res. 2012 doi: 10.1016/j.yexcr.2012.03.002. in press. [DOI] [PubMed] [Google Scholar]; Review highlighting the involvement of the reticulons in several cellular processes beyond their structural role in the ER.
- 25.Park S.H., Zhu P.P., Parker R.L., Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010;120:1097–1110. doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zurek N., Sparks L., Voeltz G. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 2011;12:28–41. doi: 10.1111/j.1600-0854.2010.01134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolley N., Sparkes I., Craddock C.P., Eastmond P.J., Runions J., Hawes C., Frigerio L. Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 2010;64:411–418. doi: 10.1111/j.1365-313X.2010.04337.x. [DOI] [PubMed] [Google Scholar]
- 28.Shibata Y., Voss C., Rist J.M., Hu J., Rapoport T.A., Prinz W.A., Voeltz G.K. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Craene J.O., Coleman J., Estrada de Martin P., Pypaert M., Anderson S., Yates J.R., 3rd, Ferro-Novick S., Novick P. Rtn1p is involved in structuring the cortical endoplasmic reticulum. Mol Biol Cell. 2006;17:3009–3020. doi: 10.1091/mbc.E06-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noueiry A.O., Ahlquist P. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Ann Rev Phytopathol. 2003;41:77–98. doi: 10.1146/annurev.phyto.41.052002.095717. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz M., Chen J., Janda M., Sullivan M., den Boon J., Ahlquist P. A positive-strand RNA virus replication complex parallels form and function of retrovirus capsids. Mol Cell. 2002;9:505–514. doi: 10.1016/s1097-2765(02)00474-4. [DOI] [PubMed] [Google Scholar]
- 32.Diaz A., Gallei A., Ahlquist P. Bromovirus RNA replication compartment formation requires concerted action of 1a's self-interacting RNA capping and helicase domains. J Virol. 2012;86:821–834. doi: 10.1128/JVI.05684-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.den Boon J.A., Chen J., Ahlquist P. Identification of sequences in Brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J Virol. 2001;75:12370–12381. doi: 10.1128/JVI.75.24.12370-12381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J., Shibata Y., Voss C., Shemesh T., Li Z., Coughlin M., Kozlov M.M., Rapoport T.A., Prinz W.A. Membrane proteins of the endoplasmic reticulum induce high-curvature tubules. Science. 2008;319:1247–1250. doi: 10.1126/science.1153634. [DOI] [PubMed] [Google Scholar]
- 35.Dawson T.R., Lazarus M.D., Hetzer M.W., Wente S.R. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol. 2009;184:659–675. doi: 10.1083/jcb.200806174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shibata Y., Shemesh T., Prinz W.A., Palazzo A.F., Kozlov M.M., Rapoport T.A. Mechanisms determining the morphology of the peripheral ER. Cell. 2010;143:774–788. doi: 10.1016/j.cell.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumann O., Walz B. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol. 2001;205:149–214. doi: 10.1016/s0074-7696(01)05004-5. [DOI] [PubMed] [Google Scholar]
- 38.Tang W.F., Yang S.Y., Wu B.W., Jheng J.R., Chen Y.L., Shih C.H., Lin K.H., Lai H.C., Tang P., Horng J.T. Reticulon 3 binds the 2C protein of enterovirus 71 and is required for viral replication. J Biol Chem. 2007;282:5888–5898. doi: 10.1074/jbc.M611145200. [DOI] [PubMed] [Google Scholar]
- 39.Teterina N.L., Gorbalenya A.E., Egger D., Bienz K., Ehrenfeld E. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J Virol. 1997;71:8962–8972. doi: 10.1128/jvi.71.12.8962-8972.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bienz K., Egger D., Rasser Y., Bossart W. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology. 1983;131:39–48. doi: 10.1016/0042-6822(83)90531-7. [DOI] [PubMed] [Google Scholar]
- 41.Schlegel A., Giddings T.H., Jr., Ladinsky M.S., Kirkegaard K. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J Virol. 1996;70:6576–6588. doi: 10.1128/jvi.70.10.6576-6588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Limpens R.W., van der Schaar H.M., Kumar D., Koster A.J., Snijder E.J., van Kuppeveld F.J., Barcena M. The transformation of enterovirus replication structures: a three-dimensional study of single- and double-membrane compartments. mBio. 2011;2:e00166-11. doi: 10.1128/mBio.00166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Used electron tomography to resolve the three-dimensional architecture of enterovirus-induced membraneous compartments and their transformation over time.
- 43•.Belov G.A., Nair V., Hansen B.T., Hoyt F.H., Fischer E.R., Ehrenfeld E. Complex dynamic development of poliovirus membranous replication complexes. J Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ultrastructural characterization of the dynamic changes in the membrane rearrangements induced by poliovirus at different time points postinfection. Branching tubular compartments seen early in infection transform into double-membrane vesicles in the late stages of replication.
- 44.Liu Y., Cheng J., Bai G.Q., Yan F.M., Wu S.H., Wang L., Zhang L.X. Screening and cloning of hepatitis C virus non-structural protein 4B interacting protein gene in hepatocytes. Zhonghua shi yan he lin chuang bing du xue za zhi = Zhonghua shiyan he linchuang bingduxue zazhi = Chin J Exp Clin Virol. 2005;19:248–251. [PubMed] [Google Scholar]
- 45.Hugle T., Fehrmann F., Bieck E., Kohara M., Krausslich H.G., Rice C.M., Blum H.E., Moradpour D. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 2001;284:70–81. doi: 10.1006/viro.2001.0873. [DOI] [PubMed] [Google Scholar]
- 46.Egger D., Wolk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mackenzie J.M., Jones M.K., Young P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 48.Wakana Y., Koyama S., Nakajima K., Hatsuzawa K., Nagahama M., Tani K., Hauri H.P., Melancon P., Tagaya M. Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem Biophys Res Commun. 2005;334:1198–1205. doi: 10.1016/j.bbrc.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 49.Rust R.C., Landmann L., Gosert R., Tang B.L., Hong W., Hauri H.P., Egger D., Bienz K. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J Virol. 2001;75:9808–9818. doi: 10.1128/JVI.75.20.9808-9818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Hsu N.Y., Ilnytska O., Belov G., Santiana M., Chen Y.H., Takvorian P.M., Pau C., van der Schaar H., Kaushik-Basu N., Balla T. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that enterovirus preferentially recruits phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ) to generate membranes rich in phosphatidylinositol-4-phosphate (PI4P) lipids, a key process regulating viral RNA replication.