Abstract
lin-4 (miR-125) microRNAs are deeply conserved across animal phylogeny. They are among the first microRNAs discovered and were implicated in regulating developmental timing in C. elegans. In this study, we report that mutations in lin-4 microRNA specifically suppress the AVM axon guidance defects in slt-1 mutants through enhancement of netrin attraction. lin-4 expression in AVM neurons rescued lin-4 mutant phenotypes in AVM axon guidance, suggesting that lin-4 acts cell autonomously in AVM to inhibit its axon attraction to netrin. lin-4 is expressed strongly in the AVM neuron when its axon guidance is underway, and is almost undetectable in the ALM neuron whose axon guidance is netrin independent. The transcription factor lin-14, a necessary target of lin-4 microRNA in the AVM neurons, stimulates netrin-mediated AVM axon ventral guidance. lin-14’s positive effect in netrin attraction is mediated by the receptor unc-40 (DCC) and its cofactor madd-2 functioning through a combination of the unc-34 (Ena) and ced-10 (Rac1)-dependent downstream pathways. lin-14 stimulates netrin-mediated axon attraction in part by enhancing UNC-40 protein expression. Our study indicates that the eventual down-regulation of lin-14 activity in the AVM neurons at the end of axon guidance is caused by lin-4 microRNA inhibition.
Introduction
During development of the nervous system, the C. elegans anterior ventral microtubule axon (AVM) is guided to the ventral midline by two cues, the UNC-6 (netrin) attractant recognized by the UNC-40 (DCC) receptor and the SLT-1 (slit) repellent recognized by the SAX-3 (robo) receptor. Upon reaching the ventral midline, the AVM axon moves anteriorly to the nerve ring (1), a neuropil generally viewed as the animal’s brain. Axons are attracted to targets, but upon arriving they must switch their responsiveness to guidance cues at the targets such that they are no longer sensitive to axon cues but rather can proceed with synapse formation. Thus, AVM axons are less able to respond to UNC-6 (netrin) as their development advances. This developmental loss of responsiveness to the axon guidance cue, in this case netrin, might be accounted for by either extrinsic inhibitory signals or intrinsic neuronal aging.
Founding members of the microRNA family were first described in C. elegans (2–4). Since then, microRNAs have been shown to be involved in such diverse events as development, metabolism, apoptosis, cell fate determination, and cancer formation. MicroRNAs down-regulate gene expression by interacting with partially complementary sequences in the 3′UTRs of their target genes (2, 3). The C. elegans genome encodes hundreds of microRNAs, about 30% of which are conserved within animals, suggesting ancient functions (5). Many of the recently discovered microRNAs are spatially expressed in the nervous system, such as lsy-6 and mir-273 (6, 7), implying an important role for microRNAs in spatial pattern formation in the nervous system. Others are temporally expressed during development of the nervous system, including lin-4 (miR-125) and let-7 (8–10), raising the possibility that microRNAs may regulate neuronal responsiveness to axon guidance cues. Although microRNAs have been clearly shown to specify left-right neuronal asymmetry and midbrain-hindbrain boundary (6, 7, 11), their roles in other aspects of neuronal development are largely unknown. Here, we report the first example of microRNA regulation of axon guidance during formation of neuronal circuit. We discovered an unexpected role for the lin-4 microRNA in axon guidance through genetic analysis of a well-characterized axon guidance event, the growth of the AVM axon to the ventral midline in C. elegans. Our results show that lin-4 microRNA functions as a potent and specific negative regulator of Netrin-mediated attraction, without effect on Slit-mediated repulsion, and provide strong evidence for lin-4 functioning through the lin-14 transcription factor in the AVM neuron to inhibit its axon attraction to Netrin. We believe, at the end of axon guidance, lin-4 microRNA signals AVM neurons to undergo a developmental loss of responsiveness to Netrin through negative regulation of LIN-14 transcription factor, leading to down-regulation of UNC-40 receptor signaling.
Results
A microRNA pathway inhibits AVM axon ventral guidance potentially through negative regulation of unc-6 (netrin) signaling
Dicer family of RNase III-related enzymes are responsible for processing most if not all premature microRNAs. In C. elegans, three nearly identical Argonaute proteins ALG-1, ALG-2, and RDE-1 were found to copurify with DCR-1 (Dicer) biochemically (12). Like Dicer, ALG-1 and ALG-2 are required for the maturation of microRNAs but only for a subgroup of them (12, 13). To ask whether any of these microRNAs regulates axon guidance, we examined AVM axon ventral guidance in alg-1, alg-2, or dcr-1 mutants.
AVM axon guidance provides a sensitive, powerful system for genetically probing how attractive and repulsive guidance signals are dynamically regulated during development. In C. elegans, ventral guidance of the AVM axon relies upon the combined action of an attractive UNC-6 (netrin) guidance cue from the ventral pole and a repulsive SLT-1 (Slit) guidance cue from the dorsal pole (Fig. 1A). Studying AVM axon guidance therefore presents an opportunity to evaluate the specific contribution of genes to either netrin or Slit guidance pathways. Because there are two parallel guidance pathways for AVM axon ventral guidance, genetic manipulations that potentiate signaling through one guidance pathway (such as ventral netrin) might suppress the defects caused by loss of the other guidance pathway (such as dorsal slit). Our analysis reveals several clues that may suggest how microRNA pathways regulate axon guidance. First, dcr-1 mutation did not affect AVM axon ventral guidance, which may be due to a simultaneous knockout of microRNAs with opposite functions in AVM axon guidance (Fig. 1C). Second, alg-2 mutant affected both netrin and slt-1-mediated axon guidance, suggesting that alg-2 mutant effects on axon guidance may not be very specific (Fig. 1C). Third, alg-1 mutation suppressed slt-1 mutant phenotypes in AVM axon ventral guidance, consistent with the model that alg-1 mutation specifically enhances netrin-mediated axon attraction, which compensates for loss-of-function in slt-1 (Fig. 1C). However it is formally possible that additional attractive forces are also enhanced in alg-1; slt-1 mutants.
Fig. 1. Elimination of alg-1 enhances netrin-dependent axon attraction, suppressing AVM axon ventral guidance defects in slt-1 mutants.
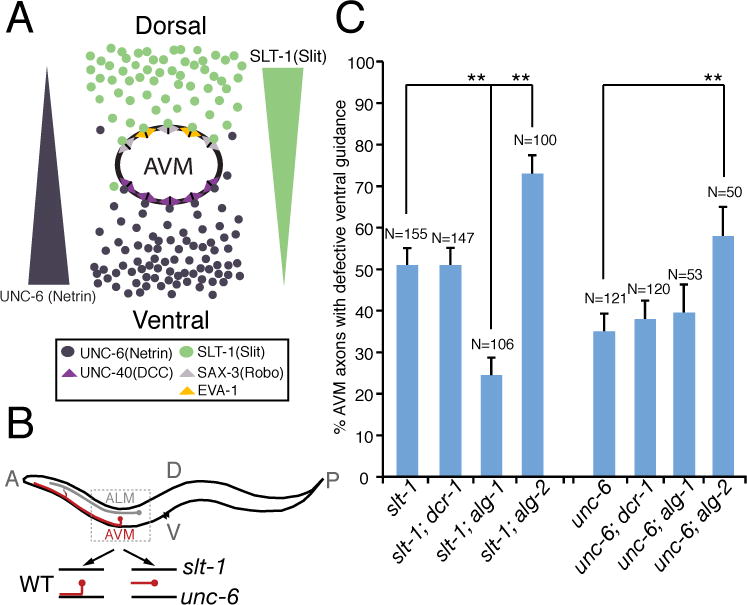
(A) Schematic drawing highlights signaling pathways that influence AVM axon ventral guidance in development. (B) Schematic diagram of wild-type and mutant AVM axons. (C) alg-1 mutation specifically suppressed AVM axon ventral guidance defects in slt-1 (slit) but not unc-6 (netrin) mutants. alg-1(gk214) and alg-2(ok304) used in this study are null alleles. dcr-1(mg375) is a reduction-of-function allele. Error bars represent standard error of the proportion. Asterisks and brackets represent P < 0.001 by Z-test for two proportions.
The effect of alg-1 mutation on AVM axon guidance may be attributed to lin-4 (miR-125) microRNA inactivation in AVM neurons
alg-1 is required for biogenesis of many microRNAs (13). We postulated that the maturation of one or more microRNAs that depends on alg-1, inhibits netrin-mediated AVM axon ventral attraction; and furthermore that this microRNA is likely to be expressed early in development in the first larval stage in order to regulate netrin-mediated AVM axon ventral attraction. Stem-loop RT-PCR was used to globally survey mature microRNA expression in C. elegans development. Spatial constraint of the stem-loop RT primer discriminates among related microRNAs that differ by as little as one nucleotide (14, Fig. 2A). Among 83 microRNAs we surveyed (Fig. 2B), including those with homologues in vertebrates, we identified 4 microRNAs (lin-4, mir-71, mir-83, and mir-90) whose abrupt initiation of expression in the first larval stage coincides with the timing of the AVM axon ventral guidance (Fig. 2B and fig. S1A). Among the 4 early-onset microRNAs we identified, only lin-4 and mir-90 are expressed in the AVM neurons (Fig. 3C–O and fig. S1B–D). For those early-onset microRNAs that are expressed in the AVM neurons, only maturation of lin-4 microRNA is affected in alg-1 mutants (Fig. 3P and fig. S1E). lin-4 encodes a heterochronic microRNA that is temporally expressed during C. elegans development (15). The rapid increase of lin-4 expression in L1 larvae is due to up-regulation of lin-4 expression in many tissues including muscles, hypodermal cells, and neurons in the head, the tail, the ventral nerve cord, the anterior body, and the mid-body regions (Fig. 3B, C). lin-4 microRNA binds to the 3′UTR of its target, lin-14, preventing its translation and permitting stage two of larval cell fate to occur (8). Mutations that reduce lin-4 or enhance lin-14 gene activities delay developmental timing of cells at the L1–L2 transition. These previous findings point to the possibility that lin-4 mutation may prolong or enhance L1-stage AVM cell fate, resulting in enhancement of axon guidance signaling in AVM neurons. In our stem-loop RT-PCR assay, mature lin-4 microRNA is almost undetectable in the embryonic stage (fig. S1A). However, its expression was dramatically increased at the first larval stage and persisted into adulthood (Fig. 2B). Similarly, a lin-4 promoter reporter became highly expressed in the AVM neurons from the late-first larval stage onwards (Fig. 3D–O, 7A). The expression of this lin-4 promoter reporter is much stronger in the AVM neurons, whose axon guidance is netrin dependent, than in the ALM neurons, whose axon guidance is netrin independent (Fig. 3D–O).
Fig. 2. Global survey of microRNA expression in C. elegans development.
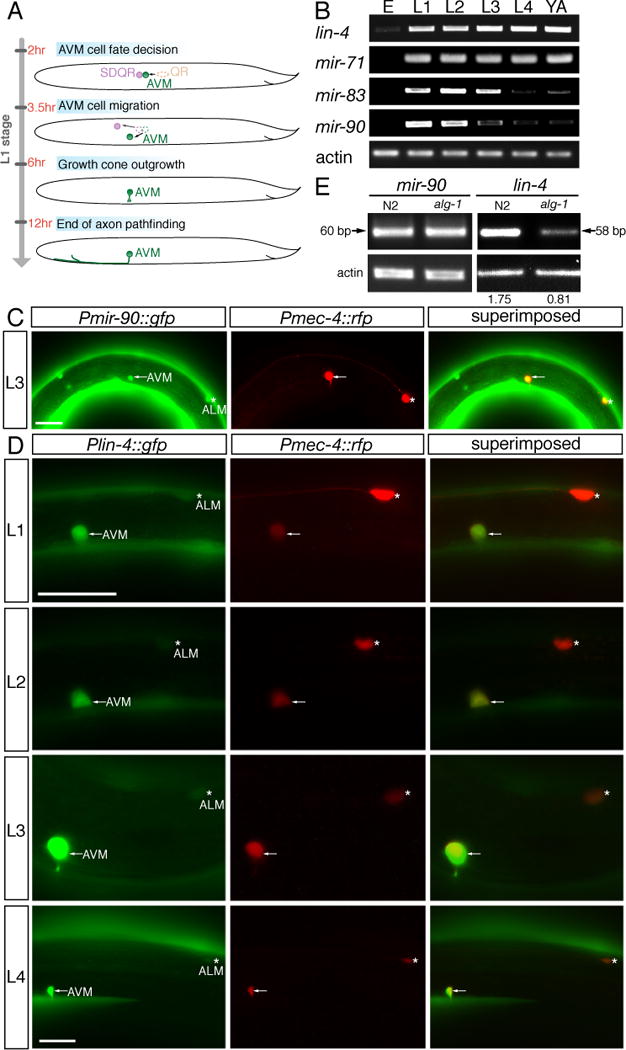
(A) Quantification of microRNAs involves two steps, stem-loop RT and conventional PCR. The stem-loop RT primer binds to the 3′ portion of the mature microRNA and is reverse transcribed with reverse transcriptase. Subsequently, the RT product is quantified using conventional PCR that includes a microRNA-specific forward primer and an universal reverse primer. The forward primer is tailed at 5′ end to increase Tm. (B) Stem-loop RT-PCR analysis of RNA isolated from populations of staged animals. Equal amount of RNA preparation from staged N2 animals was used in the RT-PCR amplification of microRNA and actin transcripts. E, L1, L2, L3, L4, and YA indicate embryonic, the first larval, the second larval, the third larval, the forth larval, and the young adult stages, respectively. MicroRNAs were clustered based on timing of their half-maximal expression.
Fig. 3. Fluorescent protein promoter reporters for identifying neuronal expression of lin-4 and stem-loop RT-PCR for detecting specifically the mature lin-4 microRNA.
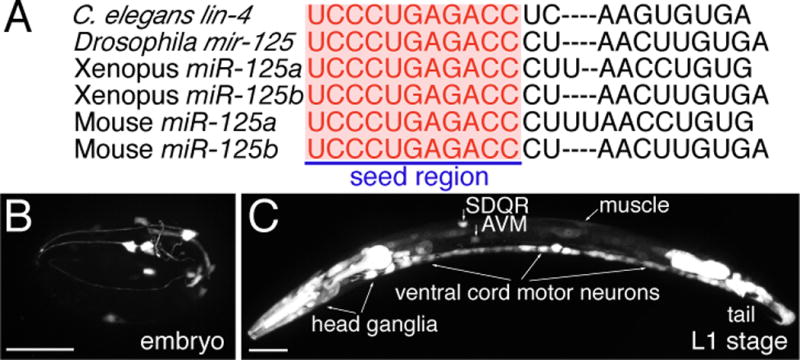
(A) Sequence alignment of C. elegans lin-4 microRNA with Drosophila melanogaster miR-125, Xenopus miR-125a and miR-125b, and Mouse miR-125a and miR-125b. Letters in red highlight identical sequences at the 5′ seed region, which is important for target mRNA recognition. (B–O) Expression of lin-4 microRNA in neurons. (B) A late embryo. (C) A late L1-stage animal, whole body. Anterior body in a late L1-stage animal (D–F), a L2-stage animal (G–I), a L3-stage animal (J–L), and a L4-stage animal (M–O). A 1.9-kb lin-4 promoter drives GFP expression in the distinct cells (B, C, D, G, J, and M). Limited lin-4 expression in the late embryonic stage in a few head neurons. Broader lin-4 expression in the late L1 stage in many neurons in the head, the body, and the tail. mec-4::dsred reporter was used to label the AVM neurons (E, H, K, and N). The superimposed images identify the GFP expressing cells as the AVM neurons (F, I, L, and O). Asterisk indicates the ALM neuron. Anterior is to left, dorsal up. Scale bar, 20 μm. (P) Measuring mature lin-4 microRNA level by stem-loop RT-PCR in alg-1 mutants versus wild-type (N2) animals. Equal amount of RNA preparation from either N2 or alg-1 mutants was used in the stem-loop RT-PCR reaction for amplifying lin-4 transcripts. Comparable amounts of actin transcripts amplified from the same RNA preparations from N2 and alg-1 mutants serve as an internal control. Value shown above each lane indicates amount of lin-4 normalized by actin.
Fig. 7. Dynamic regulation of lin-14 3′UTR activity during AVM axon guidance in the L1 stage.
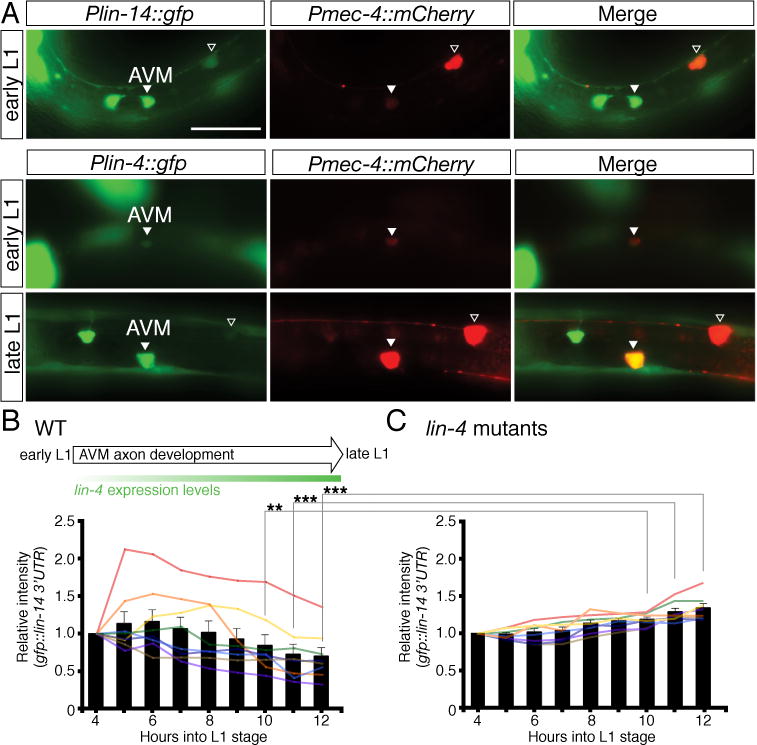
(A) Temporal expression of Plin-14::GFP and Plin-4::GFP reporters in the AVM neurons. An early L1-stage animal (top panel) showing lin-14 strongly expressed in AVM. An early L1-stage animal (middle panel) showing lin-4 weakly expressed in AVM and a late L1-stage animal (lower panel) showing lin-4 strongly expressed in AVM. The Pmec-4::mCherry reporter was used to label the AVM neurons. The merged images identify the GFP expressing cells as the AVM neurons. Arrowhead indicates the AVM neuron and the open arrowhead marks the ALM neuron. Anterior is to left, dorsal up. Scale bar, 20 μm. The expression intensity of the GFP::lin-14 3′UTR sensor in the AVM neurons was measured every hour at the first larval stage, starting at 4 hours after hatching and ending at 12 hours after hatching, in wild-type animals (B) and lin-4 mutants (C). Eight animals each were measured for wild type and lin-4 mutants. Bars represent the average intensity. ** and *** indicate intensity between wild type and lin-4 mutants is significantly different at P < 0.01 and P < 0.001, respectively. P values were calculated using a Student’s t-Test.
lin-4 microRNA is homologous to vertebrate miR-125a and miR-125b with the differences located only in the central region (Fig. 3A), a region believed to be looped out during target mRNA recognition (16). miR-125a and miR-125b have been previously implicated in neuronal development as they were first isolated from brain tissues in mice (16). Over-expression of miR-125, like lin-4, can specifically repress the expression of a mec-4::GFP::lin-14 3′UTR sensor containing seven complementary mir-125 binding sites in the AVM neurons (fig. S2 and S3).
lin-4 microRNA inhibits unc-6 (netrin) signaling during AVM axon guidance
The lin-4(e912) mutation used in this study is a loss-of-function null allele. We found that loss of lin-4 function recapitulates the alg-1 mutant phenotypes in AVM axon ventral guidance. lin-4(e912) mutation, like alg-1 mutation, alone does not display abnormal AVM axon ventral guidance. However, lin-4(e912) mutation, like alg-1 mutation, enhances netrin-mediated axon attraction, suppressing the AVM axon guidance defect in slt-1 (slit) but not unc-6 (netrin) null mutants (Fig. 1B, 1C, 4A, 4B). Recent studies by Culotti’s group identified EVA-1 as a SAX-3 (Robo) co-receptor, which binds to SLT-1 and SAX-3 on the cell surface (17). EVA-1 and SAX-3 are both required for slt-1-mediated AVM axon repulsion. However, different from EVA-1 being acting strictly as a SLT-1 receptor, SAX-3 (Robo) can also inhibit or silence UNC-40 (DCC) signaling in the absence of SLT-1. To further determine the specificity of lin-4’s function as a regulator of netrin pathway, we tested genetic interactions between lin-4 and eva-1 and between lin-4 and sax-3 null mutants. We found that lin-4 mutation significantly suppresses eva-1 mutant phenotypes as expected (Fig. 4B). However, lin-4 mutation does not suppress sax-3 mutant phenotypes (Fig. 4B). These results suggest that lin-4’s function in netrin-mediated AVM axon guidance depends on sax-3 and that lin-4 and sax-3 may inhibit UNC-40 (DCC) signaling through a similar mechanism.
Fig. 4. Enhancing netrin-mediated axon attraction by a lin-4 microRNA mutation.
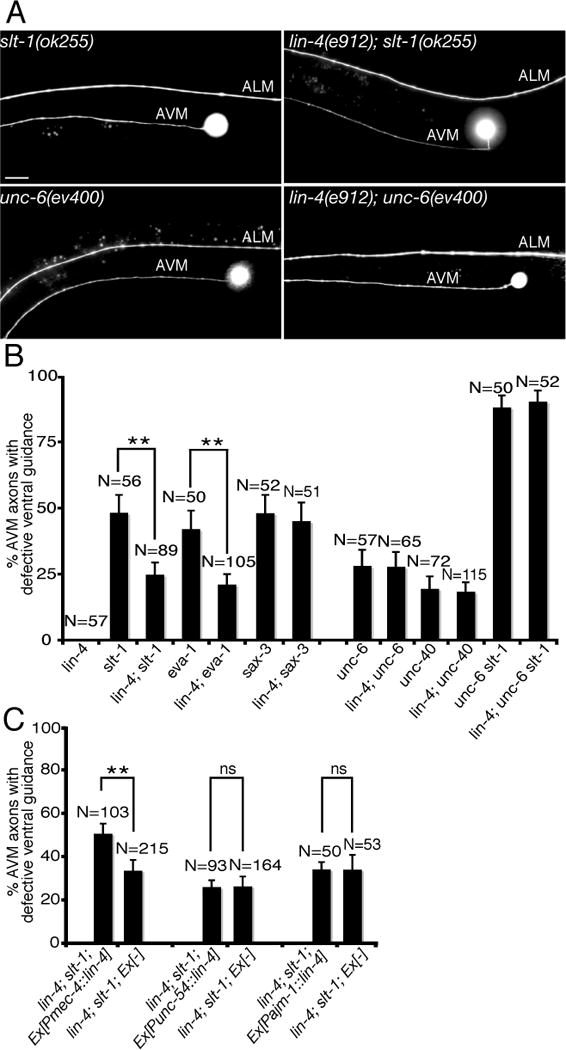
(A) Specific rescue of AVM axon guidance defects in slt-1 mutants by a loss-of-function lin-4 microRNA mutation. Anterior is to left, dorsal up. Scale bar, 20 μm. (B) Mutation in lin-4 microRNA specifically suppressed AVM axon ventral guidance defects in slt-1 (slit) and eva-1 but not unc-6 (netrin) and unc-40 (DCC) mutants. Error bars represent standard error of the proportion. Asterisks indicate that comparisons between slt-1 and lin-4; slt-1 and between eva-1 and lin-4; eva-1 are significantly different at P < 0.01 by Z-test for two proportions. (C) Cell-autonomous rescue of lin-4 mutant phenotypes in AVM axon guidance by a mec-4::lin-4 transgene. unc-54 and ajm-1 promoters were used to drive lin-4 expression in body wall muscles and hypodermal cells, respectively. Asterisks indicate that comparison is significantly different between lin-4; slt-1 with and without mec-4::lin-4 transgene at P < 0.01 by Z-test for two proportions.
The effect of lin-4 mutation on AVM axon guidance appears to be cell autonomous since expression of lin-4 microRNA in AVM neurons but not body wall muscles or hypodermal cells significantly rescued the lin-4 mutant phenotypes in suppressing AVM axon guidance defects in slt-1 mutants (Fig. 4C). The same mec-4::lin-4 transgene is sufficient to repress the expression of a mec-4::GFP::lin-14 3′UTR sensor in the AVM neurons, suggesting that the mec-4::lin-4 transgene is functional (fig. S2). Taken together, our results indicate that depletion of lin-4 microRNA in AVM neurons enhances netrin-mediated axon attraction, suppressing the AVM axon guidance defect in slt-1 mutants. Thus, lin-4 microRNA may inhibit either duration or amplitude of unc-6 (netrin) signaling in AVM axon guidance.
It was previously shown that lin-4 loss-of-function mutation affects PVM neuron specification (increase number by 40%) but not AVM neuron (0% increase) as judged by the mec-7 immunocytochemistry (18). We also found that lin-4 mutation does not have any noticeable effect on AVM cell fate. We analyzed many aspects of AVM neuronal development in lin-4 mutants, including cell migration, ventral and anterior axon growth, and expression of two AVM neuronal markers mec-4 and mir-90 and found that none of these AVM qualities is affected by the lin-4 mutation (Table S1). In fact, we consistently detected lin-4 expression in the AVM neurons only after AVM already migrated to its final position along the anterior and posterior axis and started sending out axon (Fig. 7A).
lin-14, an essential target gene of lin-4 microRNA in the AVM neurons, stimulates AVM axon ventral guidance
lin-14 encodes a transcription factor that specifies the timing and sequence of stage-specific developmental events in C. elegans (19, 20). Since lin-14 is expressed in the AVM neurons (Fig. 5B) and lin-14 3′UTR contains multiple lin-4 binding sites, we tested if lin-14 is subject to the negative regulation by lin-4 microRNA in the AVM neurons. We anticipate that, if lin-14 is the target of lin-4 microRNA, a gain-of-function lin-14 mutant allele (n355gf) that removes all seven complementary lin-4 binding sites at lin-14 3′UTR would mimic a loss-of-function lin-4 mutant allele in enhancing AVM axon ventral guidance. Our result supports this possibility by showing that lin-14(n355gf) mutation suppressed the AVM axon guidance defects in slt-1 mutants to the similar caliber as lin-4(e912lf) mutation (Fig. 4B, 5C). While suppressing the AVM axon guidance defects in slt-1 mutants, lin-14(n355gf) mutation does not suppress those in a loss-of-function unc-40 (DCC) mutant, suggesting that lin-14’s function depends on DCC (Fig. 5C). Reduced lin-14 activity suppressed lin-4 loss-of-function phenotypes in AVM axon guidance (Fig. 5D). In fact, lin-4; lin-14 slt-1 triple mutants are phenotypically indistinguishable to lin-14 slt-1 double mutants, suggesting that lin-14 acts downstream of lin-4 microRNA in the same genetic pathway for AVM axon guidance (Fig. 5D). To ask if lin-14 functions in the AVM neurons for this effect, we expressed lin-14 in the AVM neurons using a cell-type specific promoter mec-4 and found that, like the lin-4 loss-of-function phenotype, overexpressing lin-14 in the AVM neurons enhanced netrin-mediated AVM axon attraction, suppressing the defects of AVM axon guidance in slt-1 mutants (Fig. 5E, F). This result suggests that lin-14 likely functions within the AVM neurons to regulate their axon ventral guidance. To further test the effect of lin-4 microRNA on lin-14 expression in the AVM neurons, we built two sensor constructs with either the lin-14 3′UTR, containing seven lin-4 binding sites (Fig. 6A), fused to a GFP reporter or a control 3′UTR (unc-54), containing no-known lin-4 binding site, fused to a mCherry reporter, and expressed them in the AVM neurons. In L4 stage, comparing to wild-type AVM neurons, a substantially higher GFP (normalized by mCherry) signal is detected in the AVM neurons in lin-4 mutants, suggesting that lin-4 microRNA is necessary for down-regulation of lin-14 3′UTR in the AVM neurons (Fig. 6B, C). Furthermore, over-expression of lin-4 microRNA in the AVM neurons repressed the expression of GFP::lin-14 3′UTR but not GFP::unc-54 3′UTR sensors (fig. S2), suggesting that lin-4 microRNA is sufficient for down-regulation of lin-14 3′UTR in the AVM neurons.
Fig. 5. The transcription factor lin-14 enhances netrin-mediated axon attraction.
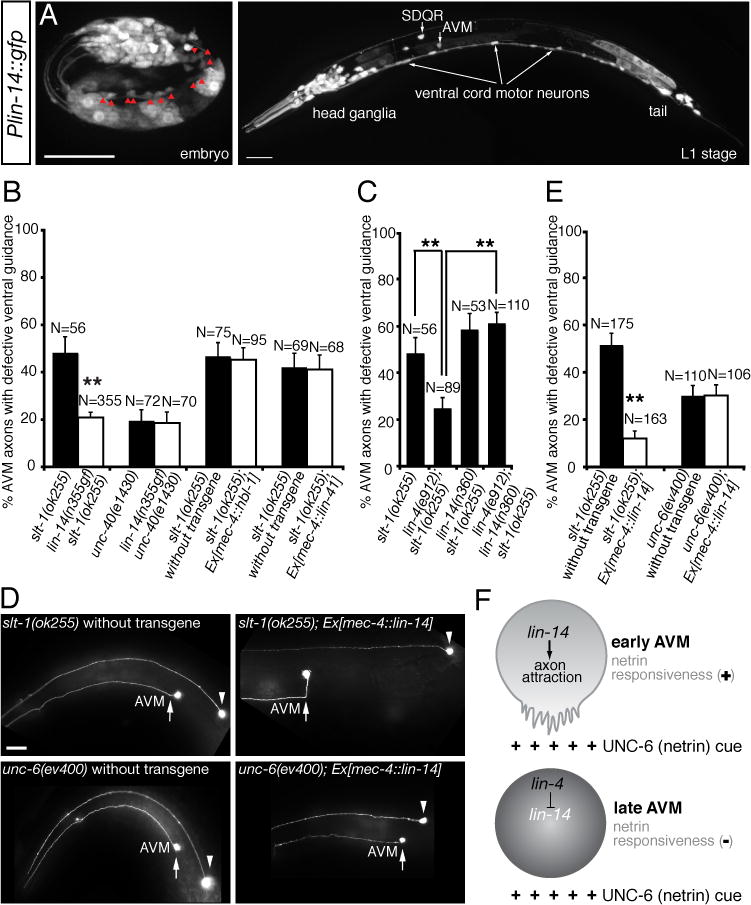
(A, B) Expression of lin-14 in neurons. (A) A late embryo. Red arrowheads indicate ventral cord motor neurons. (B) A late L1-stage animal. Scale bar, 20 μm. (C) A gain-of-function lin-14 mutant allele (n355gf) recapitulates lin-4 loss-of-function phenotype in enhancing AVM axon ventral guidance. (D) Reduced lin-14 activity suppresses lin-4 loss-of-function phenotype in AVM axon ventral guidance. (E) Specific rescue of AVM axon guidance defects in slt-1 mutants by overexpressing lin-14 in the AVM neurons. Anterior is to left, dorsal up. Scale bar, 20 μm. (F) A cell-type specific promoter-driven lin-14 expression in the AVM neurons can enhance netrin-dependent attraction, suppressing ventral guidance defects in slt-1 mutants. In all figures, error bars represent standard error of the proportion. Asterisks represent P < 0.01 by Z-test for two proportions. (G) Model of developmental switch of responsiveness to netrin by lin-4 microRNA. netrin stimulates axon attraction in early AVM neurons through the transcription factor lin-14. lin-4 microRNA expressed in later AVM neurons stops netrin-mediated axon attraction through inhibition of lin-14 expression.
Fig. 6. Down-regulation of lin-14 3′UTR by endogenous lin-4 microRNA in the AVM neurons.
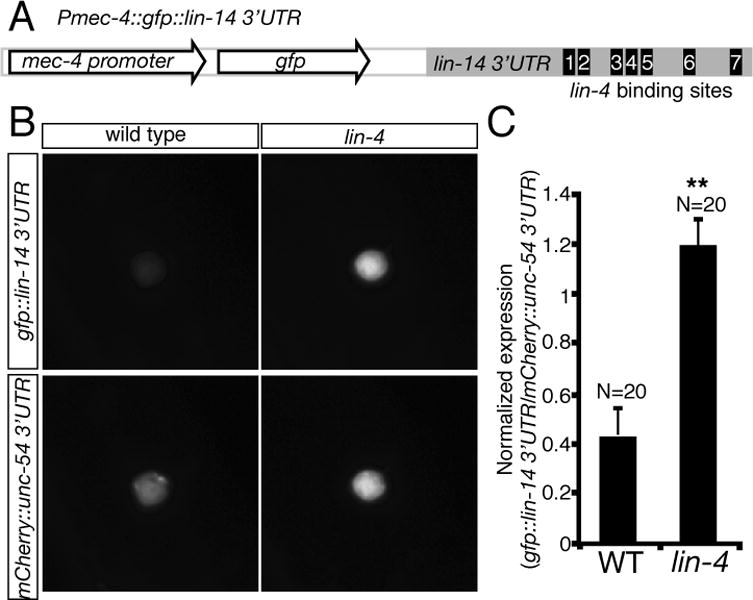
(A) Organization of Pmec-4::GFP::lin-14 3′UTR sensor construct. Seven lin-4 microRNA binding sites were computationally predicted in the lin-14 3′UTR. Sensor constructs were injected at 1ng μl−1. (B) Representative animals express a sensor with either the lin-14 3′UTR or a control (unc-54) 3′UTR. (C) Quantification of sensor expression in AVM neurons. The fluorescent intensity of Pmec-4::GFP::lin-14 3′UTR sensor is normalized by Pmec-4::mCherry::unc-54 3′UTR. When repression of a lin-14 3′UTR sensor by lin-4 occurs, a substantially higher GFP (normalized by mCherry) signal is detected in the AVM neurons in lin-4 mutants. Error bars indicate the s.e.m. Asterisks represent P < 0.001 by Student’s t-Test.
To understand the dynamic regulation of lin-14 3′UTR activity during AVM axon guidance, we analyzed the expression curve of a GFP::lin-14 3′UTR sensor in the AVM neurons in L1 stage. We found that, over time, lin-14 3′UTR activity in the AVM neurons becomes lower in wild-type animals (Fig. 7B). In contrast, lin-14 3′UTR activity in the AVM neurons remains steady and even becomes mildly higher over time in lin-4 mutants (Fig. 7C). These results suggest that the observed down-regulation of lin-14 3′UTR activity in the AVM neurons during axon guidance is caused by lin-4 microRNA inhibition.
lin-14 is not the only known target of lin-4 microRNA. hbl-1 gene, the C. elegans hunchback ortholog, also contains two lin-4 complementary elements at its 3′UTR and lin-4 microRNA is required for down-regulation of hbl-1 in the ventral nerve cord neurons (9, 10). In addition, lin-41, a member of a large family of RBCC (ring finger, B box, coiled coil) proteins, contains one lin-4 microRNA binding site at its 3′UTR (21). Over-expressing hbl-1 or lin-41 in the AVM neurons did not recapitulate the lin-4 loss-of-function mutant phenotype in suppressing the AVM axon guidance defects in slt-1 mutants, suggesting that hbl-1 and lin-41 are not the lin-4 microRNA targets in regulating AVM axon ventral guidance (Fig. 5C).
lin-14’s positive effect in netrin attraction is mediated by the receptor unc-40 and its cofactor madd-2 functioning through a combination of the unc-34 and ced-10-dependent downstream pathways
MADD-2 is a cofactor for the receptor UNC-40 (DCC) and potentiates UNC-40 activity in AVM (22, 23, Fig. 8). UNC-40 signaling in AVM is mediated by two downstream pathways, one involving UNC-34, an enabled (Ena) homolog, and the other CED-10, a Rac guanosine triphosphatase (24, 25, Fig. 8). MIG-10, a Lamellipodin homolog, defines a downstream pathway that is partially overlapped with either UNC-34 or CED-10 (26–28). To test whether lin-14’s positive effect in netrin attraction is mediated by the receptor unc-40 and its cofactor madd-2, we asked if unc-40 or madd-2 mutation could block the lin-14 over-expression effects. lin-14 over-expression is no longer able to suppress the AVM guidance defects in slt-1; unc-40 and slt-1; madd-2 double mutants (Fig. 8). These results suggest that lin-14 effect entirely depends on unc-40 receptor and its cofactor madd-2. To address whether either of the downstream pathways may mediate the lin-14 effect, we disrupted each downstream pathway in a slt-1 mutant background, where only unc-40 guidance signaling is active in AVM, and asked whether the lin-14 over-expression could still modify the AVM phenotype. The lin-14 over-expression significantly suppressed the AVM defects of slt-1; unc-34, slt-1; mig-10 and slt-1; ced-10 double mutants (Fig. 8). These results suggest that the slt-1; unc-34, slt-1; mig-10 and slt-1; ced-10 double mutants still retain a downstream pathway that is stimulated by lin-14. To test whether a combination of the downstream pathways might mediate lin-14 effect, we disrupted two downstream pathways at a time in a slt-1 mutant background, and asked whether the lin-14 over-expression could still modify the AVM phenotype. We noticed that wild type animals carrying the Pmec-4::lin-14 transgenes are sub-viable, which may explain why slt-1; ced-10; unc-34 and slt-1; mig-10; unc-34 triple mutants may not be viable when expressing the Pmec-4::lin-14 transgenes. To overcome the lethality problem, we used Pmec-7::FP4::mito to specifically disrupt unc-34 function only in the AVM neurons in the slt-1; ced-10; Ex(Pmec-4::lin-14) and slt-1; mig-10; Ex(Pmec-4::lin-14) strains. Expression of FP4-MITO depletes Ena (VASP) proteins from their normal subcellular locations, sequesters them on mitochondria, and neutralizes their function (29). In addition, we also used an RNAi approach to knockdown ced-10 function in the slt-1; unc-34; Ex(Pmec-4::lin-14) strain. The lin-14 over-expression significantly suppressed the AVM guidance defects of slt-1; mig-10; unc-34 but not slt-1; unc-34; ced-10 triple mutants (Fig. 8). Thus the slt-1; mig-10; unc-34 triple mutants retain a downstream pathway that is stimulated by lin-14, whereas the slt-1; unc-34; ced-10 triple mutants have lost the lin-14-sensitive signaling pathways. These results therefore suggest that lin-14’s positive effect in netrin attraction is mediated by the receptor unc-40 and its cofactor madd-2 functioning through a combination of the unc-34 and ced-10-dependent downstream pathways.
Fig. 8. lin-14 functions through unc-40, madd-2, unc-34 and ced-10 to promote netrin-mediated AVM axon ventral guidance.
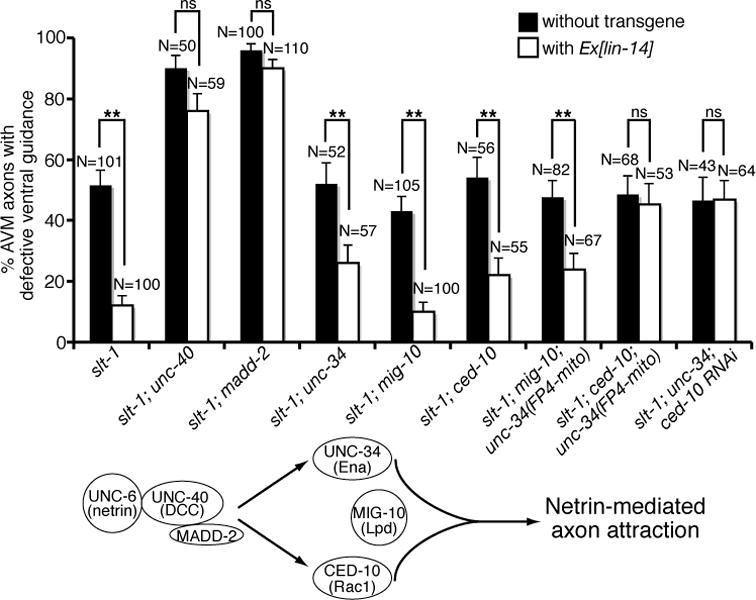
LIN-14 requires UNC-40, MADD-2, UNC-34 and CED-10 to stimulate netrin signaling. AVM was visualized with zdIs5[mec-4::gfp]. Asterisks indicate data significantly different from Ex[lin-14](-) controls (P < 0.001 by Z-test for two proportions).
lin-14 stimulates netrin-mediated axon attraction in part by enhancing UNC-40 protein expression
We show that over-expression of lin-14 in the AVM neurons can suppress AVM axon ventral guidance defects in slt-1 mutants. The effect of lin-14 over-expression can be blocked by mutations in unc-40, madd-2, or a combination of unc-34 and ced-10 genes (Fig. 8). Thus, the positive effect of lin-14 in netrin attraction is mediated by unc-40 receptor and its cofactor madd-2 functioning through two parallel downstream pathways unc-34 and ced-10 (Fig. 8). Since madd-2, unc-34 and ced-10 transduce guidance signal that is originated from the unc-40 receptor, we wonder whether unc-40 is the prime target of lin-14 regulation in the netrin pathway. LIN-14 encodes a transcription factor with unknown function in neurons (19, 20). To ask whether lin-14 may affect unc-40 promoter activity or protein expression, we analyzed the effect of lin-14 on unc-40 promoter activity and protein expression in the anterior touch neurons. We found that lin-14 over-expression does not affect the expression intensity of either a 3.5-kb or a 5-kb unc-40 promoter::GFP reporter in the AVM neurons (fig. S4). In fact, lin-14 did not affect the promoter activity of various unc-40 pathway genes we analyzed (fig. S4). To determine if UNC-40 proteins can be regulated by lin-14, we generated three transgenic lines that express UNC-40::GFP fusion proteins in touch neurons at different amounts. Expression of UNC-40::GFP proteins at low amount in touch neurons results in 100% of cases showing UNC-40 proteins localized to the perinuclear region in vesicle-like compartments (Fig. 9A, B). Medium amount of UNC-40 expression reduces percentage of touch neurons showing restricted UNC-40 perinuclear localization and increases percentage of touch neurons with a broader UNC-40 distribution (Fig. 9B). High amount of UNC-40 expression causes 100% of AVM and ALM neurons expanding UNC-40 protein distribution to the whole cell (Fig. 9B). Interestingly, over-expression of lin-14 can also expand UNC-40 protein distribution from the perinuclear region to the whole cell (Fig. 9A, B). Our results thus suggest that lin-14 stimulates netrin-mediated axon attraction in part by enhancing UNC-40 protein expression. Indeed, over-expression of unc-40::GFP in the AVM neurons recapitulated the effect of lin-14 over-expression, suppressing AVM axon ventral guidance defects in slt-1 but not unc-6 mutants (Fig. 9C).
Fig. 9. Effects of lin-14 on unc-40 protein distribution in the anterior touch neurons.
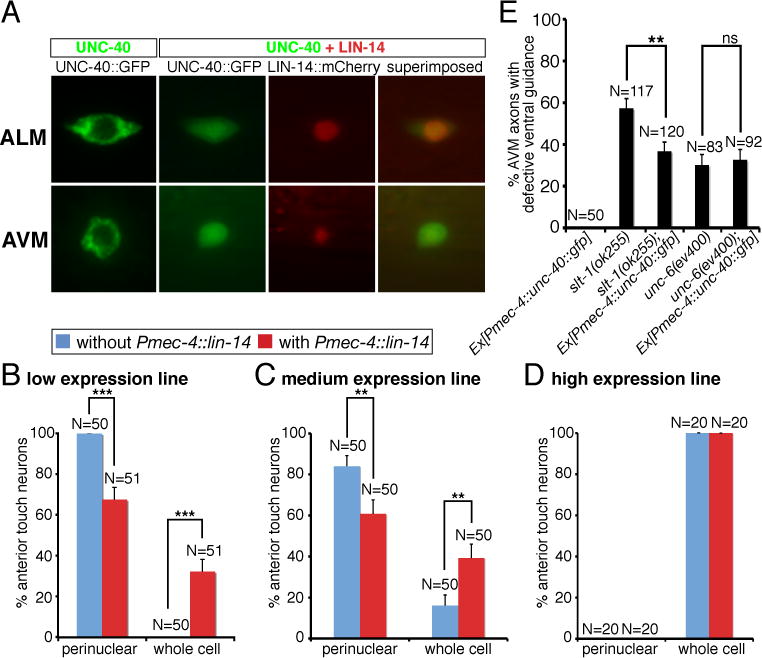
(A) Distribution of UNC-40::GFP and LIN-14::mCherry fusion proteins in the ALM and AVM neurons. (B) Quantification of anterior touch neurons showing UNC-40 protein distribution either restricted to the perinuclear region or expanded to the whole cell, depending on UNC-40 dosage and LIN-14 amount. Using the expression intensity of the Mos[Pmec-4::unc-40::GFP] single copy insertion line as a reference, we estimated 1 copy is the low expression line, 1.2 copies is the medium expression line, and 9.5 copies is the high expression line. (C) Over-expression of unc-40::GFP in the AVM neurons suppressed the AVM axon ventral guidance defects in slt-1 but not unc-6 mutants. In all figures, error bars represent standard error of the proportion. Asterisks and brackets represent P < 0.01 by Z-test for two proportions.
Discussion
Roles for microRNAs in regulating neuronal development have been suggested before by the observation of defects in asymmetric neuronal development in C. elegans mutant in specific microRNAs, like lsy-6 and mir-273, and by in vivo studies of neuronal morphogenesis in vertebrate microRNA mutants (6, 7, 11), but whether microRNAs participate in axon guidance is unknown. Local translational regulation of axon guidance molecules at the developing growth cones by microRNAs has been long speculated. But our discovery here is a conceptual departure. We showed that a developmental timing microRNA represses the expression of a transcription factor to inhibit Netrin-mediated axon attraction. We believe this microRNA signals the neuron to undergo a developmental switch of responsiveness to netrin as its axon guidance comes to an end.
We discovered an unexpected role for the lin-4 microRNA in axon guidance through genetic analysis of a well-characterized axon guidance event, the growth of the AVM axon to the ventral midline in C. elegans. Our results show that lin-4 microRNA functions as a potent and specific negative regulator of Netrin-mediated attraction, without effect on Slit-mediated repulsion, and provide strong evidence that lin-4 represses the expression of lin-14 transcription factor in the AVM neuron to inhibit its axon attraction to Netrin. We further show that lin-14’s positive effect in netrin attraction is mediated by the receptor unc-40 and its cofactor madd-2 functioning through a combination of the unc-34 and ced-10-dependent downstream pathways. In this study, we identified the molecular mechanisms by which lin-4 regulates netrin-mediated AVM axon attraction, it remains unknown, however, what and how signal turns on lin-4 expression in the AVM neurons.
lin-4 promoter GFP reporter is broadly expressed in C. elegans neurons during development of the nervous system and its expression is broader and generally stronger in the L1 stage than in the embryonic stage (Fig. 3B, C). Notably, it is expressed in lateral ganglion (ADL, AWB, AWC, AFD), ventral nerve cord (DA, DB, DD, VD), anterior-body region (SDQR), tail (PQR), touch neurons (AVM, PVM, ALM, PLM), and HSN neurons. lin-14 promoter GFP reporter is also broadly expressed in C. elegans neurons. Its strong expression in neurons starts in the late embryonic stage (Fig. 5A). Analysis of expression patterns of lin-4 and lin-14 promoter reporters reveals overlapping expression of two genes in several neurons, including AVM, ALM, PVM, PLM, DD, VD, DA, DB, SDQR, HSN, and PQR. These results suggest that the lin-4 and lin-14 relationship may be extended beyond AVM neurons. Some of these neurons are known to be guided by unc-40 alone signaling (AVM, PVM, HSN, and ADL) while others are guided by unc-40 + unc-5 signaling (SDQR, DD, VD, DA and DB).
While lin-4 and lin-14 mutations affect the timing of HSN axon outgrowth, they do not affect the timing of AVM axon outgrowth. Thus, the effect of lin-4 and lin-14 mutations on AVM axon guidance is likely direct and not due to the temporal decoupling of axon outgrowth and axon guidance. We analyzed HSN axon outgrowth in wild-type animals and lin-4 loss-of-function mutants. We found that, in wild-type animals, HSN axon outgrowth was absent in L3 stage in most of cases (fig. S5A). However, axon outgrowth was observed in most of the wild-type HSN neurons in L4 and young adult stages (fig. S5A). In contrast, HSN neurons did not grow out axons in L3, L4, or young adult stage in lin-4 loss-of-function mutants (fig. S5A). These results suggest that lin-4 is required for HSN axon outgrowth. A similar observation has been made by a recent study in which lin-4 was identified as an important factor that promotes HSN axon outgrowth, but whether it plays a role in HSN axon guidance is unclear (30, 31).
When we determined the effects of lin-4 loss-of-function and lin-14 gain-of-function mutations on the timing of AVM axon outgrowth, we found that AVM axon outgrowth is not affected by either lin-4 or lin-14 mutations. 5 hours into L1 stage, axonal marker (Pmec-4::GFP) was not visible in AVM neurons in wild type, lin-4, and lin-14 mutants. 7 hours into L1 stage, AVM axon outgrowth (both ventral and anterior projections) was detected in most of cases in wild type, lin-4, and lin-14 mutants (fig. S5B). 10 hours into L1 stage, all strains we analyzed (wild type, lin-4, and lin-14 mutants) showed 100% AVM axon outgrowth (fig. S5B). Thus, the timing of AVM axon outgrowth is not altered in lin-4 and lin-14 mutants. These results suggest that the effect of lin-4 and lin-14 mutations on AVM axon guidance is likely direct and not due to the altered timing of AVM axon outgrowth.
There is an emerging view that microRNAs are important in specifying the transition of cells from totipotency to commitment. Our results suggest that lin-4 microRNA may signal the transition of AVM neurons from early phase of differentiation featured by responsiveness to axon guidance cues (netrin) to later phase of differentiation featured by loss of responsiveness to axon guidance cues (Fig. 5G). lin-14 transcription factor is a lin-4 microRNA target, stimulating netrin-mediated AVM axon guidance in part by enhancing UNC-40 protein expression. The fact that lin-4 microRNA acts in the AVM neurons to negatively regulate netrin-mediated axon attraction suggests that lin-4 microRNA either inhibits amplitude or restricts duration of unc-40 (DCC) signaling in AVM neurons. Given that developmental mechanisms are frequently conserved across animal phylogeny, general rules of microRNA regulation of axon guidance in C. elegans are likely to be applicable in higher organisms.
Materials and Methods
Genetics and strains
C. elegans strains were cultured using standard methods (32). All strains were grown at 20°C unless otherwise specified. A strain list appears as Table S2.
Transgenic animals
“Germline” transformation of C. elegans was performed using standard techniques (33). For example, the Pmec-4::lin-14 transgene was injected at 10 ng/μl along with the coinjection marker Podr-1::rfp at 40 ng/μl. Transgenic lines were maintained by following Podr-1::rfp fluorescence.
PCR fusion reaction
mec-4::lin-4 was generated by PCR fusion, in which two separate primary PCR products were fused by a secondary PCR. Template used was genomic DNA.
| lin-4_G1 | CTATCAAGTTATAGAGGGATCATGCTTCCGGCCTGTTCCCTG |
| lin-4_G1_R | CAGGGAACAGGCCGGAAGCATGATCCCTCTATAACTTGATAG |
| lin-4_G2 | AGATCTGCTCAAACCGTCCTG |
| mec-4_P1 | CCAAGCTTCAATACAAGCTC |
Constructs
Pmec-4::lin-14 and Pmec-4::hbl-1 were constructed by PCR amplification of genomic fragment of lin-14 or hbl-1, followed by digestion with NheI and KpnI and ligation to Pmec-4-pSM vector. Pmec-4::GFP::lin-143′UTR was constructed by PCR amplification of a 1.8-kb genomic fragment of lin-143′UTR, followed by digestion with EcoRI and SpeI and ligation to Pmec-4-GFP-pSM vector. Plin-4::GFP was constructed by PCR amplification of a 1.9-kb lin-4 promoter fragment from genomic DNA, followed by digestion with FseI and XmaI and ligation to GFP-pSM vector. Plin-14::GFP was constructed by PCR amplification of a 4-kb lin-14 promoter fragment from genomic DNA, followed by digestion with FseI and AscI and ligation to GFP-pSM vector. Punc-40::GFP was constructed by PCR amplification of either a 3.5-kb or a 5-kb unc-40 promoter fragment from genomic DNA, followed by digestion with FseI and AscI and ligation to GFP-pSM vector. Pmadd-2::GFP was constructed by PCR amplification of a 3-kb madd-2 promoter fragment from genomic DNA, followed by digestion with FseI and AscI and ligation to GFP-pSM vector. Pced-10::GFP was constructed by PCR amplification of a 3-kb ced-10 promoter fragment from genomic DNA, followed by digestion with FseI and AscI and ligation to GFP-pSM vector. Pmig-10::GFP was constructed by PCR amplification of a 3.5-kb mig-10 promoter fragment from genomic DNA, followed by digestion with FseI and AscI and ligation to GFP-pSM vector. To generate Pmec-4::mig-10::GFP, a DNA fragment containing the mig-10 cDNA was inserted at the 5′ end of GFP in a GFP-pSM vector containing a 1-kb fragment of the mec-4 promoter between SphI and SalI sites. To generate Pmec-4::unc-40::GFP, a DNA fragment containing the unc-40 cDNA was inserted at the 5′ end of GFP in a GFP-pSM vector containing a 1-kb fragment of the mec-4 promoter between NheI and KpnI sites. For MosSCI experiments, Pmec-4::unc-40::GFP sequence was cut out of pSM using the FseI and SpeI restriction sites, and was cloned into a pCFJ151 MosSCI insertion vector (34) that had been modified to include a FseI site in the MCS (35).
| lin-14_F_NheI | GCTAGCTAGCTTGTCCTTCTGCCCAATCAAG |
| lin-14_R_KpnI | CGGGGTACCTCTATTGTGGACCTTGAAGAG |
| hbl-1_F_NheI | GCTAGCTAGCGAAAAAGGATTAGTGGTCCTG |
| hbl-1_R_KpnI | CGGGGTACCTTATTGGTGTCTGGCTTGGTAC |
| lin-143′UTR_F_EcoRI | CGGAATTCACATCAGTCTCTTCACCCATC |
| lin_143′UTR_R_SpeI | GGACTAGTGTAAGGTTCAGAGATGCATC |
| lin-4_P_F_FseI | GTCTGGCCGGCCCATGTCCTGCCCATTCCGTAG |
| lin-4_P_R_XmaI | TCCCCCCGGGCTCATAAACCAACCAAAACTC |
| mig-10_P_F_FseI | GTCTGGCCGGCCGACAAGCAATCCATCATCATC |
| mig-10_P_R_AscI | CAGCGGCGCGCCGCATAAAAGAGCACAAATCAG |
| unc-34_P_F_FseI | GTCTGGCCGGCCAGCCCACCAAAATTACAGTAC |
| unc-34_P_R_AscI | CAGCGGCGCGCCCTGGCTCAAAAAAGTGCAATC |
| unc-40_P_F_FseI | CAGCGGCGCGCCCTTCTGTCGAATTATCGCATTC |
| unc-40_P_R_AscI | GTCTGGCCGGCCCATGGGCATTTATCATCACTG |
| madd-2_P_F_FseI | GTCTGGCCGGCCTTTCCGTGCGGTTGCTTAGAG |
| madd-2_P_R_AscI | CAGCGGCGCGCCTTGAGTATTGTCAGGTGGAAG |
| ced-10_P_F_FseI | GTCTGGCCGGCCACCGTATCCAATGGGAGCTTC |
| ced-10_P_R_AscI | CAGCGGCGCGCCGAGCTCCGCGAGCACAAGCAC |
Stem-loop reverse transcription-PCR
We quantified the mature lin-4 microRNA level by modifying the microRNA assay developed previously (14). Reverse transcription reactions contained purified total RNA, 50 nM stem-loop RT primer, 1X RT first strand buffer, 0.25 mM each of dNTPs, 10 mM MgCl2, 0.1 M DTT, 200 U SuperScript III reverse transcriptase and 40 U RNase inhibitor. The mixture of RNA template, dNTPs and RT primer was incubated for 5 min at 65°C. The mixture was then placed on ice for at least 1 min before adding RT buffer, DTT, MgCl2, SuperScript III and RNase inhibitor. The reaction was incubated for 50 min at 50°C before heat inactivation at 85°C for 5 min. PCR was conducted using 0.25μl RT products as template in 20μl PCR for 17 cycles.
| RT_primer_lin-4 | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCACACTT |
| RT_lin-4_F | CGGCGGTCCCTGAGACCTCAA |
| Universal reverse primer | CTGGTGTCGTGGAGTCGGCAATTC |
| actin-F | AATTGATGTTCGTCGTGGACTC |
| actin-R | TCTGAAGTCTCCCATCACTGAG |
MosSCI integrations
Mos single-copy integrants were generated using the direct insertion protocol described in Frokjaer-Jensen et al. (2008). 50 EG4322 ttTi5605; unc-119(ed3) worms were injected with rab-3::mCherry, myo-2::mCherry, myo-3::mCherry, pJL43.1 (a vector containing the Mos1 transposase under the control of the germline promoter glh-2), and a vector containing the mec-4 promoter::unc-40::GFP sequence to be inserted flanked by sequences homologous to the insertion site. Animals that were rescued for the unc-119 phenotype (array-positive) were allowed to starve out twice, and then unc-119 rescued animals that lacked the three mCherry coinjection markers (integrant-positive, array negative) were cloned out from separate plates to find independent integrated lines. These lines were outcrossed twice to wild-type animals, and the presence of the intact insertion was verified by PCR.
Microscopy
Axonal processes of AVM neurons were visualized with the integrated mec-4::gfp transgene zdIs5 at L4 stage. The observer was not blind to the genotype. Animals were placed on 5% Noble Agar pads in M9 buffer containing 10 mM sodium azide and examined with a Plan-NEOFLUAR 40x, 1.4 NA oil-immersion objective on a Zeiss AxioImager. Images for axons and body morphology were captured using a Zeiss Axio camera. Fluorescence intensity was measured using the Zeiss Axiovision Rel 4.7 image analysis software.
Supplementary Material
Acknowledgments
We thank F. Ciamacco, B. Bayne, and K. Campbell for technical assistance; We also thank B. Lesch for the MosSCI technique, A. Fire for C. elegans vectors, the C. elegans Genetic Center for C. elegans strains, and the WormBase for readily accessible information; V. Cleghon for critical reading of the manuscript; and C. Bargmann, M. Tessier-Lavigne, V. Ambros, and members of the Chang and Chuang labs for helpful discussions. This work was funded by grants from the following: the Whitehall Foundation Research Awards (C.C. and C.-F.C.), the March of Dimes Foundation Research Award (C.C.), the Alfred P. Sloan Research Fellowship (C.-F.C.), and the CIHR grant RMF-82501 (C.C.).
Footnotes
Author contributions: Y.Z. and H.C. performed experiments and analyzed data; D.D. analyzed axonal phenotypes in lin-4; slt-1 and lin-4; unc-6 strains; and C.C. and C.-F.C. designed experiments, analyzed data and wrote the paper.
Competing interests: The authors declare that they do not have any competing financial, personal, or professional interest.
References
- 1.Chang C, Yu TW, Bargmann CI, Tessier-Lavigne M. Inhibition of netrin-mediated axon attraction by a receptor protein tyrosine phosphatase. Science. 2004;305:103–106. doi: 10.1126/science.1096983. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 5.Ruvkun G. The perfect storm of tiny RNAs. Nat Med. 2008;14:1041–1045. doi: 10.1038/nm1008-1041. [DOI] [PubMed] [Google Scholar]
- 6.Johnston RJ, Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 7.Chang S, Johnston RJ, Jr, Frokjaer-Jensen C, Lockery S, Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- 8.Boehm M, Slack F. A developmental timing microRNA and its target regulate life span in C. elegans. Science. 2005;310:1954–1957. doi: 10.1126/science.1115596. [DOI] [PubMed] [Google Scholar]
- 9.Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- 10.Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C. elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- 11.Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 12.Duchaine TF, Wohlschlegel JA, Kennedy S, Bei Y, Conte D, Jr, Pang K, Brownell DR, Harding S, Mitani S, Ruvkun G, Yates JR, 3rd, Mello CC. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 13.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 14.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- 16.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 17.Fujisawa K, Wrana JL, Culotti JG. The slit receptor EVA-1 coactivates a SAX-3/Robo mediated guidance signal in C. elegans. Science. 2007;317:1934–1938. doi: 10.1126/science.1144874. [DOI] [PubMed] [Google Scholar]
- 18.Mitani S, Du H, Hall DH, Driscoll M, Chalfie M. Combinatorial control of touch receptor neuron expression in Caenorhabditis elegans. Development. 1993;119:773–783. doi: 10.1242/dev.119.3.773. [DOI] [PubMed] [Google Scholar]
- 19.Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- 20.Hristova M, Birse D, Hong Y, Ambros V. The Caenorhabditis elegans heterochronic regulator LIN-14 is a novel transcription factor that controls the developmental timing of transcription from the insulin/insulin-like growth factor gene ins-33 by direct DNA binding. Mol Cell Biol. 2005;25:11059–11072. doi: 10.1128/MCB.25.24.11059-11072.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 22.Hao JC, Adler CE, Mebane L, Gertler FB, Bargmann CI, Tessier-Lavigne M. The tripartite motif protein MADD-2 functions with the receptor UNC-40 (DCC) in Netrin-mediated axon attraction and branching. Dev Cell. 2010;18:950–960. doi: 10.1016/j.devcel.2010.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alexander M, Selman G, Seetharaman A, Chan KK, D’Souza SA, Byrne AB, Roy PJ. MADD-2, a homolog of the Opitz syndrome protein MID1, regulates guidance to the midline through UNC-40 in Caenorhabditis elegans. Dev Cell. 2010;18:961–972. doi: 10.1016/j.devcel.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 24.Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- 25.Gitai Z, Yu TW, Lundquist EA, Tessier-Lavigne M, Bargmann CI. The netrin receptor UNC-40/DCC stimulates axon attraction and outgrowth through enabled and, in parallel, Rac and UNC-115/AbLIM. Neuron. 2003;37:53–65. doi: 10.1016/s0896-6273(02)01149-2. [DOI] [PubMed] [Google Scholar]
- 26.Chang C, Adler CE, Krause M, Clark SG, Gertler FB, Tessier-Lavigne M, Bargmann CI. MIG-10/lamellipodin and AGE-1/PI3K promote axon guidance and outgrowth in response to slit and netrin. Curr Biol. 2006;16:854–862. doi: 10.1016/j.cub.2006.03.083. [DOI] [PubMed] [Google Scholar]
- 27.Quinn CC, Pfeil DS, Chen E, Stovall EL, Harden MV, Gavin MK, Forrester WC, Ryder EF, Soto MC, Wadsworth WG. UNC-6/netrin and SLT-1/slit guidance cues orient axon outgrowth mediated by MIG-10/RIAM/lamellipodin. Curr Biol. 2006;16:845–853. doi: 10.1016/j.cub.2006.03.025. [DOI] [PubMed] [Google Scholar]
- 28.Quinn CC, Pfeil DS, Wadsworth WG. CED-10/Rac1 mediates axon guidance by regulating the asymmetric distribution of MIG-10/lamellipodin. Curr Biol. 2008;18:808–813. doi: 10.1016/j.cub.2008.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goh KL, Cai L, Cepko CL, Gertler FB. Ena/VASP proteins regulate cortical neuronal positioning. Curr Biol. 2002;12:565–569. doi: 10.1016/s0960-9822(02)00725-x. [DOI] [PubMed] [Google Scholar]
- 30.Olsson-Carter K, Slack FJ. A developmental timing switch promotes axon outgrowth independent of known guidance receptors. PLoS Genet. 2010;6:e1001054. doi: 10.1371/journal.pgen.1001054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsson-Carter K, Slack FJ. The POU transcription factor UNC-86 controls the timing and ventral guidance of Caenorhabditis elegans axon growth. Dev Dyn. 2011;240:1815–1825. doi: 10.1002/dvdy.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 34.Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lesch BJ, Bargmann CI. The homeodomain protein hmbx-1 maintains asymmetric gene expression in adult C. elegans olfactory neurons. Genes Dev. 2010;24:1802–1815. doi: 10.1101/gad.1932610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


