Abstract
Stem/progenitor cells often generate distinct cell types in a stereotyped birth order, and over time lose competence to specify earlier-born fates by unknown mechanisms. In Drosophila, the Hunchback transcription factor acts in neural progenitors (neuroblasts) to specify early-born neurons, in part by indirectly inducing the neuronal transcription of its target genes, including the hunchback gene. We used in vivo immuno-DNA FISH and found that the hunchback gene moves to the neuroblast nuclear periphery, a repressive subnuclear compartment, precisely when competence to specify early-born fate is lost, and several hours and cell divisions following termination of its transcription. hunchback movement to the lamina correlated with downregulation of the neuroblast nuclear protein, Distal antenna (Dan). Either prolonging Dan expression or disrupting lamina interfered with hunchback repositioning and extended neuroblast competence. We propose that neuroblasts undergo a developmentally-regulated subnuclear genome reorganization to permanently silence Hunchback target genes that results in loss of progenitor competence.
INTRODUCTION
Stem and progenitor cells typically produce different cell types in a stereotyped birth order during development. Over time, progenitors can lose competence to generate earlier-born cell fates. How stem/progenitor competence is lost, and how it can be experimentally regained, is a fundamental question that has important implications for understanding development and for using stem cells therapeutically. Progressive loss of competence has been well characterized in mammalian and insect neural progenitors, where single progenitors make an ordered series of distinct neural progeny (reviewed in Cepko et al., 1996; Kwan et al., 2012; Pearson and Doe, 2004; Sousa-Nunes et al., 2010). For example, mammalian cortical progenitors have an early competence window to generate projection neuron subtypes and a later competence window to generate glia (reviewed in McConnell, 1989; Okano and Temple, 2009). Yet virtually nothing is known about how competence is regulated in any mammalian or insect stem/progenitor cell lineage.
Drosophila embryonic neural progenitors (neuroblasts) provide an excellent model system for investigating progenitor competence. Neuroblasts sequentially express the transcription factors Hunchback (Hb) > Krüppel (Kr) > Pdm1/2 > Castor > Grainyhead, and generate specific neuronal or glial cell types during each window of gene expression (Baumgardt et al., 2009; Benito-Sipos et al., 2011; Brody and Odenwald, 2000; Isshiki et al., 2001; Kambadur et al., 1998; Kao et al., 2012; Karlsson et al., 2010; Mettler et al., 2006; Touma et al., 2012; Tran and Doe, 2008)(Figure 1A). The best characterized of these factors is Hb, which is a zinc finger transcription factor homologue of mammalian Ikaros (Elliott et al., 2008; Kim et al., 1999). hb transcription is activated by an unknown mechanism in newly formed neuroblasts and is repressed 1–2 divisions (~1hr) later by the repressor Seven-up (Svp) (Kanai et al., 2005; Kohwi et al., 2011; Mettler et al., 2006) (Figure 1A). Neurons born from the Hb-expressing neuroblasts maintain hb transcription via a neuron-specific cis-regulatory element (Hirono et al., 2012).
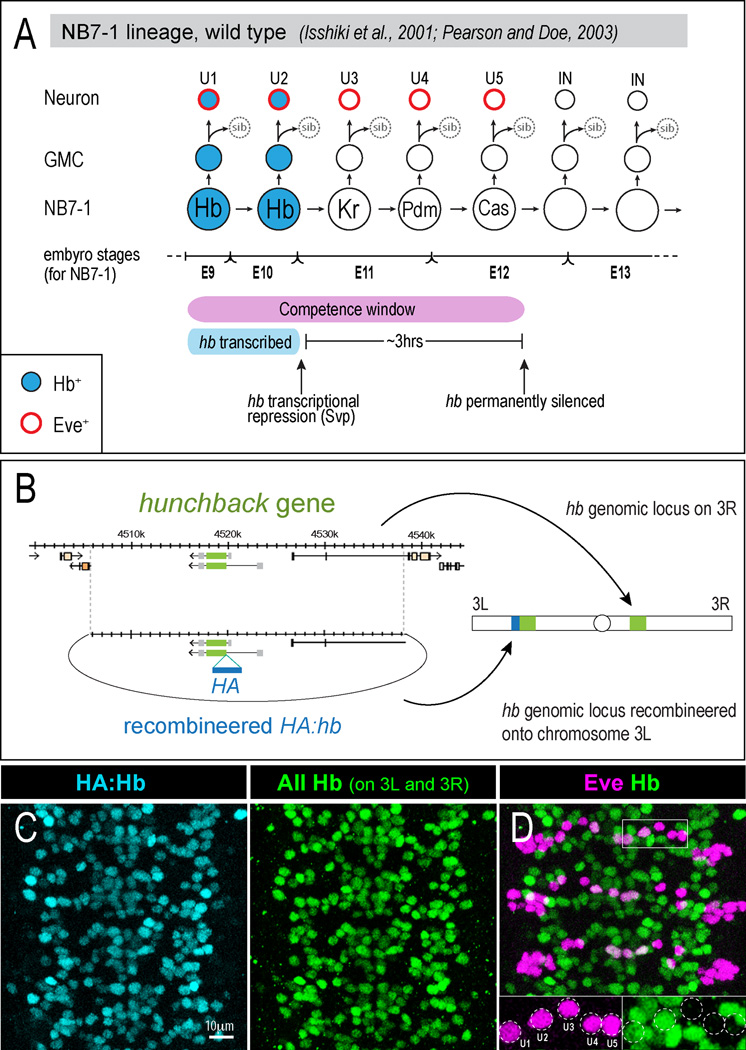
Figure 1. Temporal identity within the NB7-1 lineage and defining the early competence window.
(A) Wild type NB7-1 cell lineage. Hb expression is shown in blue; subsequent temporal identity gene expression is indicated for NB7-1 but not shown in the progeny. hb transcription is terminated after the neuroblast second division by the transcriptional repressor Seven-up (Svp). The hb gene becomes permanently silenced after the neuroblast fifth division (Pearson and Doe, 2003). Embryonic stage transitions (bottom line) refer to NB7-1 only.
(B) We used recombineering (Venken et al., 2006) to make a 32kb genomic construct with an HA-epitope in frame with hb coding sequences. The ΦC31 system (Bischof et al., 2007; Groth et al., 2004) was used for site-specific integration into an attP site on the left arm of chromosome 3.
(C) Immunostaining for HA (blue, to detect the HA:Hb protein) shows the same pattern as staining for both HA:Hb and endogenous Hb (green).
(D) Embryos containing both the endogenous hb gene and the recombineered HA:hb gene have wild type numbers of Eve+ Hb+ U1/U2 neurons (see insets).
Hb is necessary and sufficient to specify early-born neuronal identity in multiple neuroblast lineages (Cleary and Doe, 2006; Isshiki et al., 2001; Kanai et al., 2005; Novotny et al., 2002; Tran and Doe, 2008). This has been best studied in neuroblast 7-1 (NB7-1). NB7-1 produces the Even-skipped (Eve)-positive U1-U5 motoneurons during its first five cell divisions, and then ~20 Eve-negative interneurons during its subsequent divisions (Figure 1A)(Bossing et al., 1996; Pearson and Doe, 2003; Schmid et al., 1999). hb mutants lack early-born U1/U2 neurons, and prolonged hb expression in NB7-1 can generate ectopic U1/U2 neurons (Isshiki et al., 2001; Novotny et al., 2002). Importantly, NB7-1 loses competence to make U1/U2 neurons in response to ectopic Hb after its fifth division (Cleary and Doe, 2006; Pearson and Doe, 2003). We refer to this five-division interval as the neuroblast "early competence window." Furthermore, ectopic Hb is unable to induce U1/U2 identity when misexpressed specifically in post-mitotic neurons (Pearson and Doe, 2003). Thus, Hb acts in the neuroblast to specify early-born U1/U2 neuronal identity, and NB7-1 loses competence to respond to Hb after its fifth cell division.
Here we investigate the molecular mechanism by which neuroblast competence to specify Hb+ early-born neuronal identity is lost over time. We find that the hb genomic locus moves from the nuclear interior to the nuclear periphery, a transcriptionally repressive subnuclear compartment, nearly synchronously within the entire neuroblast population. Strikingly, the timing of hb gene repositioning to the nuclear periphery correlates with the end of the NB7-1 early competence window, which occurs 3 divisions (~3 hours) after hb transcription has ceased. Depletion of Lamin, an intermediate filament component of the nuclear envelope, reduces hb positioning at the nuclear periphery and extends neuroblast competence. Therefore, the hb gene undergoes two distinct levels of transcriptional regulation in NB7-1: hb transcription is terminated after the second cell division, and then after the fifth division the hb locus becomes repositioned to the nuclear periphery, where it becomes permanently silenced and ends the neuroblast early competence window. We further show that the Pipsqueak domain protein, Distal antenna (Dan), is downregulated in neuroblasts concurrent with hb movement to the nuclear periphery; prolonging Dan expression in neuroblasts prevents hb gene repositioning to the lamina and extends neuroblast competence.
RESULTS
Recombineered HA-tagged hb mimics endogenous hb expression and is a specific marker for U1/U2 neuronal identity
Previously, heat shock-induced pulses of ectopic Hb expression were used to reveal that NB7-1 is competent to specify Hb+ U1/U2 neurons during its first five divisions, referred to as the “early competence window” (pink bar, Figure 1A; Pearson and Doe, 2003). In contrast, using the Gal4/UAS system to continuously express ectopic Hb in NB7-1 (en-gal4 UAS-hb) resulted in the generation of over 20 extra neurons with U1/U2-like identity (Isshiki et al., 2001; Pearson and Doe, 2003). This result suggested that Hb could extend the neuroblast early competence window. However, the extra U1/U2-like neurons were only assayed in the embryo, when some Gal4/UAS-induced Hb protein may still have been present. Therefore, it is unknown if the ectopic U1/U2 neurons are able to express endogenously-transcribed hb, a key molecular characteristic of early-born identity, or if they are able to maintain Hb expression into larval stages after ectopic Hb protein has declined. Since Hb functions as both the inducer of early-born identity (in the neuroblast) as well as the responder of cell fate (in the neuronal progeny), we wanted to develop a definitive molecular marker for U1/U2 neuronal identity, and to assay neuronal identity in larval stages. The only known positive marker for U1/U2 identity is endogenous hb expression. Hence, we generated a hemagglutinin (HA)-tagged recombineered (Venken et al., 2006) genomic construct (HA:hb) that contains 32kb of genomic DNA from the hb locus (Figure 1B). Immunostaining for HA showed that HA:Hb precisely mimicked the endogenous hb expression pattern (Figure 1C) and did not generate extra U1/U2 neurons when integrated into the genome in addition to the existing endogenous hb gene (Figure 1D). Furthermore, HA:hb rescued a hb null mutant to viability (data not shown). Thus, immunostaining for HA in the HA:hb genotype can be used as a proxy for endogenous hb transcription, and is a specific, positive marker for early-born U1/U2 neuronal identity. We can now ectopically express untagged Hb protein and use HA staining as a molecular read-out for endogenous hb gene expression and U1/U2 neuronal identity.
U1/U2 identity can be induced by Hb only during the neuroblast competence window
Using our HA:Hb reporter, we revisited the question of whether Hb can extend the NB7-1 early competence window. We ectopically expressed untagged Hb protein throughout the NB7-1 lineage (en-gal4 UAS-hb) and assayed for the number of U1/U2 neurons generated (Figure 2). We stained for Eve (to identify all U neurons), HA:Hb (to detect authentic U1/U2 neurons), and Hb (to detect both endogenous Hb and the ectopically expressed Hb). Consistent with our previous results, we found all Eve+ U neurons were also Hb+ in the embryo (Figure 2A, green; Pearson and Doe, 2003). Importantly, however, only 5.4 ± 0.4 neurons were HA:Hb+, and thus authentic U1/U2 neurons (Figure 2A, blue). The HA:Hb+ U1/U2 neurons were all clustered at the dorsal side of the CNS (data not shown), indicating that they were the earliest-born neurons in the lineage (Pearson and Doe, 2003). Consistent with their bona fide U1/U2 neuronal identity, these were the only neurons that maintained endogenous Hb expression into larval stages (Figure 2B), which is well after the en-gal4 driver turns off and ectopic Hb protein is no longer present. We conclude that Hb can specify U1/U2 neuronal identity, but only during the NB7-1 five division competence window.
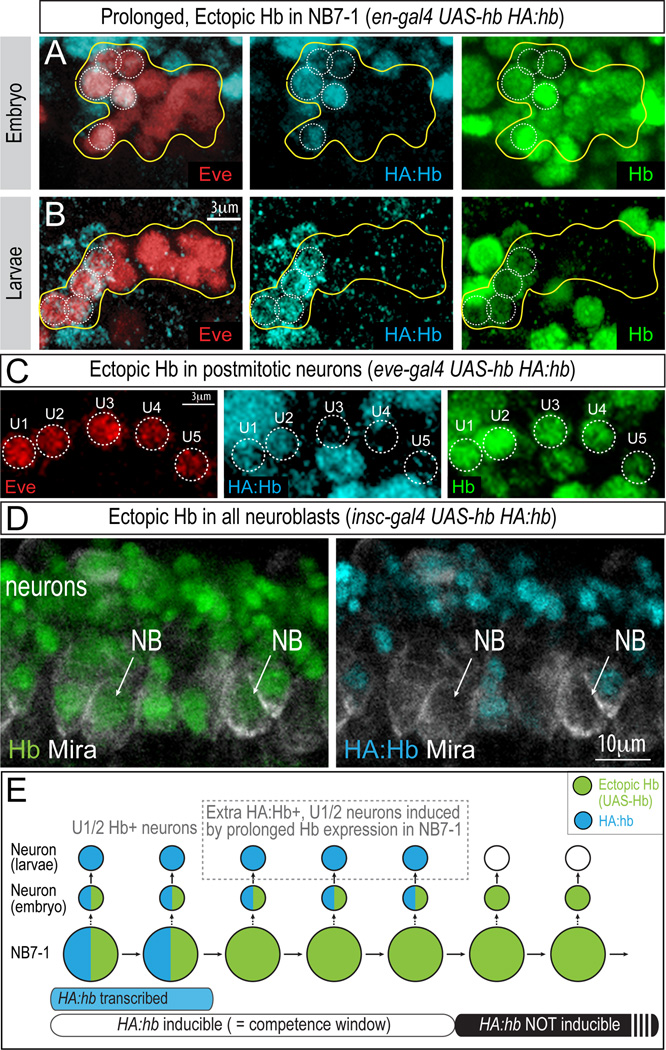
Figure 2. Hb does not directly activate hb transcription, but is required within the neuroblast for subsequent expression of HA:Hb in neuronal progeny born during the competence window.
(A) Prolonged Hb expression in NB7-1 results in all Eve+ neurons being Hb+, but only ~5 Eve+ neurons are HA:Hb+. Stage 14 embryo (en-gal4 UAS-hb HA:hb).
(B) Only ~5 Eve+ neurons are HA:Hb+ (blue) in 2nd instar larvae (en-gal4 UAS-hb HA:hb).
(C) Ectopic Hb protein in post-mitotic neurons (green) does not induce HA:Hb expression (blue). Only early-born U1/U2 neurons express HA:Hb. Ventral view, stage 16 (eve-gal4 UAS-hb HA:hb).
(D) Ectopic Hb protein in neuroblasts (green; NBs, arrows) does not induce expression of HA:Hb expression (blue) within neuroblasts. Lateral view, stage 11 (insc-gal4 UAS-hb HA:hb). Also see Figure S1.
(E) Schematic summary. Blue indicates endogenous HA:Hb, whereas green indicates Hb protein generated from UAS-hb ectopic overexpression. Blue bar at the bottom indicates active hb transcription in the neuroblast; white bar indicates the NB7-1 early competence window, when ectopic Hb in the neuroblast can induce HA:Hb expression in neuronal progeny.
Having shown that ectopic Hb can induce U1/U2 neuronal identity only during the early neuroblast competence window, we next asked whether ectopic Hb acts in neuroblasts or neurons to induce endogenous Hb expression. We observed that ectopic Hb expression specifically in post-mitotic U1-U5 neurons (eve-gal4 UAS-hb) was unable to induce HA:Hb expression (Figure 2C), consistent with previous observations (Pearson and Doe, 2003), suggesting that Hb must act in the neuroblast to specify U1/U2 identity. Interestingly, ectopic expression of Hb protein in neuroblasts did not directly induce HA:Hb expression within the neuroblasts themselves (Figure 2D), but rather indirectly induced subsequent HA:Hb expression in the neurons derived from the neuroblast (Figure 2A; summarized in Figure 2E). Similarly, Gal4/UAS-induced expression of Hb in neuroblasts was not able to directly activate hb transcription (Figure S1)(Grosskortenhaus et al., 2005). We conclude that ectopic Hb protein in the neuroblast can induce expression of HA:hb and hb in its neuronal progeny, but importantly this occurs only during the early competence window. Thus, the hb gene transitions through three states during the NB7-1 lineage: during the first two divisions the hb gene is actively transcribed (Figure 2E, blue bar); during the next three divisions the hb gene is not transcribed but can be induced by ectopic Hb protein (Figure 2E, white bar); and following the fifth division the hb gene becomes permanently silenced and is refractory to induction by ectopic Hb protein (Figure 2E, black bar).
The hb gene shows a near-synchronous movement to the nuclear lamina within the entire neuroblast population
The above data led us to hypothesize that the hb locus must undergo a transition within the neuroblast after the fifth division that renders it non-responsive to Hb protein, thus ending the neuroblast early competence window. A strength of the Drosophila neuroblast system is that one can image neuroblasts in vivo as they progress through their cell lineages. Therefore, we tracked the subnuclear position of the hb genomic locus within the population of wild type neuroblasts at different developmental stages in vivo. We used immunostaining to detect neuroblasts and DNA fluorescence in situ hybridization (DNA FISH, Bantignies et al, 2011) to detect the hb genomic locus within these cells in intact embryos. Our goal was to determine if the hb gene becomes repositioned into a repressive subnuclear domain – e.g., DAPI-intense foci or the nuclear lamina (Zhao et al., 2009) – concurrent with loss of neuroblast competence.
We generated a fluorescently labeled DNA probe spanning 10kb surrounding the hb coding region (this sequence is also present in the 32kb HA:hb genomic transgene). After probe hybridization, we immunostained embryos with antibodies to the pan-neuroblast marker Worniu and the nuclear envelope protein Lamin (Figure 3A-C) or stained with DAPI (not shown). We could detect up to two hb gene foci per neuroblast nucleus at all stages of development examined (average 1.35; n=926 foci). Duplication of the hb locus (HA:hb on chromosome 3L, endogenous hb on chromosome 3R) increased the number to up to four foci per neuroblast (average 2.12; n=823 foci), confirming the specificity of the DNA FISH probe. We did not observe an obvious relationship between hb gene position and DAPI-intense foci at any embryonic stage. In contrast, we observed a striking movement of the hb gene from the nuclear interior to the nuclear lamina nearly simultaneously within the entire neuroblast population at stage 12 (blue bar, Figure 3D). Quantifying the position of the hb gene at various distances from the nuclear lamina at progressively later embryonic stages (9–15) revealed a gradual movement from the nuclear interior towards the nuclear lamina between stages 9–11 (white bar, Figure 3D). We conclude that hb gradually moves closer to the nuclear periphery as the neuroblasts age, with a final, near-synchronous localization to the nuclear envelope.
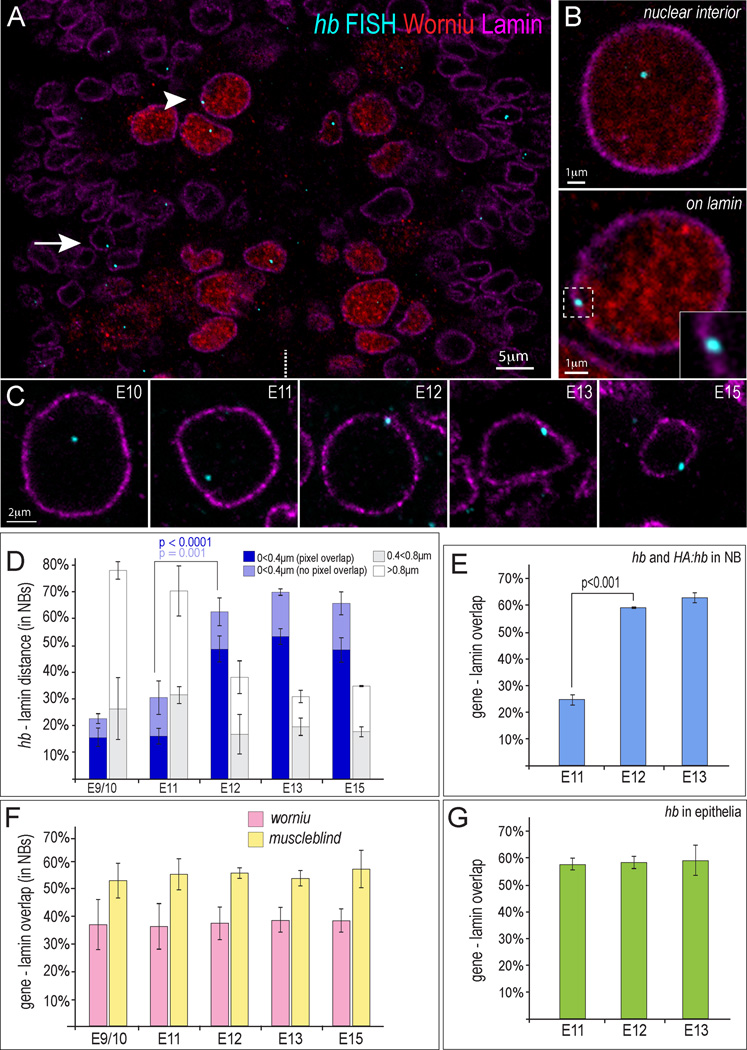
Figure 3. Coordinated movement of the hb genomic locus in the neuroblast population to the nuclear lamina at stage 12.
(A) A single optical plane of a stage 12 embryo (ventral view) hybridized with DNA probe against hb genomic locus (blue) and immunostained for the Worniu neuroblast marker (red) and Lamin nuclear envelope marker (magenta). Arrowhead, neuroblast; arrow, epithelial cell. Midline, white dotted line.
(B) Representative high-mag images of a single optical plane through a neuroblast showing the hb gene positioned in the neuroblast nuclear interior (top) or at the nuclear periphery (bottom). Inset enlarges the region indicated by white dotted box.
(C) Representative images of single neuroblasts indicating position of the hb gene (blue) within the nucleus at all analyzed stages (Worniu not shown).
(D) % hb loci positioned at the indicated distances from nuclear Lamin within neuroblasts at the indicated embryonic stages. E9/10=5 embryos (225 hb alleles), E11=3(246), E12=5(395), E13=3(285), and E15=3(227). Dark blue bar indicates pixel overlap between the hb FISH signal and the lamin signal (P value for stage 11–12 is <0.0001). Light blue indicates the hb FISH signal was detected within 0.4µm of lamin, but no pixels overlapped (P value for all signals detected <0.4um from lamin for stage 11–12 is =0.001).
(E) % hb loci with Lamin-FISH pixel overlap in HA:hb embryos. Both hb and HA:hb genes are detected with the FISH probe, yet a similar shift of the loci to the nuclear periphery is observed as that of hb alone (see D), indicating that HA:hb locus subnuclear position is regulated the same as the hb locus. Numbers scored: s11=3 embryos (277 alleles), s12=3(274), s13=3(272).
(F) % worniu (pink) or muscleblind loci (yellow) with overlapping pixels with the lamin signal. Numbers scored for worniu, muscleblind, respectively are: s9/10=5 embryos (259 worniu alleles), 3 embryos (145 muscleblind alleles); s11=4(155), 4(168); s12=5(272), 3(129); s13=4(215), 3(156); and s15=4(143), 3(135).
(G) Percentage of hb alleles with overlapping pixels with the lamin signal in epithelial cells (green). Numbers scored: s11=3(210), s12=3(232), s13=3(221). Error bars = SD.
We performed several additional experiments to better understand hb subnuclear repositioning in neuroblasts. Our hb FISH probe detected both the endogenous hb gene and the HA:hb 32kb recombineered gene, and in embryos containing both of these genes we observed the same repositioning of hb gene FISH signals to the nuclear periphery at stage 12 (Figure 3E); thus the subnuclear positioning of hb and HA:hb are similarly regulated. This result shows that the cis-regulatory DNA sequence for repositioning to the nuclear lamina must lie within the 32kb span of the HA:hb genomic construct, and such lamina-targeting mechanism can act on distinct chromosomal regions (hb on chromosome 3R; HA:hb on chromosome 3L). In contrast, the worniu and muscleblind genes (continuously or never expressed in all neuroblasts, respectively) showed no change in neuroblast subnuclear position over the same interval (Figure 3F). Similarly, the hb gene did not undergo repositioning in epithelial cells, which do not express hb (Figure 3G). These results show that gene repositioning is gene-specific (hb and HA:hb) and cell type-specific (neuroblasts). We never detected more than ~70% of any loci positioned to within 0.4μm of the lamina (Figure 3D-G); this may be the maximum percentage possible due to the dynamics of chromosome movements within the nucleus (Chubb et al., 2002; Heun et al., 2001; Marshall et al., 1997; Peric-Hupkes and van Steensel, 2010). We conclude that hb and HA:hb genes show a developmentally coordinated repositioning to the nuclear periphery within the entire neuroblast population at stage 12. The timing of this event was unexpected, because gene targeting to the lamina is typically associated with transcriptional repression (Guelen et al., 2008;Meister et al., 2010; Peric-Hupkes et al., 2010; Pickersgill et al., 2006; Shevelyov et al., 2009; Towbin et al., 2010; Williams et al., 2006), and termination of hb transcription occurs much earlier at stage 10 (see Figure 1A).
Repositioning of the hb locus to the nuclear lamina correlates tightly with the end of the NB7-1 competence window
To determine how closely the timing of hb gene repositioning to the nuclear lamina correlates with the end of the neuroblast early competence window, we focused on NB7-1, for which the length of the competence window has been well defined (Cleary and Doe, 2006; Pearson and Doe, 2003). One major advantage of the Drosophila neuroblast model system is that neuroblasts can be individually identified by their stereotyped position within the neuroblast array at early embryonic stages (Doe, 1992). This allowed us to track the position of the hb genomic locus not just among the neuroblast population, but also at the resolution of a single, identified neuroblast. Neuroblast positions become less stereotyped after stage 11, however, making it difficult to identify NB7-1 when its competence window ends at late stage 12 (Pearson and Doe, 2003). Therefore, we used the split-gal4 system (Luan et al., 2006) to generate a new tool for marking NB7-1 throughout neurogenesis. Briefly, we used recombineering to make a transgene that expresses the Gal4 DNA-binding domain under the control of a gooseberry enhancer, and a second transgene that expresses the Gal4 VP16 transactivation domain under the control of an achaete enhancer (Figure 4A). gooseberry and achaete are expressed in distinct subsets of neuroblasts, but only NB7-1 expresses both genes. Thus, combining the two transgenes resulted in the formation of a functional Gal4 protein in NB7-1 (Figure 4A-C). Using the NB7-1 split-gal4 line, we were able to label and unequivocally identify NB7-1 at all embryonic stages. Combined with immuno-DNA FISH, we were able to track the position of the hb gene specifically in NB7-1 throughout neurogenesis. We found that in NB7-1 the hb gene is initially localized to the nuclear interior, but became repositioned to the nuclear periphery by stage 13 (Figure 4D-E). This is slightly later than the timing of hb repositioning to the periphery in the entire neuroblast population (see above), but it precisely correlates with the loss of NB7-1 competence by stage 13 (Pearson and Doe, 2003).
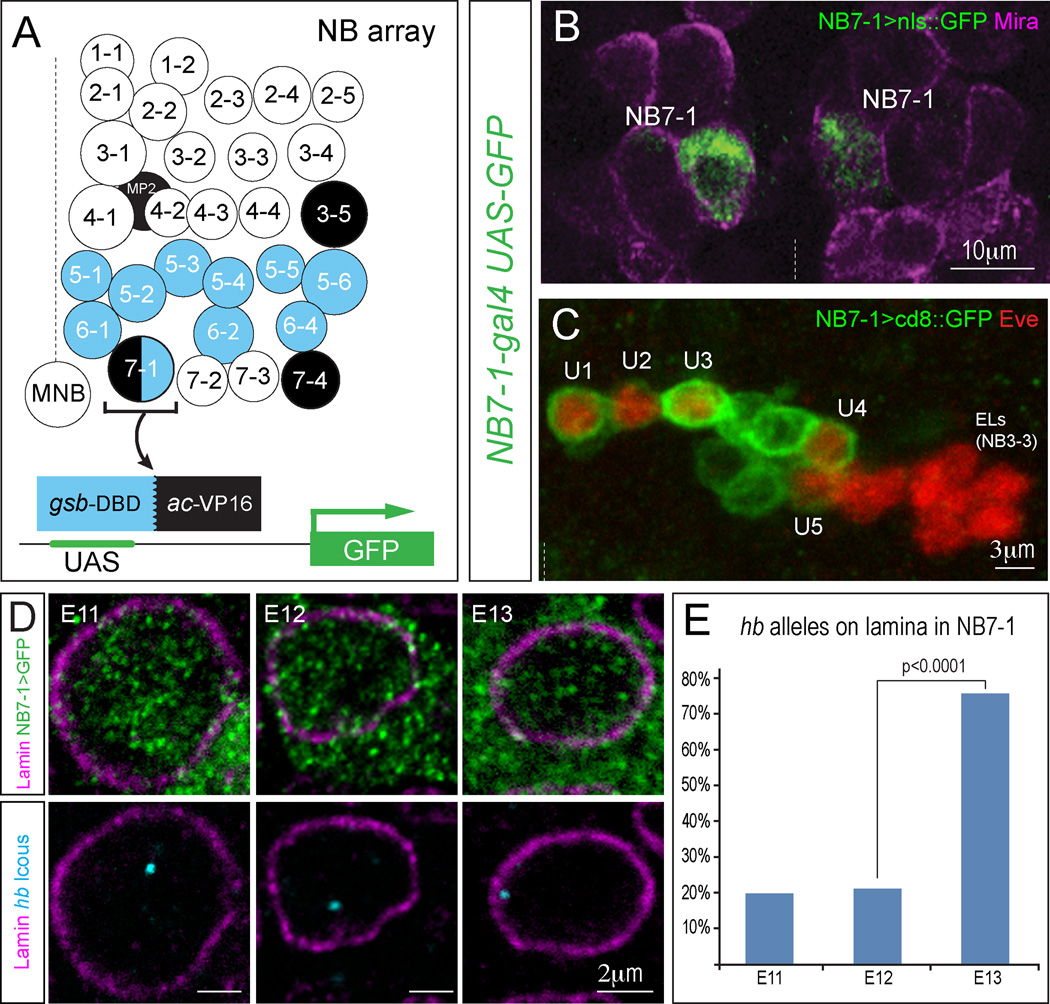
Figure 4. Movement of hb to the nuclear lamina is concurrent with loss of competence in NB7-1.
(A) Schematic of all neuroblasts present in each hemisegment, with gooseberry (blue) or achaete (black) expression indicated; only NB7-1 expresses both. Infrequently, NB6-1 is also labeled, but these can be identified by position and were excluded from the hb FISH quantifications.
(B) The NB7-1 split-gal4 driver was used to drive nuclear-localized GFP expression (green) and co-stained for the neuroblast marker Miranda (magenta) to show expression in the bilateral NB7-1 at stage 11.
(C) The split-gal4 driver marks the neuronal progeny of NB7-1 in one hemisegment at stage 16. All five U motoneurons born from NB7-1 marked by Eve expression (red) are labeled by cd8:GFP (green), but not the Eve+ EL neurons derived from NB3-3.
(D) NB7-1 split-gal4 driver was used to drive GFP expression (green) to identify NB7-1 at different stages. Note that GFP protein fills the cytoplasm, and can be detected outside of the nuclear membrane border (magenta). DNA FISH was used to detect hb subnuclear gene position (blue) in NB7-1.
(E) % hb loci with overlapping pixels with the lamin signal in NB7-1. Hemisegments scored (number of embryos examined): s11=45(9), s12=42(8), s13=53(8). Increased hb positioning at the nuclear periphery is observed by stage 13, precisely when NB7-1 is no longer competent to specify early-born fate. P value from Chi-squared test. Midline denoted in B and C with dashed line.
Depletion of Lamin decreases hb positioning at the nuclear periphery, reduces hb silencing, and extends neuroblast competence
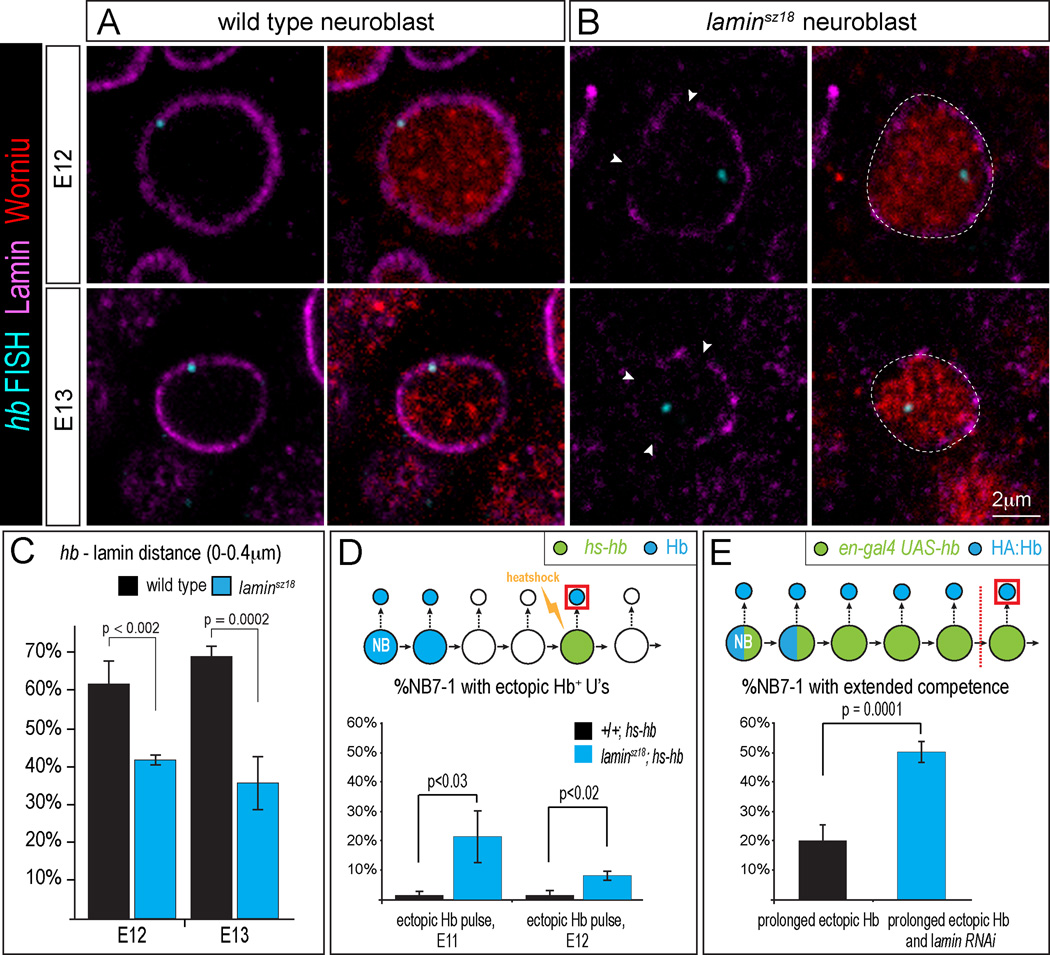
Our data show that at the end of the neuroblast early competence window, the hb gene becomes repositioned to the nuclear lamina, a nuclear compartment that many recent studies have shown is a highly repressive domain (reviewed in Peric-Hupkes and van Steensel, 2010; Rajapakse and Groudine, 2011; Shevelyov and Nurminsky, 2012). To determine if hb positioning at the nuclear periphery is required for permanent hb gene silencing and termination of the neuroblast early competence window, we used a laminsz18 null mutation, or expression of a lamin RNAi transgene, to reduce Lamin levels in neuroblasts. We first assayed homozygous laminsz18 null mutants. Due to perdurance of maternal gene products, Lamin protein levels are high in early embryos and gradually decline only at stages 12–13 (Figure S2). Whereas wild type neuroblasts have uniform Lamin staining around the nuclear envelope at stages 12–13 (Figure 5A), lamin sz18 mutants at the same stages show gaps in Lamin staining at the nuclear envelope (Figure 5B, arrowheads). In these laminsz18 mutant neuroblasts, we observed a decrease in hb positioning at the nuclear periphery (Figure 5B; quantified in Figure 5C). Next, we tested whether the reduction of hb positioning at the nuclear periphery affects hb silencing and/or the length of the competence window. We asked whether extra Hb+ U1/U2 neurons could be produced in response to brief ectopic Hb expression by transiently expressing a pulse of ectopic Hb protein in NB7-1 late in its competence window, in either wild type or laminsz18 mutant embryos. We observed very few ectopic Hb+ U1/U2 neurons in wild type, as expected (Figure 5D, black bars). In contrast, laminsz18 mutants given the same Hb pulse showed a significant increase in the percentage of NB7-1 lineages with ectopic Hb+ U1/U2 neurons (Figure 5D, blue bars). We conclude that lowering Lamin protein levels reduces hb gene silencing, thereby allowing more Hb+ U1/U2 neurons to be generated within the early competence window.
Figure 5. Decrease in Lamin levels reduces hb silencing and extends neuroblast competence.
(A-B) Wild type neuroblasts have continuous Lamin at the nuclear envelope (A), but laminsz18 mutants have breaks in Lamin staining at the nuclear envelope (B, arrowheads). Also see Figure S2.
(C) laminsz18 mutants show reduced hb positioning at the nuclear periphery. Number of embryos scored (number of hemisegments examined): wild type, s12=5(395), s13=3(285); laminsz18 mutants, s12=3(191), s13=5(285). Error bars=SD.
(D) % hemisegments with extra Hb+ U1/U2 neurons at stage 16. Either wild type (black bar) or laminsz18 mutants (blue bar) received a transient pulse of ectopic Hb at the indicated embryonic stage (E11, E12). Number of embryos scored (number of hemisegments examined): wild type, s11=5(72), s12=4(56); laminsz18 mutants, s11=3(47), s12=4(72). Error bars=SE.
(E) % hemisegments that have >5 HA:Hb+ neurons in embryos with prolonged ectopic Hb expression either with lamin RNAi, blue bar, N=11 embryos (188 hemisegments) or without lamin RNAi, black bar, N=11 embryos (194 hemisegments). Above each graph is a schematic of the experiment. Error bars=SE. Also see Figure S3.
We next determined if reduced Lamin levels could extend neuroblast competence. We used a UAS-laminRNAi transgene to reduce Lamin levels, and probed for the length of the early competence window by continuously expressing ectopic Hb in NB7-1 and assaying the number of HA:Hb+ U1/U2 neurons generated. Prolonged expression of ectopic Hb in wild type neuroblasts (en-gal4 UAS-hb) gave >5 HA:Hb+ neurons in 20.3±5.1% (SE) of the lineages (Figure 5E, black bar); in contrast, overexpression of Hb and the lamin RNAi transgene (en-gal4 UAS-hb UAS-laminRNAi) more than doubled the number of lineages with >5 HA:Hb+ neurons to 50.3±3.6% (SE) (p = 0.0001; Figure 5E, blue bar). Theoretically, reduced Lamin levels could result in a derepression of Hb, but we did not observe Hb derepression in the laminsz18 mutant neuroblasts at stage 12 (Figure S3). We conclude that hb positioning at the nuclear periphery is required for efficient hb gene silencing and the loss of neuroblast competence.
The Distal antenna (Dan) protein regulates hb subnuclear positioning and neuroblast competence
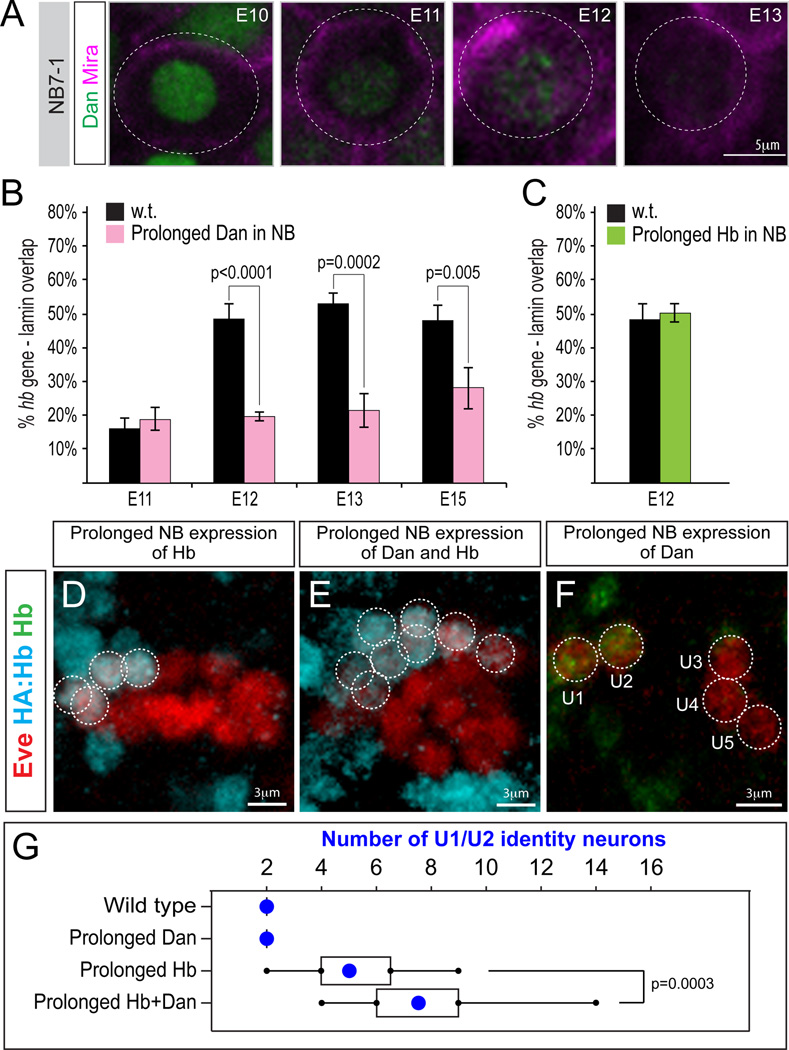
We previously showed that Dan, a Pipsqueak-motif nuclear protein, is detected in all neuroblasts from stage 9–11, and is downregulated nearly simultaneously in all neuroblasts during stage 12 (Kohwi et al., 2011; Figure S4), with one of the final neuroblasts to downregulate Dan being NB7-1 at late stage 12 (Figure 6A). Thus, downregulation of Dan is tightly correlated with hb gene movement to the nuclear lamina both within the whole neuroblast population, as well as within NB7-1. Dan is part of a protein superfamily defined by an N-terminal Pipsqueak DNA-binding domain, which includes the CENP-B centromere protein and Pogo transposase (Siegmund and Lehmann, 2002). CENP-B and Pogo form oligomers to bring distal DNA elements together in nuclear space through their DNA binding domains, and then perform catalytic functions using their C-terminal domains (Siegmund and Lehmann, 2002). Dan contains a single Pipsqueak domain and can form oligomers in vitro (Curtiss et al., 2007), but lacks known C-terminal catalytic domains (Siegmund and Lehmann, 2002).
Figure 6. Prolonging Dan expression blocks hb gene movement to the nuclear lamina and extends neuroblast competence.
(A) In NB7-1, Dan nuclear protein (green) is detectable until late stage 12 (magenta, neuroblast marker Miranda; Engrailed positional marker is not shown). Also see Figure S4.
(B) % hb loci with overlapping pixels with the lamin signal in wild type (black) or neuroblasts with prolonged Dan expression (insc-gal4/UAS-dan; pink).
(C) % hb loci with Lamin-FISH pixel overlap at stage 12 in wild type (black) or neuroblasts with prolonged Hb expression (insc-gal4/UAS-hb; green).
(D-F) Embryos overexpressing Hb, Hb plus Dan, or Dan alone stained for the U neuron marker Eve (red) and HA:Hb (blue; D,E) or Hb (green; F). Also see Figure S5.
The tight temporal correlation of Dan downregulation and hb repositioning to the lamina – plus the role of other Dan protein superfamily members in genome organization – led us to hypothesize that Dan downregulation may be required for movement of the hb gene to the nuclear lamina. Indeed, prolonged Dan expression in all neuroblasts (inscutable-gal4 UAS-dan) blocked movement of the hb gene to the nuclear lamina, and the proportion of hb loci at the nuclear periphery remained low from stage 9/10 through stage 15 (Figure 6B, pink bars). In contrast, prolonged expression of ectopic Hb in the neuroblast had no effect on the timing of hb gene repositioning to the nuclear lamina (Figure 6C, green bars), consistent with our conclusion that Hb specifies early-born neuronal identity but has no effect on the competence window. We conclude that Dan has the ability to regulate subnuclear gene organization in neuroblasts.
Prolonged Dan expression in neuroblasts prevented the hb genomic locus from repositioning to the nuclear lamina. If hb gene movement to the nuclear lamina terminates neuroblast competence, as we propose, then prolonged Dan expression should extend neuroblast competence. Wild type neuroblasts have a ~5-division competence window, and neuroblasts with prolonged ectopic Hb expression (HA:hb en-gal4 UAS-hb) cannot generate more than ~5 HA:Hb+ U1/U2 neurons (Figure 6D; quantified in G). When we prolonged expression of Dan together with Hb (HA:hb en-gal4 UAS-hb UAS-dan), we observed an increase to ~8 HA:Hb+ U1/U2 neurons (Figure 6E; quantified in G), significantly more than with misexpression of Hb alone. Thus, prolonged expression of Dan extended NB7-1 competence to respond to ectopic Hb. In contrast, prolonged expression of Dan alone in neuroblasts (en-gal4 UAS-dan) did not increase the number of neurons with Hb+ earlyborn identity (Figure 6F; quantified in G). This shows that prolonged expression of Dan in neuroblasts does not change the timing of hb transcription or directly specify U1/U2 neuronal identity. In light of these results, we note that it is theoretically possible that the reduction in hb gene silencing observed in our lamin mutant analysis could have been due to prolonged expression of Dan in the neuroblast, rather than through reduction in hb gene positioning at the nuclear periphery. However, laminsz18 mutant neuroblasts showed no change in the levels or timing of Dan expression (Figure S3, bottom panels). We conclude that prolonged expression of Dan, but not Hb, can maintain hb in the nuclear interior and extend neuroblast competence.
If Dan is sufficient to extend competence and maintain hb in the nuclear interior, is it necessary for these events? We assayed a mutant (dan danr) lacking both Dan and its close relative, Dan-related (Danr), and observed no change the length of the competence window or in the timing of hb subnuclear position (Figure S5), providing additional support for the tight relationship between hb subnuclear position and competence, and raising the possibility that Dan may function redundantly to regulate competence (see Discussion). Taken together, our data highlight the independent regulation of neuronal fate specification and progenitor competence: Hb potently induces early-born neuronal identity but has no effect on the competence window, whereas Dan can extend competence but cannot induce early-born cell fates.
DISCUSSION
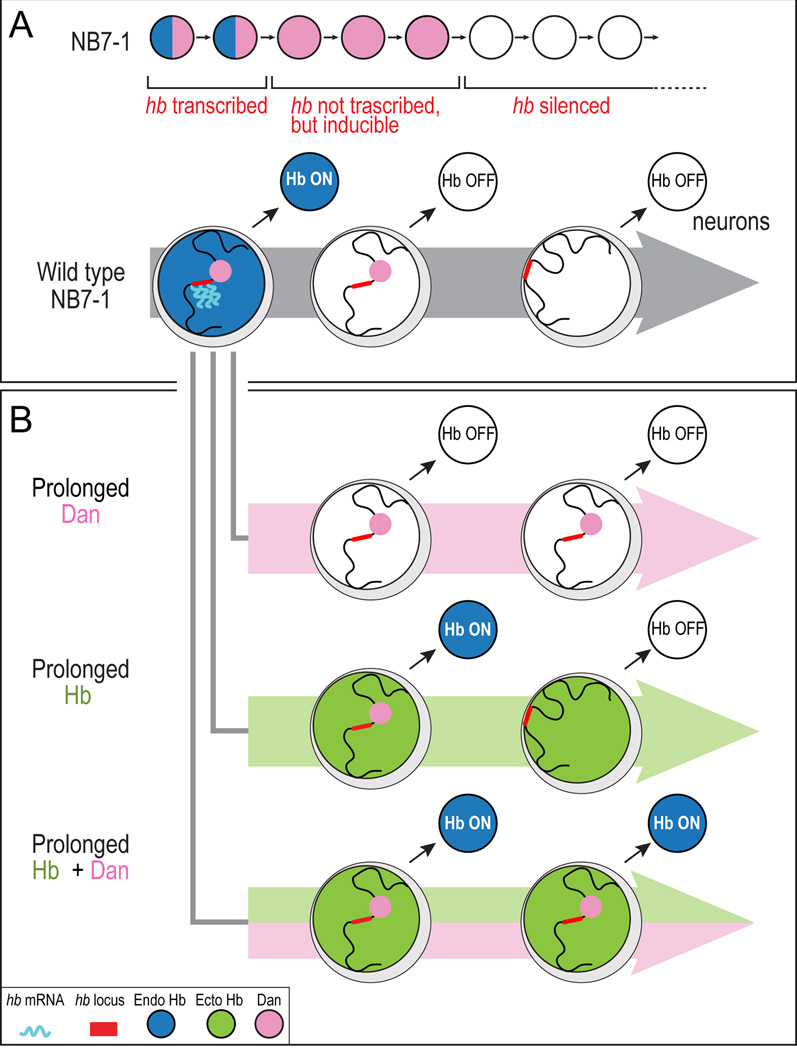
We find that the Drosophila embryo undergoes a reorganization of genome architecture that is gene-, cell type-, and developmental stage-specific. As neuroblasts age, the hb genomic locus becomes repositioned to the nuclear periphery, which marks the end of the neuroblast competence window to specify early-born cell fates. Why can’t ectopic Hb induce early-born fates after the close of the competence window? We propose that hb is just one of many genes that move to the nuclear lamina at the end of the early competence window – that a genome-wide reorganization shifts the neuroblasts into a state where Hb is unable to regulate the same targets it could during the competence window. In support of this model, we have shown that misexpression of Hb in the NB5-6 lineage has no effect on the activation of late-born cell fate factors (data not shown), consistent with a new genome organization that is refractory to Hb-induced early-born neuronal identity.
Our data lead us to propose that neural progenitors undergo a developmentally regulated reorganization of genome architecture as they age, potentially changing the palette of genes available to specify cell fate in aging progenitors. We propose the following three-step model for neuroblast competence. (1) In the newly formed NB7-1, the hb gene is in the nuclear interior and is transcriptionally active; this is the time when early-born Hb+ U1/U2 neurons are generated (Figure 7A, left). (2) After the second division of NB7-1, the transcriptional repressor Svp terminates hb transcription (Kanai et al., 2005; Mettler et al., 2006); however, the hb gene remains accessible in the nuclear interior where ectopic Hb protein can indirectly induce hb neuronal transcription to generate extra U1/U2 neurons (Figure 7A, middle). (3) After the fifth division of NB7-1, Dan protein is downregulated, resulting in the movement of hb (and probably many other Hb target genes) to the nuclear lamina; at this point, ectopic Hb in the neuroblast can no longer induce transcription of hb, and the competence window is closed (Figure 7A, right).
Figure 7. Neuroblast temporal identity and neuroblast competence are independently regulated.
(A) Neuroblast competence model.
(B) Dan and Hb independently regulate neuroblast temporal identity and competence. Note that ectopic Hb protein cannot induce endogenous Hb expression within the neuroblast. Rather, endogenous Hb is induced in the neurons born during the early competence window.
Our results are consistent with growing evidence from multiple organisms that repositioning of a gene to the nuclear lamina can cause transcriptional repression (Guelen et al., 2008; Peric-Hupkes and van Steensel, 2010; Pickersgill et al., 2006). For example, forced tethering of reporter genes to Lamin can repress reporter expression (Finlan et al., 2008; Meister et al., 2010; Reddy et al., 2008; Shevelyov et al., 2009; Williams et al., 2006), and Lamin depletion can de-repress silent genes (Shevelyov et al., 2009; Towbin et al., 2010). An important difference of our work, however, is that hb movement to the lamina occurs ~3h after termination of hb transcription, when hb undergoes an additional level of repression to become permanently silenced. Does lamina targeting induce permanent hb gene silencing, or vice versa? Depletion of the nuclear envelope protein Lamin displaces hb away from the lamina, decreases hb silencing, and increases neuroblast competence; this strongly suggests that lamina targeting is an early and essential step in permanent hb gene silencing and the loss of neuroblast competence. However we cannot rule out the possibility that hb positioning at the nuclear periphery might maintain, rather than establish, the permanently silenced state.
Our results show that neuroblast cell fate specification and neuroblast competence are independently regulated temporal programs. Prolonged expression of Dan can extend the competence window but cannot induce neuronal identity (Figure 7B, top). Conversely, Hb can specify early-born neuronal identity but cannot extend the competence window (Figure 7B, middle). Importantly, co-expression of Dan and Hb act synergistically: Dan extends the neuroblast competence window, and Hb "fills" this window with U1/U2 neurons, thereby specifying more early-born identity neurons than prolonging Hb alone (Figure 7B, bottom).
The mechanism and function of Dan is poorly understood. Why might Dan be sufficient, but not necessary, to promote neuroblast competence? One common finding is that dan and dan related (danr) genes show weak double mutant phenotypes, but strong misexpression phenotypes, in all tissues examined. For example, dan danr double mutants can live to adulthood with weak antennal and eye defects (Curtiss et al., 2007; Emerald et al., 2003), and show no change in the neuroblast competence window (this work). These findings are consistent with a redundant protein or pathway that can compensate for loss of Dan/Danr. A second model arises from comparing the dan danr mutant and Dan overexpression phenotypes. dan danr double mutants have a slight delay hb transcription termination at stage 10 (distinct from permanent hb silencing at stage 12) (Kohwi et al., 2011), but no effect on hb gene movement to the nuclear periphery (this work). Conversely, prolonged Dan expression has no effect on hb transcription, but blocks hb gene movement and extends the competence window. Thus, during the competence window Dan may promote a genome organization that allows timely access of the Svp transcriptional repressor to the hb locus, whereas following the competence window, Dan must be eliminated to allow a new genome organization to form. This model emphasizes the role of Dan in maintaining a genome organization in which hb and Hb target genes can be efficiently regulated. A third, not mutually exclusive, model is that Dan binds DNA via its Pipsqueak domain to competitively inhibit other Pipsqueak-like factors from recruiting hb and other loci to the nuclear lamina. Indeed, the founding member of the Pipsqueak-motif family, Pipsqueak, is a GAGA-binding factor (Huang et al., 2002), and recent work has shown an enrichment for GAGA motifs in Lamin-associated DNA sequences (Zullo et al., 2012). Consistent with this model, Dan protein is dispersed throughout the nucleoplasm (Figure 6A) where it could associate with hb and other loci.
What is the normal function of a competence window? Competence windows could provide both flexibility and limitations on the production of neural diversity. They could serve as a substrate for natural selection by allowing variation in neuronal subtype numbers through fluctuations in the length over which a progenitor is exposed to a temporal identity cue. Conversely, a competence window could also prevent stochastic fluctuations in the expression of a temporal identity cue from generating a neuronal subtype at a completely inappropriate time (e.g., an early-born fate at the end of a progenitor lineage), thereby limiting potentially deleterious effects. Another potential function of competence windows is that successive competence windows may allow the same cell fate determinant to generate different cell types. Indeed, during spinal cord development Olig2 first promotes neurogenesis and later induces oligodendrogenesis (reviewed in Rowitch, 2004), and retinal and cortical progenitors have been shown to progress through multiple competence states during which they can specify only distinct cell fates (McConnell, 1989; Okano and Temple, 2009). In support of this model, we previously showed that Dan has two waves of neuroblast expression, one at stage 9-late 12 and a second at stage 13–16 (Kohwi et al., 2011). Bimodal Dan expression may produce two competence windows in which the same temporal identity factors can access different genomic targets to generate additional neuronal diversity. For example, during the first Dan expression window Hb, Kr, Pdm, Castor, and Svp specify U1-U5 motoneuron identities in the NB7-1 lineage (Isshiki et al., 2001; Kanai et al., 2005; Kohwi et al., 2011; Mettler et al., 2006; Novotny et al., 2002); during the second Dan expression windows Kr, Castor, and Svp are re-expressed and a different population of neurons is produced (Benito-Sipos et al., 2011; Isshiki et al., 2001; Kanai et al., 2005; Kohwi et al., 2011).
Mammalian Ikaros, a Hb homologue, is expressed in young progenitors where it specifies early-born retinal ganglion cell (RGC) identity (Elliot et al., 2008). As with Hb, re-expression of Ikaros in older progenitors in vivo cannot induce specification of the early-born retinal ganglion cells (RGCs), although Ikaros misexpression in late retinal progenitors cultured in vitro can activate RGC-specific genes (Elliot et al., 2008). Perhaps in vitro cultured progenitors are lacking an extrinsic cue that closes the competence window, allowing Ikaros to generate more early-born neurons. Interestingly, Dan downregulation could also be regulated by an extrinsic cue, because Dan downregulation occurs nearly simultaneously in the entire neuroblast population, despite each neuroblast being at a different stage of its cell lineage.
Mammalian retinal and cortical progenitor cells change competence over time, generating an ordered series of distinct neural cells (Cepko et al., 1996; Livesey and Cepko, 2001; McConnell, 1989). In the future, it would be interesting to determine whether the genes expressed in early-born cortical or retinal cell types (Kwan et al., 2012; Leone et al., 2008; Trimarchi et al., 2007) are repositioned to the nuclear lamina in mammalian neural progenitors as competence to specify these cells is lost over time. Determining the mechanisms underlying loss of competence and identifying the molecular players in this process would have important implications for understanding normal brain development, adult tissue homeostasis, and tissue repair.
EXPERIMENTAL PROCEDURES
Fly stocks
We used recombineering (Venken et al., 2009) to make HA:hb transgenes containing three hemagglutinin tags (HA) in frame with the hb coding sequence and ~32kb of genomic DNA, and ΦC31-mediated integration was targeted to attP sites on the second or third chromosome (Bischof et al., 2007; Groth et al., 2004). We also used recombineering to make the gooseberry-DBD; achaete-VP16 split-gal4 stocks, which were inserted into attP sites on the second and third chromosomes. We used the following previously reported fly stocks: UAS-laminRNAi (TRIP consortium), engrailed-gal4 (Harrison et al., 1995; Isshiki et al., 2001; Pearson and Doe, 2003); inscutable-gal4 (1407-gal4, Bloomington Stock Center, Bloomington, IN); UAS-hb chromosomes II and III (Wimmer et al., 2000); hsp70-hb chromosome III (Struhl et al., 1992); UAS-dan (Emerald et al., 2003).
Immunostaining, RNA in situ hybridization and immuno-DNA FISH
Standard methods were used for antibody staining (Carney et al., 2012; Kohwi et al., 2011); larvae overexpressing Hb do not hatch and were manually devitellinized. The following primary antibodies were used: Dan rat 1:400 (gift from Jennifer Curtiss; New Mexico State Univ.), Eve mouse 3C10-c 1:50 (Developmental Studies Hybridoma Bank, DHSB, Iowa City, Iowa), Eve guinea pig 1:500 (gift from John Reinitz, Asian Distribution Center for Segmentation Antibodies, Mishima, Japan), GFP chicken 1:500 (Aves Labs, Tigard, Oregon), HA mouse 1:500 (Roche), Lamin rabbit R836 1:2000 (gift from Paul Fisher), Lamin mouse AD95-c 1:100 (DSHB), Hb rabbit 1:200 (Doe lab), Miranda rat 1:500 (Doe lab), Worniu rat 1:1 (Doe lab). Fluorescent secondary antibodies were from Jackson Lab Immunoresearch (West Grove, PA) or Invitrogen (Eugene, OR). Standard methods were used for RNA in situ hybridization (Grosskortenhaus et al., 2005).
We followed the immuno-DNA-FISH protocol described in Bantignies et al. (2011). We used the DNA FISH Tag kit (Invitrogen, #F32948) to generate DNA probes. Primers used to generate DNA FISH probes are listed in Supplemental Experimental procedures. DNA-FISH embryos were imaged on Zeiss 700 confocal with a 0.4μm step size. Pinholes were adjusted to equalize the resolution of all wavelengths in the x, y, and z axes. To measure gene position within the nucleus, we selected the optical section with the strongest FISH signal and measured the shortest distance from the center of the FISH signal to the nuclear lamina using ImageJ. FISH foci that were positioned within 0.4µm of the lamin fluorescent signal were further separately scored as either Lamin-FISH “pixel overlapping” or “no pixel overlapping.” Neuroblast FISH signals at stage 11–12 (the transition stages when hb gene movement to the lamina occurs) were initially scored by the first author, then scored blindly (with regard to stage) by a second person. The results were statistically indistinguishable.
Heat-shock Hb overexpression
Standard methods were used for heat shock Hb experiments (Pearson and Doe, 2003; Cleary and Doe, 2006) using fly stocks with a single hs-hb insertion on chromosome 3. The pulse of ectopic Hb is delivered at stage 11 or 12, but the NB7-1 progeny neurons are scored at stage 16. Some +/+; hs-hb and laminsz18; hs-hb embryos were fixed and stained just prior to heat shock to ensure the embryonic stages were synchronized between the genotypes.
Supplementary Material
Highlights.
Drosophila neuroblasts can make Hb+ neurons only during the early competence window
hb gene repositioning to the nuclear periphery ends the early competence window
Repositioning of the hb locus occurs 3 cell divisions after end of hb transcription
Dan and laminB regulate hb subnuclear positioning and neuroblast competence
Acknowledgements
We thank Hui Zong, Rui Galvão, Chong Liu, Eric Selker, Mike Cleary, and Doe lab members for discussion and comments on the manuscript; Keiko Hirono for blind scoring of the hb FISH data; Frédéric Bantignies for technical advice; Jennifer Curtiss for antibody and fly stocks; and Josh Udovich for technical assistance. We thank Janet Hanawalt and Marina Hirsch for copy editing. This work was supported by a Leukemia & Lymphoma Society career development award and a K99/R00 NIH grant from NICHD (HD072035) to MK, and funding from the NIH (HD27056) and HHMI to CQD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Polycomb-dependent regulatory contacts between distant Hox loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Benito-Sipos J, Ulvklo C, Gabilondo H, Baumgardt M, Angel A, Torroja L, Thor S. Seven up acts as a temporal factor during two different stages of neuroblast 5–6 development. Development. 2011;138:5311–5320. doi: 10.1242/dev.070946. [DOI] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179:41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Dev Biol. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Carney TD, Miller MR, Robinson KJ, Bayraktar OA, Osterhout JA, Doe CQ. Functional genomics identifies neural stem cell sub-type expression profiles and genes regulating neuroblast homeostasis. Dev Biol. 2012;361:137–146. doi: 10.1016/j.ydbio.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb JR, Boyle S, Perry P, Bickmore WA. Chromatin motion is constrained by association with nuclear compartments in human cells. Current biology : CB. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- Cleary MD, Doe CQ. Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 2006;20:429–434. doi: 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss J, Burnett M, Mlodzik M. distal antenna and distal antenna-related function in the retinal determination network during eye development in Drosophila. Dev Biol. 2007;306:685–702. doi: 10.1016/j.ydbio.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CQ. Molecular markers for identified neuroblasts and ganglion mother cells in the Drosophila central nervous system. Development. 1992;116:855–863. doi: 10.1242/dev.116.4.855. [DOI] [PubMed] [Google Scholar]
- Elliott J, Jolicoeur C, Ramamurthy V, Cayouette M. Ikaros confers early temporal competence to mouse retinal progenitor cells. Neuron. 2008;60:26–39. doi: 10.1016/j.neuron.2008.08.008. [DOI] [PubMed] [Google Scholar]
- Emerald BS, Curtiss J, Mlodzik M, Cohen SM. Distal antenna and distal antenna related encode nuclear proteins containing pipsqueak motifs involved in antenna development in Drosophila. Development. 2003;130:1171–1180. doi: 10.1242/dev.00323. [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosskortenhaus R, Pearson BJ, Marusich A, Doe CQ. Regulation of temporal identity transitions in Drosophila neuroblasts. Dev Cell. 2005;8:193–202. doi: 10.1016/j.devcel.2004.11.019. [DOI] [PubMed] [Google Scholar]
- Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Laroche T, Shimada K, Furrer P, Gasser SM. Chromosome dynamics in the yeast interphase nucleus. Science. 2001;294:2181–2186. doi: 10.1126/science.1065366. [DOI] [PubMed] [Google Scholar]
- Hirono K, Margolis JS, Posakony JW, Doe CQ. Identification of hunchback cis-regulatory DNA conferring temporal expression in neuroblasts and neurons. Gene Expr Patterns. 2012;12:11–17. doi: 10.1016/j.gep.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DH, Chang YL, Yang CC, Pan IC, King B. pipsqueak encodes a factor essential for sequence-specific targeting of a polycomb group protein complex. Mol Cell Biol. 2002;22:6261–6271. doi: 10.1128/MCB.22.17.6261-6271.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Kambadur R, Koizumi K, Stivers C, Nagle J, Poole SJ, Odenwald WF. Regulation of POU genes by castor and hunchback establishes layered compartments in the Drosophila CNS. Genes Dev. 1998;12:246–260. doi: 10.1101/gad.12.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai MI, Okabe M, Hiromi Y. seven-up Controls switching of transcription factors that specify temporal identities of Drosophila neuroblasts. Dev Cell. 2005;8:203–213. doi: 10.1016/j.devcel.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Kao CF, Yu HH, He Y, Kao JC, Lee T. Hierarchical deployment of factors regulating temporal fate in a diverse neuronal lineage of the Drosophila central brain. Neuron. 2012;73:677–684. doi: 10.1016/j.neuron.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson D, Baumgardt M, Thor S. Segment-specific neuronal subtype specification by the integration of anteroposterior and temporal cues. PLoS Biol. 2010;8:e1000368. doi: 10.1371/journal.pbio.1000368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Hiebert LS, Doe CQ. The pipsqueak-domain proteins Distal antenna and Distal antenna-related restrict Hunchback neuroblast expression and early-born neuronal identity. Development. 2011;138:1727–1735. doi: 10.1242/dev.061499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan KY, Sestan N, Anton E. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–1546. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone DP, Srinivasan K, Chen B, Alcamo E, McConnell SK. The determination of projection neuron identity in the developing cerebral cortex. Current opinion in neurobiology. 2008;18:28–35. doi: 10.1016/j.conb.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Luan H, Peabody NC, Vinson CR, White BH. Refined spatial manipulation of neuronal function by combinatorial restriction of transgene expression. Neuron. 2006;52:425–436. doi: 10.1016/j.neuron.2006.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW. Interphase chromosomes undergo constrained diffusional motion in living cells. Current biology : CB. 1997;7:930–939. doi: 10.1016/s0960-9822(06)00412-x. [DOI] [PubMed] [Google Scholar]
- McConnell SK. The determination of neuronal fate in the cerebral cortex. Trends in neurosciences. 1989;12:342–349. doi: 10.1016/0166-2236(89)90041-6. [DOI] [PubMed] [Google Scholar]
- Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010;24:766–782. doi: 10.1101/gad.559610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettler U, Vogler G, Urban J. Timing of identity: spatiotemporal regulation of hunchback in neuroblast lineages of Drosophila by Seven-up and Prospero. Development. 2006;133:429–437. doi: 10.1242/dev.02229. [DOI] [PubMed] [Google Scholar]
- Novotny T, Eiselt R, Urban J. Hunchback is required for the specification of the early sublineage of neuroblast 7-3 in the Drosophila central nervous system. Development. 2002;129:1027–1036. doi: 10.1242/dev.129.4.1027. [DOI] [PubMed] [Google Scholar]
- Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Curr Opin Neurobiol. 2009;19:112–119. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Regulation of neuroblast competence in Drosophila. Nature. 2003;425:624–628. doi: 10.1038/nature01910. [DOI] [PubMed] [Google Scholar]
- Pearson BJ, Doe CQ. Specification of temporal identity in the developing nervous system. Annu Rev Cell Dev Biol. 2004;20:619–647. doi: 10.1146/annurev.cellbio.19.111301.115142. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D, van Steensel B. Role of the nuclear lamina in genome organization and gene expression. Cold Spring Harb Symp Quant Biol. 2010;75:517–524. doi: 10.1101/sqb.2010.75.014. [DOI] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Rajapakse I, Groudine M. On emerging nuclear order. The Journal of cell biology. 2011;192:711–721. doi: 10.1083/jcb.201010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–247. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–419. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126:4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Shevelyov YY, Lavrov SA, Mikhaylova LM, Nurminsky ID, Kulathinal RJ, Egorova KS, Rozovsky YM, Nurminsky DI. The B-type lamin is required for somatic repression of testis-specific gene clusters. Proc Natl Acad Sci U S A. 2009;106:3282–3287. doi: 10.1073/pnas.0811933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevelyov YY, Nurminsky DI. The nuclear lamina as a gene-silencing hub. Curr Issues Mol Biol. 2012;14:27–38. [PubMed] [Google Scholar]
- Siegmund T, Lehmann M. The Drosophila Pipsqueak protein defines a new family of helix-turnhelix DNA-binding proteins. Dev Genes Evol. 2002;212:152–157. doi: 10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Cheng LY, Gould AP. Regulating neural proliferation in the Drosophila CNS. Current opinion in neurobiology. 2010;20:50–57. doi: 10.1016/j.conb.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Touma JJ, Weckerle FF, Cleary MD. Drosophila Polycomb complexes restrict neuroblast competence to generate motoneurons. Development. 2012;139:657–666. doi: 10.1242/dev.071589. [DOI] [PubMed] [Google Scholar]
- Towbin BD, Meister P, Pike BL, Gasser SM. Repetitive transgenes in C. elegans accumulate heterochromatic marks and are sequestered at the nuclear envelope in a copy-number- and lamin-dependent manner. Cold Spring Harbor symposia on quantitative biology. 2010;75:555–565. doi: 10.1101/sqb.2010.75.041. [DOI] [PubMed] [Google Scholar]
- Tran KD, Doe CQ. Pdm and Castor close successive temporal identity windows in the NB3-1 lineage. Development. 2008;135:3491–3499. doi: 10.1242/dev.024349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Stadler MB, Roska B, Billings N, Sun B, Bartch B, Cepko CL. Molecular heterogeneity of developing retinal ganglion and amacrine cells revealed through single cell gene expression profiling. The Journal of comparative neurology. 2007;502:1047–1065. doi: 10.1002/cne.21368. [DOI] [PubMed] [Google Scholar]
- Venken KJ, Carlson JW, Schulze KL, Pan H, He Y, Spokony R, Wan KH, Koriabine M, de Jong PJ, White KP, et al. Versatile P[acman] BAC libraries for transgenesis studies in Drosophila melanogaster. Nat Methods. 2009;6:431–434. doi: 10.1038/nmeth.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Williams RR, Azuara V, Perry P, Sauer S, Dvorkina M, Jorgensen H, Roix J, McQueen P, Misteli T, Merkenschlager M, et al. Neural induction promotes large-scale chromatin reorganisation of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- Wimmer EA, Carleton A, Harjes P, Turner T, Desplan C. Bicoid-independent formation of thoracic segments in Drosophila. Science. 2000;287:2476–2479. doi: 10.1126/science.287.5462.2476. [DOI] [PubMed] [Google Scholar]
- Zhao R, Bodnar MS, Spector DL. Nuclear neighborhoods and gene expression. Current opinion in genetics & development. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H. DNA sequent-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.