Abstract
To identify the evolutionary genetic novelties that contributed to shape human-specific traits such as the use of a complex language, long-term planning and exceptional learning abilities is one of the ultimate frontiers of modern biology. Evolutionary signatures of functional shifts could be detected by comparing noncoding regions that are highly conserved across mammals or primates and rapidly accumulated nucleotide substitutions only in the lineage leading to humans. As gene loci densely populated with human-accelerated elements (HAEs) are more likely to have contributed to human-specific novelties, we sought to identify the transcriptional units and genomic 1 Mb intervals of the entire human genome carrying the highest number of HAEs. To this end, we took advantage of four available data sets of human genomic accelerated regions obtained through different comparisons and algorithms and performed a meta-analysis of the combined data. We found that the brain developmental transcription factor neuronal PAS domain-containing protein 3 (NPAS3) contains the largest cluster of noncoding-accelerated regions in the human genome with up to 14 elements that are highly conserved in mammals, including primates, but carry human-specific nucleotide substitutions. We then tested the ability of the 14 HAEs identified at the NPAS3 locus to act as transcriptional regulatory sequences in a reporter expression assay performed in transgenic zebrafish. We found that 11 out of the 14 HAEs present in NPAS3 act as transcriptional enhancers during development, particularly within the nervous system. As NPAS3 is known to play a crucial role during mammalian brain development, our results indicate that the high density of HAEs present in the human NPAS3 locus could have modified the spatiotemporal expression pattern of NPAS3 in the developing human brain and, therefore, contributed to human brain evolution.
Keywords: NPAS3, brain, evolution, humans, schizophrenia
Introduction
The identification of the genetic basis underlying the phenotypic differences that characterize Homo sapiens is one of the major goals of current evolutionary studies. Distinctive human traits such as the use of a complex language, long-term planning, and exceptional learning capacities are considered to be the result of a series of molecular and phenotypic changes in the developmental program of our brain that occurred along the last 6 My of human lineage evolution. The genes that contributed to peculiar human traits have probably acquired novel mutations in their coding or regulatory sequence leading to the origin of differential protein functional domains or changes in their spatial or temporal expression, respectively. By comparing sequences from ortholog loci of several vertebrate genomes, including the human and chimpanzee, it is possible to identify brain genes and genomic regions showing accelerated evolution in the human lineage as candidates to reveal the molecular basis of the particular configuration of the human brain. So far, most studies have focused in comparing protein-coding regions and a few genes linked to speech or brain size with signatures of positive selection in the human lineage have been identified, suggesting that they might have played a role during recent human evolution (Enard et al. 2002; Evans, Anderson, Vallender, Choi, et al. 2004; Evans, Anderson, Vallender, Gilbert, et al. 2004; Evans et al. 2005; Mekel-Bobrov et al. 2005). More recently, other examples including the characterization of the gene SRGAP2 and its human-specific paralogs in the maturation of dendritic spines (Charrier et al. 2012; Dennis et al. 2012) have highlighted the importance of performing functional studies of human-specific genes that evolved as a consequence of duplication and deletions in the human lineage (Fortna et al. 2004) to investigate the evolutionary history of our brain.
As protein-coding regions account for only one-third of all conserved sequences in the human genome and several studies performed in other species showed that lineage-specific evolution of regulatory sequences appeared to have driven phenotypic changes (for a review, see Carroll [2005, 2008]), detection of human-specific accelerated conserved noncoding regions would likely be highly informative. In fact, almost 40 years ago, King and Wilson (1975) proposed the idea that evolution of noncoding regulatory sequences played a major role in shaping the unique features of the human brain although a proof of this concept is still pending. As gene loci densely populated with human-accelerated elements (HAEs) are more likely to have contributed to human-specific novelties, we sought to identify the transcriptional units and genomic 1 Mb intervals of the entire human genome carrying the highest number of HAEs. To this end, we took advantage of four recent independent genome-wide studies performed to identify accelerated conserved nonprotein-coding regions in the human genome (Pollard, Salama, King, et al. 2006; Pollard, Salama, Lambert, et al. 2006; Prabhakar et al. 2006; Bush and Lahn 2008; Lindblad-Toh et al. 2011) that altogether detected approximately 1,800 genomic human fast evolving regions. In this work, we performed a meta-analysis of these four public databases to identify clusters of human genomic-accelerated elements. Although these studies searched for conserved genomic regions with accelerated substitution rates specific for the human branch, each of them yielded partially overlapping output sequences probably because they used different type and number of compared species and different algorithms to assess acceleration. Our meta-analysis revealed that most transcriptional units and 1 Mb intervals carry none, 1, 2, or 3 HAEs, and only very limited cases showed a concentration of HAEs greater than 7. A maximum and exceptional case corresponds to the NPAS3 (neuronal PAS domain-containing protein 3) locus that contains 14 HAEs, the largest cluster of noncoding-accelerated elements in the human genome. NPAS3 encodes a transcription factor of the bHLH-PAS family (Brunskill et al. 1999) and is mainly expressed in the developing central nervous system of mice and humans as well as in the adult brain (Brunskill et al. 1999; Gould and Kamnasaran 2011). Analysis of NPAS3-deficient mice showed that this gene plays an important role during brain development and the control of neurosignaling pathways (Erbel-Sieler et al. 2004; Brunskill et al. 2005). In addition, NPAS3 was identified as a candidate gene for schizophrenia in family (Kamnasaran et al. 2003; Pickard et al. 2005, 2006) and association studies (Pickard et al. 2008; Huang et al. 2010). To explore the possibility that the human-specific genetic changes present in NPAS3 played a role in brain evolution, we tested the ability of the individual 14 HAEs clustered in the NPAS3 locus to act as transcriptional enhancers using a transgenic zebrafish reporter expression assay. We found that 11 out of the 14 HAEs present in NPAS3 drive reporter gene expression to the developing central nervous system, suggesting that the accelerated evolution of the these regulatory sequences could have had an impact on the expression of NPAS3 in humans.
Results
Genome-Wide Cluster Map of Human-Accelerated Nonprotein-Coding Elements
To identify clusters of HAEs in the human genome, we used four publicly available databases (Pollard, Salama, King, et al. 2006; Pollard, Salama, Lambert, et al. 2006; Prabhakar et al. 2006; Bush and Lahn 2008; Lindblad-Toh et al. 2011). We assembled the 1,820 regions identified in these works into a single data set, where overlapping sequences present in two or more original databases were counted as single elements (fig. 1A). This nonredundant secondary data set contains 1,629 HAEs. Next, we determined the clustering of HAEs in gene intervals, as reported by the largest RefSeq for each gene, and in 1 Mb genomic intervals (fig. 1B). This analysis revealed that the vast majority of genes or 1 Mb genomic intervals lack HAEs. From the 19,897 annotated human RefSeq genes (human genome assembly 2006; NCBI36/hg18), only 433 (2.5%) carry at least one HAE (supplementary table S1, Supplementary Material online, and fig. 1B). An analysis of distribution shows that most HAE-containing RefSeq genes carry only 1, 2, or 3 HAEs (416 of 496, 84%, fig. 1B) and only 7 human RefSeq genes from the entire human genome contain 6 or more HAEs (table 1 and fig. 1C). Thus, genes carrying a large cluster of HAEs are highly uncommon. Similarly, after fractioning the human genome in 1 Mb intervals (∼3,000), we found that only 14 (0.5%) carry 6 or more HAEs (fig. 1C and table 1).
Fig. 1.
Clustering of HAEs in the human genome. (A) Venn diagram showing the overlap among the HAEs from the different databases: HARs (Pollard, Salama, King, et al. 2006; Pollard, Salama, Lambert, et al. 2006); HACNS (Prabhakar et al. 2006); HTBE (Bush and Lahn 2008); and 29-mammals HARs (2× HAR; Lindblad-Toh et al. 2011). In addition, there are 65 overlapping elements between the 2× HAR and the HACNS databases and 3 elements between the HAR and the HTBE databases that are not included in the diagram. Number of HACNS and 2× HAR elements in the diagram is larger than the number of elements in the published data sets due to multiple overlap at HACNS56/2× HAR20 and HACNS11 loci. (B) Frequency histogram showing the distribution of HAEs in clusters of 1–14 per gene (obtained as merged RefSeq intervals) and per 1 Mb length genomic intervals (only window 2 [win 2] shown). (C) Schematic drawing of the location of the HAEs clusters in the chromosomes of the human genome. On the right of the chromosomes the blue bars indicate the density of HAEs as generated by the density track in Ensembl (www.ensembl.org, last accessed February 28, 2013). Clusters of HAEs in RefSeq genes containing more than 6 elements are indicated by red arrows. In addition, the name of the gene including the cluster is indicated. Green arrows indicate the largest HAE clusters by megabase (only clusters of more than 8 HAEs are shown) that are not indicated by red arrows.
Table 1.
Genes and Genomic Regions Containing more than 6 HAEs.
| No. of HAEs | RefSeq Position (hg18) | Gene Symbol | HAEs by 100 kb |
|---|---|---|---|
| 14 | chr14: 32478210–33340702 | NPAS3 | 1.62 |
| 8 | chr3: 59710076–61212173 | FHIT | 0.53 |
| chr16: 6009133–7703341 | RBFOX1 | 0.47 | |
| chr16: 76691052–77804065 | WWOX | 0.72 | |
| 7 | chr11: 130745581–131711926 | NTM | 0.72 |
| 6 | chr7: 145444386–147749021 | CNTNAP2 | 0.26 |
| 6 | chr13: 95541093–96289813 | HS6ST3 | 0.80 |
| No. of HAEs | Mb Position (hg18) | Genes in Interval | HAEs by 100 kb |
| 14 | chr14: 32250032–33250032 | AKAP6, NPAS3 | 1.4 |
| 12 | chr14: 32500032–33500032 | NPAS3, EGLN3 | 1.2 |
| 9 | chr14: 32000032–33000032 | AKAP6, NPAS3 | 0.9 |
| chr9: 16000016–17000016 | BNC2 | 0.9 | |
| chr2: 21250021–22250021 | — | 0.9 | |
| chr14: 32750032–33750032 | NPAS3, EGLN3 | 0.9 | |
| 8 | chr2: 21000021–22000021 | APOB, LOC645949 | 0.8 |
| chr4: 182000182–183000182 | LINC00290 | 0.8 | |
| chr4: 182250182–183250182 | LINC00290 | 0.8 | |
| chr13: 70500070–71500070 | DACH1 | 0.8 | |
| chr16: 6500006–7500006 | RBFOX1 | 0.8 | |
| chr2: 21500021–22500021 | LOC645949 | 0.8 | |
| chr9: 15750015–16750015 | C9orf93, BNC2 | 0.8 | |
| chr13: 95750095–96750095 | HS6ST3, OXGR1, MBNL2 | 0.8 | |
| chr16: 6750006–7750006 | RBFOX1 | 0.8 | |
| chr3: 60250060–61250060 | FHIT | 0.8 | |
Interestingly, we have found one RefSeq corresponding to the NPAS3 gene, that contains the exceptional amount of 14 HAEs, the largest number for a single gene in the entire human genome (fig. 1C and table 1). NPAS3 is followed by WWOX (chr16: 76691052–77804065), FHIT (chr3: 59710076–61212173), and RBFOX1 (chr16: 6009133–7703341) that contain 8 HAEs each (fig. 1C and table 1). As the transcriptional units of NPAS3, WWOX, FHIT, and RBFOX1 are rather large, encompassing 0.9, 1.1, 1.5, and 1.7 Mb, respectively, we tested whether large genes are highly enriched in HAEs. From the 92 transcriptional units larger than 800 kb present in the human genome, 63 (68%) contain at least one HAE, indicating that large genes have a significantly higher probability of containing accelerated elements (supplementary table S1, Supplementary Material online; Fisher’s exact test P < 0.00001), in agreement with the fact that the frequency of distribution of conserved noncoding elements in the human genome is rather uniform. Actually, large genes accumulate an expected number of noncoding conserved sequences as identified with the PhastCons program (Fisher’s exact test not significant; supplementary table S1, Supplementary Material online).
Analysis of 1 Mb sliding windows (every 250 kb) also showed that the 1 Mb region more densely populated with HAEs is that carrying NPAS3 (table 1). Furthermore, the second HAE-dense 1 Mb window, containing 12 HAEs, also includes NPAS3 (table 1). In addition, there are four 1 Mb regions containing nine HAEs, two of which include NPAS3 as well. The other two fragments are located in chromosome 9 (chr9: 16000016–17000016) that includes the 0.5 Mb Basonuclin 2 gene (BNC2) and a gene desert region located in chromosome 2 (table 1 and fig. 1B and C). Finally, there are ten 1 Mb regions containing eight HAEs (table 1 and fig. 1B and C).
To analyze more carefully the density of HAEs in NPAS3, we have divided the number of HAEs by the length of the transcriptional unit for each gene containing HAEs. The results indicate that NPAS3 displays 1.6 HAEs every 100 kb of length ranking first as the densest HAE-populated gene among the top HAE-carrying genes (table 1).
The Accelerated Evolution of the NPAS3 Locus Is Restricted to Its Noncoding Regions
To gain insight into the evolution of the NPAS3 locus, we analyzed the human-specific substitutions in its coding and noncoding regions. Although NPAS3 is the human gene with the largest cluster of HAEs, its encoded protein is highly conserved among mammals showing an average of approximately 95% of identity that increases to 98.7% among primates (supplementary table S2, Supplementary Material online). NPAS3 identity between human and chimpanzee is 99.8 %, with only two amino acid substitutions (A559P and A787G) over 940 residues. In addition, we have analyzed the evolution of the protein-coding sequence of NPAS3 in the human and primate lineages to detect acceleration or positive selection. Our results show that NPAS3 did not undergo any process of positive selection in the lineage leading to humans (table 2). Altogether, these data reveal that while the coding region of NPAS3 has been evolving under strong purifying selection, a significant portion of conserved intronic noncoding sequences underwent a recent human-specific accelerated evolutionary process.
Table 2.
NPAS3 Coding Sequence Analysis.
| NPAS3 | Log Likelihood | |||||
|---|---|---|---|---|---|---|
| M0 Model | −14,902.86123 | |||||
| Branch-Site Models | LRT | P | Parameters Estimation | |||
| Primates | ||||||
| Alternative hypothesis | −10,044.620481 | 0.000000 | 0.0000 | p0 = 0.97012 | p1 = 0.02988 | p2 = 0.00000 |
| Null hypothesis | −10,044.620481 | ω0 = 0.01766 | ω1 = 1.00000 | ω2 = 1.00000 | ||
| Human | ||||||
| Alternative hypothesis | −10,044.620473 | 0.000000 | 0.0000 | p0 = 0.97012 | p1 = 0.02988 | p2 = 0.00000 |
| Null hypothesis | −10,044.620473 | ω0 = 0.01766 | ω1 = 1.00000 | ω2 = 1.00000 | ||
Note.—LRT (likelihood ratio test) = 2 × [LnLHA (alternative hypothesis) − LnLH0 (null hypothesis)]; P, P value of the chi-square test.
To further compare the evolution of coding and noncoding sequences in the NPAS3 genomic region, we performed an analysis of the accelerated and conserved sequences in the NPAS3 genomic region using phyloP (http://compgen.bscb.cornell.edu/phast/, last accessed February 28, 2013). We selected the genomic portion that spans from the last base of AKAP6 to the first base of EGLN3, the 5′ and 3′ adjacent genes to NPAS3, respectively. That selection renders a genomic interval of 1091152 base pairs that covers from base 32372020 to base 33463171 of chromosome 14 in the human genome (NCBI36/hg18). We split this genomic region into four nonoverlapping partitions: NPAS3 coding sequence, HAEs elements, noncoding 17-way most conserved PhastCons elements (without HAEs), and noncoding nonconserved genomic regions. We obtained for each of these partitions an alignment with four selected primate species: human, chimpanzee, orangutan, and rhesus macaque. We used only primate species to obtain reliable alignments in the nonconserved noncoding regions. Using the phyloP program, from the PHAST software package, we computed P values of conservation and acceleration for our test alignments under the Siepel, Pollard, and Haussler (SPH) method (Siepel et al. 2006). The P values of conservation and acceleration were calculated in each case for a subtree holding only the human branch and a supertree made up of the remaining species: chimpanzee, orangutan, and rhesus. The neutral model of evolution with which to compare all the test alignments was derived from the primates.mod neutral model used in UCSC’s conservation track. This analysis confirmed our results obtained using PAML showing that the coding region of NPAS3 is highly conserved (P = 0.001 in the human branch and P = 3.54 × 10−28 in the supertree) and does not show any signatures of acceleration in the human lineage (P = 0.99). In addition, we found that the PhastCons regions containing HAEs show a significantly higher probability to fit an accelerated model of evolution in the human lineage (P = 1.47 × 10−4) than a conservation model (P = 0.99), whereas they show signatures of high conservation in the nonhuman primates (P = 1.16 × 10−65). On the other hand, conserved noncoding PhastCons regions not containing HAEs show a higher probability of fitting a conservation model than an acceleration model in the whole set of species (2.29 × 10−97). In addition, nonconserved noncoding regions do not fit a conservation model neither in the supertree (P = 0.82) or the subtree (P = 0.99) and show signatures of acceleration and neutral evolution in the subtree (P = 7.5 × 10−7) and the supertree (P = 0.187), respectively.
Functional Characterization of NPAS3 Accelerated Elements in Transgenic Zebrafish
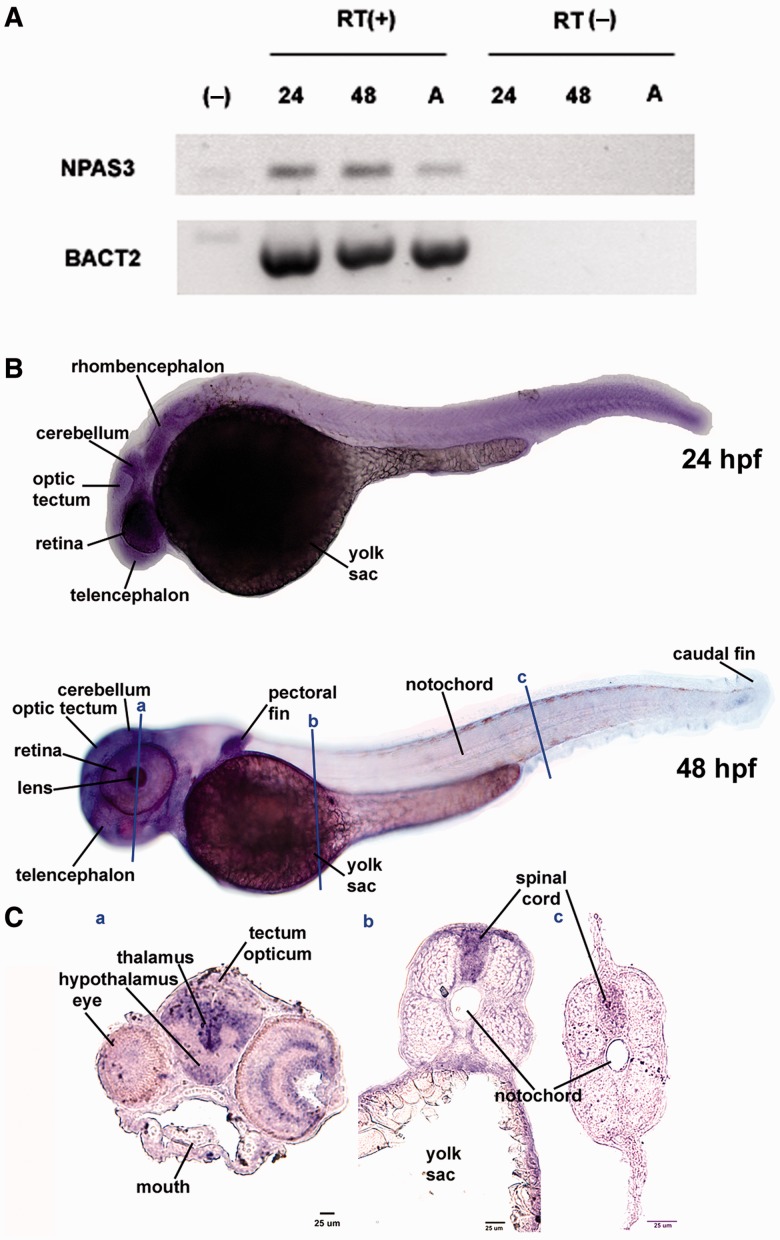
As NPAS3 is expressed in highly proliferative zones of the anterior developing mouse and human brain (Brunskill et al. 1999; Gould and Kamnasaran 2011; Allen Brain Atlas, www.brain-map.org, last accessed February 28, 2013), the HAEs present in NPAS3 could have played a role in human brain evolution. To test this possibility, we investigated the ability of the 14 HAEs of NPAS3 to act as transcriptional enhancers during brain development by using a reporter expression assay in transgenic zebrafish. Although transgenic zebrafish have been shown to be an efficient and reliable expression system in which to test putative human transcriptional enhancers (Fisher, Grice, Vinton, Bessling, McCallion 2006), we considered of importance to first determine whether NPAS3 is expressed in the developing zebrafish brain. To this end, we performed reverse transcriptase-polymerase chain reaction (RT-PCR) and in situ hybridization at several developmental stages using two different primer sets and two digoxigenin-labeled antisense riboprobes designed against the zebrafish NPAS3 mRNA. Our RT-PCR results (fig. 2A) indicate that NPAS3 is strongly expressed during early stages of zebrafish development and in the adult brain and our in situ hybridization studies showed that NPAS3 is ubiquitously expressed at 24 h postfertilization (hpf) and a day later (48 hpf) the expression pattern is restricted to specific structures and tissues, including the anterior developing brain (fig. 2B). At 48 hpf, NPAS3 is highly expressed along the central nervous system, including the brain and the spinal cord (fig. 2C). In the brain, we observed high expression in diencephalic structures including the thalamus, hypothalamus, and optic tectum (fig. 2C) and also in the telencephalon (fig. 2B). In addition, we observed expression in the eye, heart and particularly, a high expression in the branchial arcs and the roof of the mouth (fig. 2B and C). Thus, these results together with previous reports obtained in mouse and human embryos (Brunskill et al. 1999; Gould and Kamnasaran 2011) indicate that the developmental expression pattern of NPAS3 is conserved across vertebrates and confirm that the zebrafish is a suitable in vivo model in which to study the regulation of NPAS3 expression.
Fig. 2.
Expression of the NPAS3 gene during zebrafish development. (A) RT-PCR analysis of NPAS3 expression in zebrafish embryos and adult brain. RT-PCR products of NPAS3 and control gene b-actin2 are shown: (−), negative control; 24, 24 hpf; 48, 48 hpf; A, adult brain with (+) or without (−) reverse transcriptase. (B) Whole mount in situ hybridization studies show the spatial expression pattern of the gene NPAS3 at 24 hpf (top) and 48 hpf (bottom). (C) In situ hybridization on coronal cryostat slices showing a detail of the NPAS3 expression pattern in 48 hpf zebrafish embryos.
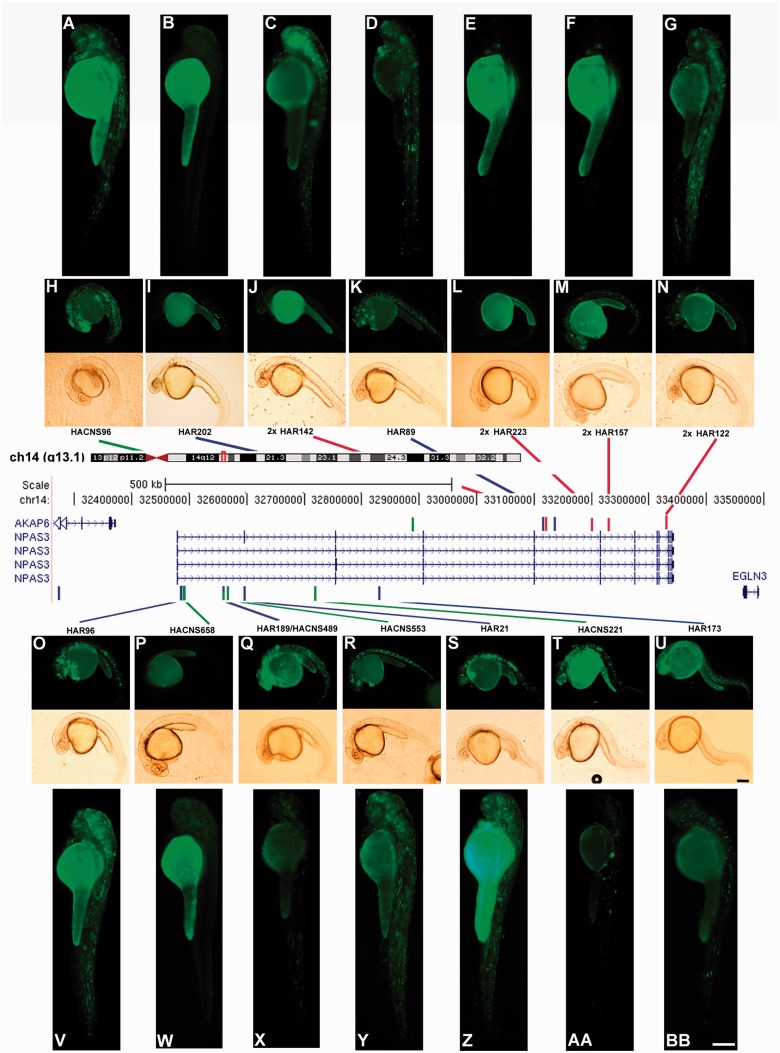
To generate transgenic zebrafish, we subcloned each of the 14 HAEs in an Tol2-based expression vector upstream of a mouse c-fos minimal promoter (Dorsky et al. 2002) followed by EGFP coding sequences (Kawakami et al. 2004; Fisher, Grice, Vinton, Bessling, Urasaki, et al. 2006). Expression analyses performed at 24 and 48 hpf revealed that 11 of the 14 HAEs present in NPAS3 drove distinctive and reproducible EGFP expression patterns at 24 and 48 hpf (fig. 3 and table 3). From the 11 HAEs capable of driving EGFP expression in a reproducible manner, only 6 are conserved in vertebrates, including zebrafish. Moreover, two out of the three HAEs that do not drive consistent EGFP expression are conserved in zebrafish (supplementary table S3, Supplementary Material online). The control vector produced essentially no signal with occasional specimens carrying irregular arrays of green fluorescent cells (table 3). Altogether, these results demonstrate that most NPAS3 HAEs may act as transcriptional enhancers during brain development and confirm previous findings that the capability of a sequence to behave as an enhancer does not necessarily depend on sequence conservation (Fisher, Grice, Vinton, Bessling, McCallion 2006).
Fig. 3.
The NPAS3 gene contains the largest number of HAEs. In the central panel, schematic drawing modified from the UCSC genome browser, showing the NPAS3 locus in genomic context on chromosome 14q13.1. Below, the four splicing variants of the NPAS3 gene are shown. The coloured bars indicate the location of the 14 HAEs. Blue bars indicate HARs (Pollard, Salama, King, et al. 2006; Pollard, Salama, Lambert, et al. 2006); green bars indicate HACNS (Prabhakar et al. 2006); and red bars show 29-mammals HARs (2× HAR; Lindblad-Toh et al. 2011). The overlapping HAE is shown by a green–blue bar. On top and below of the central panel, fluorescent and bright field microphotographs showing representative transgenic zebrafish of each HAE expressing EGFP at 24 hpf (H, I, J, K, L, M, N, O, P, Q, R, S, T, and U) and 48 hpf (A, B, C, D, E, F, G, V, W, X, Y, Z, AA, and BB). Small boxes (U and BB) represent 250 μm.
Table 3.
Expression Analysis in Transgenic Zebrafish at 24 and 48 h Postfertilization.
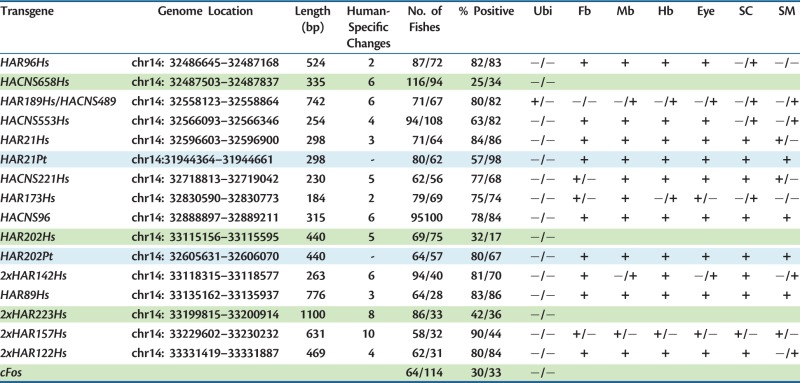 |
Note.—Green cells indicate HAEs in addition to control transgene cFos that do not show expression in transgenic zebrafishes. Light blue cells indicate chimpanzee transgenes. Ubi, ubiquitous; Fb, forebrain; Mb, midbrain; Hb, hindbrain; SC, spinal cord; SM, somitic muscle. Positive (+) and negative (−) expression.
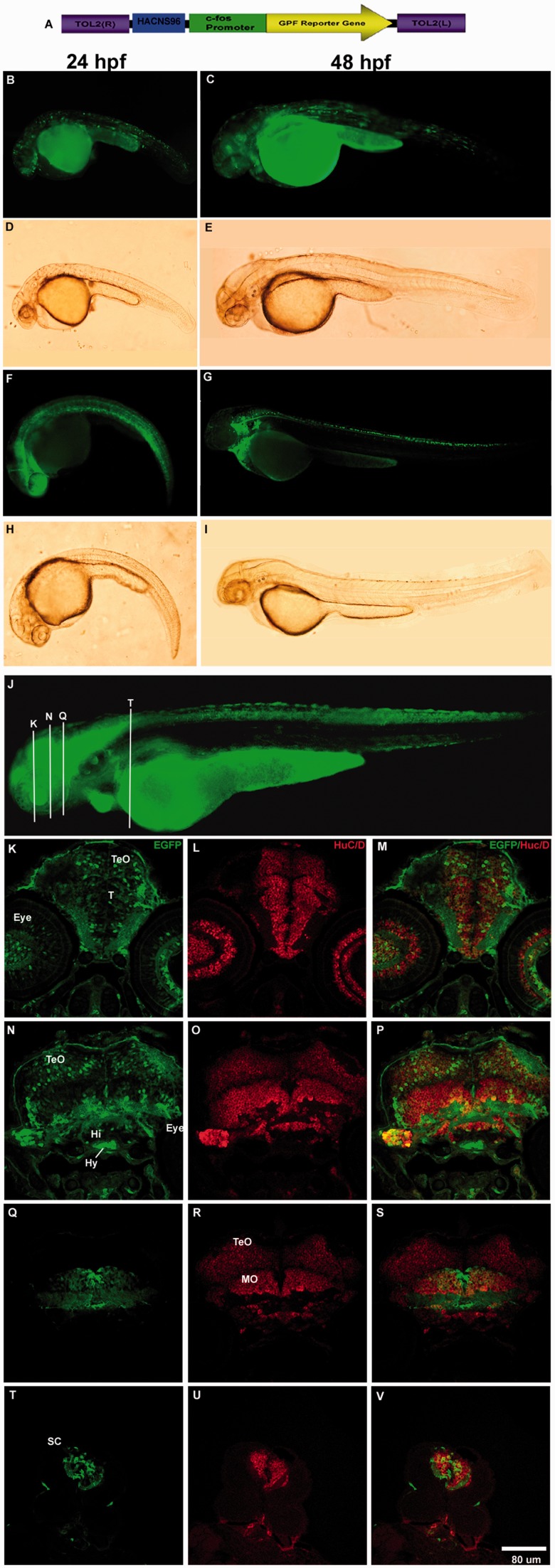
To validate the reproducibility of transient expression experiments, we generated transgenic zebrafish lines for selected HAEs (HAR21, HAR96, chimp HAR202, human HAR202, and HACNS96) and found that the expression patterns observed in transient transgenic zebrafish at 24 and 48 h is consistent with those observed in the stable transgenic lines (figs. 4 and 5 and supplementary fig. S1, Supplementary Material online).
Fig. 4.
HACNS96 drives EGFP expression to the developing nervous system in stable and transient transgenic zebrafish lines at 24, 48, and 72 hpf. (A) Diagram of the transgene structure based on the Tol2 system. Fluorescent (B, C, F, and G) and bright field (D, E, H, and I) photomicrographs of selected whole mount transient or stable transgenic zebrafish embryos expressing the reporter protein EGFP under control of the human HACNS96 sequence at 24 hpf (B, D, F, and H) and 48 hpf (C, E, G, and I). (J) Representative transgenic zebrafish expressing EGFP under the control of HANCS96 at 72 hpf where white bars indicate approximately the location of coronal cryostat sections showing a detail of the EGFP expression pattern (K, N, Q, and T), cells expressing the early neuronal marker HuC/D (L, O, R, and U) and the resulting overlay (M, P, S, and V). TeO, tectum opticum; T, midbrain tegmentum; DT, dorsal tegmentum; Po, preoptic region; MO, medulla oblongata; OC, otic capsule; SC, spinal cord; Hi, hypothalamus; Hy, hypophysis.
Fig. 5.
EGPF expression driven by HAR96 in stable and transient transgenic zebrafish lines at 24 and 48 hpf. (A) Diagram of the transgene structure based on the Tol2 system. Lower panel, fluorescent (B, C, F, and G) and bright field (D, E, H, and I) photomicrographs of selected whole mount transient or stable transgenic zebrafish embryos expressing the reporter protein EGFP under control of the human HAR96 sequence at 24 hpf (B, D, F, and H) and 48 hpf (C, E, G, and I). Coronal cryostat sections showing a detail of the EGFP expression pattern at three levels (white vertical bars in G) of the 48 hpf developing zebrafish (J, K, and L). Below, cells expressing EGFP (M and P) the early neuronal marker HuC/D (N and Q) and the resulting overlay (O and R) in the 48 hpf developing zebrafish (sections levels are approximately indicated in G). TeO, tectum opticum; T, midbrain tegmentum; DT, dorsal tegmentum; Po, preoptic region; MO, medulla oblongata; OC, otic capsule; SC, spinal cord; Hi, hypothalamus; Hy, hypophysis.
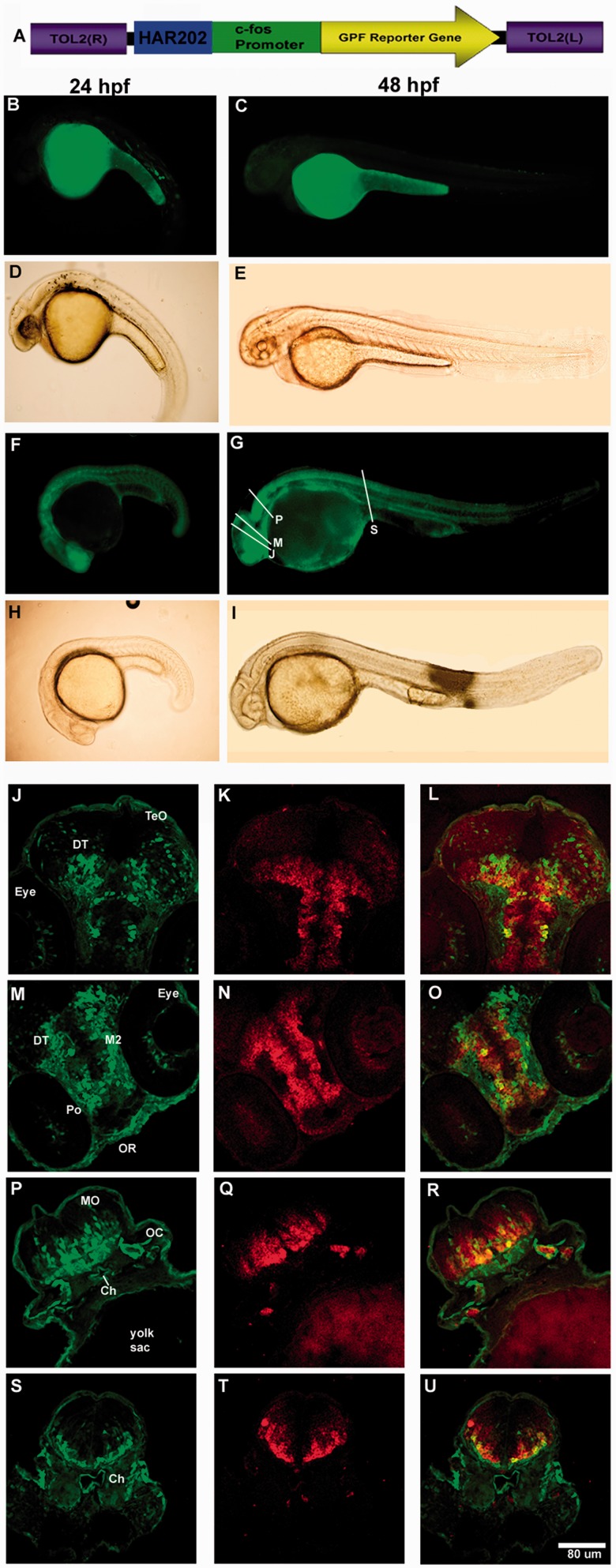
The three HAEs incapable to drive reporter gene expression in transgenic zebrafish probably do not code for transcriptional enhancers. Alternatively, the accelerated substitutions accumulated in the lineage leading to humans disrupted enhancer function in a human-specific manner probably modifying the transcription factor repertoire that can bind to the sequence. Interestingly, we found that all human-specific substitutions present in each one of the 14 NPAS3-HAEs produce either a gain or loss of putative transcription factor binding sites (TFBS; supplementary table S3, Supplementary Material online), suggesting that they could have contributed to changes in the spatiotemporal expression domain of NPAS3 in the human brain. To test this latter possibility, we selected one of these three elements, HAR202, which is highly conserved throughout vertebrates, including the zebrafish genome (fig. 6B). Thus, we tested in transgenic zebrafish the expression pattern driven by a transgene containing human HAR202 (HAR202-Hs) in parallel with a similar transgene carrying its chimpanzee ortholog (HAR202-Pt). Interestingly, transient and stable expression analysis at 24 and 48 hpf revealed that although HAR202-Hs did not generate any reproducible expression, HAR202-Pt drove a consistent EGFP expression pattern in the developing nervous system of transgenic zebrafish (table 3 and fig. 6). To find an explanation for the apparent loss of expression in the human HAR202 enhancer, we analyzed the TFBS gain and loss compared with the chimpanzee sequence. We found that two TFBS were lost in the human HAR202 sequence, STAT5 and BCL6 (supplementary table S3, Supplementary Material online). Both transcription factors are expressed in the developing brain (Markham et al. 2007; Leamey et al. 2008) and although the function of BCL6 in the developing brain has not been elucidated yet, it has been shown that STAT5 plays a key role in the generation of cortical interneurons and in axon guidance in the spinal cord and the telencephalon during brain development (Markham et al. 2007). More experiments will be needed to probe if the loss of these TFBS is the mechanism underlying human HAR202 lack of expression and also the functional impact that this could have had in the expression of NPAS3 in the human brain.
Fig. 6.
Differences in expression driven by human and chimpanzee HAR202 orthologs. (A) Diagram of the transgene structure based on the Tol2 system. Below, fluorescent (B, C, F, and G) and bright field (D, E, H, and I) photomicrographs of selected whole mount transgenic zebrafish embryos expressing the reporter protein EGFP under control of the human (HAR202-Hs) or the chimpanzee (HAR202-Pt) sequences at 24 and 48 hpf. Coronal cryostat sections showing a detail of cells expressing EGFP (J, M, P, and S), cells expressing the early neuronal marker HuC/D (K, N, Q, and T) and the resulting overlay (L, O, R, and U) in the 48 hpf developing zebrafish (sections levels are approximately indicated in G). TeO, tectum opticum; T, midbrain tegmentum; DT, dorsal tegmentum; Po, preoptic region; MO, medulla oblongata; OC, otic capsule; SC, spinal cord; Hi, hypothalamus; Hy, hypophysis.
In addition, to analyze comparatively the expression patterns driven by human and chimpanzee orthologs of additional HAEs, we selected HAR21 because the human version of this sequence drove strong expression of the reporter gene EGFP in transgenic zebrafish. We found that the chimpanzee version of HAR21 (HAR21-Pt) drove a consistent EGFP expression pattern in the developing nervous system of transgenic zebrafish (table 3) showing that this sequence behaves in a similar manner to the human sequence in the developmental stages analyzed. This result does not discard the fact that different temporal and spatial expression patterns could be driven by these two sequences at other developmental stages. More studies will be necessary to assess this possibility.
HAEs Expression Analysis in the Developing Zebrafish Embryo
To analyze in more detail the expression pattern driven by selected NPAS3-HAEs (HAR202 chimp, HAR96, HAR21, and HANCS96), we performed an expression analysis of EGFP in coronal cryostat sections of developing zebrafish at 24, 48, and 72 hpf. Although, in general, we observed that the four selected HAEs drove EGFP expression to the developing nervous system of zebrafish embryos at the three stages analyzed, we found some differences in the expression pattern driven by each enhancer. The four transgenes showed a wide expression pattern in the developing nervous system including the telencephalon, the optic tectum, the midbrain, the hindbrain, and the spinal cord (figs. 4–6 and supplementary fig. S1, Supplementary Material online). The four transgenes also drove expression to the developing eye (figs. 4–6 and supplementary fig. S1, Supplementary Material online). We observed that four transgenes drove the expression of EGFP to early born neurons labeled with HuC/D expression (Park et al. 2000). In fact, we observed in HACNS96 transgenic zebrafish that EGFP partially colocalized with HuC/D expressing cells in the tectum optic and in the most lateral part the tegmentum (TeO and T; fig. 4K–M). In addition, we observed high EGFP-HuC/D colocalization in the ventral part of the medulla oblongata (MO) in the hindbrain (fig. 4Q–S) and in the spinal cord (SC; fig. 4T–V). We also observed that the chimp HAR202 (fig. 6P–U) and the HAR96 (fig. 5M–R) sequences drove expression to early born neurons in the ventral and lateral parts of the hindbrain and spinal cord. On the other hand, we observed that HAR202Pt (fig. 6J–L) as well as HAR96 (fig. 5J, K) drove expression of EGFP to fewer cells in the tectum opticum compared with HANCS96Hs (fig. 4K–M). Besides, we observed that the HAR21hs enhancer drove a wide EGFP expression pattern in the developing central nervous system including forebrain, midbrain, hindbrain, and spinal cord (supplementary fig. 1, Supplementary Material online).
Discussion
In this article, we identified the genes and genomic regions containing the largest clusters of HAEs in the human genome. In general, we found that very large genes have a significantly higher probability of containing accelerated elements in agreement with the fact that they carry more noncoding conserved sequences. Among the genes displaying the largest cluster of HAEs, we identified NPAS3 as an extreme case containing 14 noncoding HAEs. This surprisingly great number of HAEs for a single gene contrasts with the expected difference between the human and chimpanzee NPAS3 protein. In fact, during the evolution of the human lineage, the coding portion of NPAS3 has been highly conserved showing only 2 amino acid substitutions over 940 residues compared with the chimpanzee ortholog. In addition, we found that the coding region of NPAS3 did not undergo accelerated evolution in the human or primate lineages.
It has been suggested that phenotypic divergence between humans and chimpanzees is largely due to changes in regulatory sequences rather than in the protein-coding sequences (King and Wilson 1975). As the 14 HAEs we detected in NPAS3 could be regulatory elements, we tested their ability to drive expression of the reporter fluorescent protein EGFP in transgenic zebrafish embryos. We previously validated the use of this expression system by showing that the only NPAS3 gene present in the zebrafish genome is expressed in the developing nervous system, making transgenic zebrafish a suitable vertebrate model where to study the regulation of NPAS3 expression. We found that at least 11 HAEs have the capability to drive the expression of the reporter gene EGFP in transgenic zebrafish embryos at 24 and 48 hpf in a reproducible manner. Thus, our data suggest that NPAS3 could have had a significant impact in human brain evolution. In addition, our data demonstrate that reporter expression in transgenic zebrafish is a useful live vertebrate model to rapidly evaluate HAEs as putative transcriptional enhancers in agreement with has been initially shown for human conserved noncoding elements (Fisher, Grice, Vinton, Bessling, McCallion 2006). For the HAEs HAR21, HAR96, HAR202, and HACNS96, we also generated stable transgenic zebrafish lines and found that EGFP expression is highly restricted to brain and spinal cord structures where NPAS3 is normally expressed in vertebrates (Brunskill et al. 1999; Gould and Kamnasaran 2011) as we consistently observed in transient transgenic embryos, indicating that evaluation of reporter gene expression following embryo microinjection is likely to provide reliable results.
To further assess the functional impact of the accelerated evolution of the NPAS3-HAEs, we compared the expression ability of the human and chimpanzee HAR202 ortholog sequences. We selected this element because the human HAR202 failed to yield EGFP expression in transgenic zebrafish embryos. As chimpanzee HAR202 produced a reproducible expression pattern at 24 and 48 hpf in the developing brain, it is tempted to speculate that the molecular evolution of HAR202 in the human lineage led to a loss of regulatory function. The differential expression properties of the human and chimp HAR202 orthologs probably accounts for the first documented functional difference of a brain enhancer between these two closely related primates.
These results are consistent with previous results that suggested that the fast evolution of HAR202 was possibly driven by relaxation of constraint (Kostka et al. 2012). It has been described that human-accelerated regions (HARs) show a strong W to S bias (Pollard, Salama, King, et al. 2006; Prabhakar et al. 2006; Bush and Lahn 2008) and that a significant number of these HARs are located in the final band of chromosome arms (Pollard, Salama, King, et al. 2006) where a higher recombination rate has been described (Lander et al. 2001). It has been suggested that regions of high recombination rate are associated with increases in W to S substitutions through a process known as biased gene conversion (Duret et al. 2002; Galtier and Duret 2007; Duret and Arndt 2008; Duret and Galtier 2009). We observed that 8 out of the 14 NPAS3-HAEs showed a strong AT (weak or W) to GC (strong or S) substitution bias (supplementary table S3, Supplementary Material online), whereas only one HAE (HAR21) showed an S to W bias. Although NPAS3 is not located near the tip of chromosome 14, we cannot rule out the possibility that biased gene conversion may have contributed to the evolution of NPAS3 HAEs. On the other hand, a recent study developed to quantify the nonlinear interacting effects of selection and biased gene conversion on the substitution rates and patterns of HARs showed that more than one-half (55%) have substitution rates that significantly exceed local estimates of the neutral rate, suggesting that these regions may have been shaped by positive selection rather than by relaxation of constraint (Kostka et al. 2012). Regarding NPAS3-HAEs, the mentioned study found that HAR21 shows signatures of positive selection, whereas the fast evolution of HAR89, HAR96, and HAR202 is better explained by relaxation of constraint (Kostka et al. 2012). In addition, HAR173 and HAR189 show signatures of bias gene conversion (Kostka et al. 2012). It is interesting to note that regarding HAR202 our comparative expression results between the human and chimpanzee ortholog sequences suggest a loss of function of this element, which is consistent with the relaxation of constraint showed by the analysis of Kostka et al. (2012). On the other hand, we found that the chimpanzee version of HAR21 (HAR21-Pt) drove a consistent EGFP expression pattern in the developing nervous system of transgenic zebrafish in a similar manner to the human sequence in the developmental stages analyzed. This result does not discard the fact that different temporal and spatial expression pattern could be driven by these sequences at other developmental stages. More studies will be necessary to assess this possibility.
Analysis of NPAS3-deficient mice showed that this gene plays an important role during brain development and the control of neurosignaling pathways (Erbel-Sieler et al. 2004). In addition, NPAS3 was identified as a candidate gene for schizophrenia after analyzing a chromosome abnormality in a small family with schizophrenia and mental retardation (Kamnasaran et al. 2003; Pickard et al. 2005, 2006). NPAS3 is located in a schizophrenia susceptibility locus (chr 14q13) previously identified by linkage (Chiu et al. 2002) and association studies, including a genome-wide association study, that found multiple NPAS3 single-nucleotide polymorphisms and mental illness risk haplotypes (Pickard et al. 2008; Huang et al. 2010). The translocation found in schizophrenia was predicted to generate a truncated NPAS3 protein retaining DNA-binding activity but lacking the dimerization and transactivation domains, suggesting a loss-of-function model as one mechanism by which genetic variation in NPAS3 increases risk for mental illness (Chiu et al. 2002). In support of the loss of NPAS3 function in schizophrenia, mouse knockout models for NPAS3 and related protein NPAS1 present some behavioural, neuroanatomical, and molecular traits observed in schizophrenic subjects such as hyperlocomotion, memory impairment, reduced prepulse inhibition, reduced corpus callosum and hippocampal volume, enlarged ventricles, abnormal dopamine signaling, and reduced reelin expression (Erbel-Sieler et al. 2004; Brunskill et al. 2005; Pieper et al. 2005).
Surprisingly, we found other genes linked to schizophrenia and autism spectrum disorder (ASD) among the top HAEs-carrying genes. In fact, association, linkage, gene expression, imaging data, and knockout mouse models support the role of contactin-associated protein-like 2 (CNTNAP2), which carries six HAEs in its transcriptional unit in ASD (Alarcón et al. 2008; Peñagarikano et al. 2011; Peñagarikano and Geschwind 2012; Tan et al. 2012) and schizophrenia (Friedman et al. 2008; Ji et al. 2012). In addition, we found that the RNA-binding protein FOX-1 homolog, RBFOX1 (also termed A2BP1 or FOX1) contain the surprising amount of eight HAEs in its transcriptional unit. RBFOX1 gene disruption has been associated with a variety of neurodevelopmentally based brain disorders, including autism (Barnby et al. 2005; Martin et al. 2007; Sebat et al. 2007) and schizophrenia (Xu et al. 2008).
Finally, this report provides an example of how the accelerated evolution of conserved nonprotein-coding elements can produce a significant functional impact by losing TFBS or gaining new ones. In fact, our in silico analysis showed that all HAEs involve putative TFBS losses or gains, suggesting that the human-specific nucleotide changes could have led to modifications in the spatiotemporal expression domain of NPAS3 in the developing human brain. Moreover, we found that two TFBS were lost in the human HAR202 sequence, STAT5 and BCL6 (supplementary table S3, Supplementary Material online). Both BCL6 and STAT5 are expressed in the developing brain (Markham et al. 2007; Leamey et al. 2008) and although the function of BCL6 in the developing brain has not been elucidated yet, it has been shown that STAT5 plays a key role in the generation of cortical interneurons and in axon guidance in the spinal cord and the telencephalon during brain development (Markham et al. 2007). Further studies will be necessary to assess whether the loss of these TFBS is the mechanism underlying human HAR202 lack of expression and if this human-specific evolutionary process is related to the involvement of NPAS3 as a susceptibility locus for schizophrenia.
Materials and Methods
Analysis of HAEs Clustering
To identify clusters of genomic-accelerated elements in the human genome, we used four publicly available databases containing a total of 1,820 HAEs including 202 HARs (Pollard, Salama, King, et al. 2006; Pollard, Salama, Lambert, et al. 2006), 992 human-accelerated noncoding sequences (HACNS; Prabhakar et al. 2006), 63 human terminal branch elements (HTBE; Bush and Lahn 2008), and 563 29-mammals HARs (2× HAR; Lindblad-Toh et al. 2011). Genomic coordinates originally published as hg17 intervals (HAR and HACNS) were manually converted to hg18 intervals according to the UCSC Genome Browser (www.genome.ucsc.edu, last accessed February 28, 2013) using BLAT. The resulting hg18 intervals from the four selected databases were concatenated into one data set that we named HAEs where overlapping elements in two or more original data sets were converted into one element. We analyzed the clustering of the human-accelerated sequences joining the HAEs data set with a data set of RefSeq intervals or four data sets of 1-Mb length genomic intervals that overlapped every 250 kb using Galaxy tools (http://main.g2.bx.psu.edu/, last accessed February 28, 2013). RefSeq intervals were obtained from the USCS table browser. To avoid redundancies, two or more overlapping RefSeq genes were converted into one genomic interval. Occurrences of each record (RefSeq or 1 Mb region) were counted to generate the number of HAEs per RefSeq or in 1 Mb.
To obtain the percentage of conserved sequences that underwent a process of accelerated evolution, we obtained the conserved noncoding regions in the NPAS3 locus from the UCSC table browser (as identified in 17 way_most_conserved multi-species genomic alignments by the PhastCons program with a conservation score ≥400). Conserved regions that overlapped spliced ESTs, interspersed repeats, low-complexity DNA, and duplicated blocks were eliminated using the following UCSC browser’s tracks as filters: spliced ESTs, repeat masker, and self-chain. A total of 400 conserved regions were obtained. We calculated the number of bases covered by these 400 PhastCons regions (101,140 bp) and by the 14 PhastCons conserved elements that contain the 14 NPAS3 HAEs. A list of Ref Seq genes with a length ≥800 kb was obtained filtering them by length using Galaxy tools. The genomic coordinates were obtained manually according to the UCSC Genome Browser using BLAT and redundancies were eliminated merging the overlapping genomic intervals.
Zebrafish Husbandry
All experiments including zebrafish were performed in wild-type AB strain from the Zebrafish International Resource Center from the University of Oregon, according to approved protocols by the institutional Animal Studies Committee. Adult zebrafish were maintained at 28 °C in a 14/10 h light/dark cycle in a completely automatic Aquatic Habitats aquarium.
In Vivo HAE Transcription Enhancer Assay in Zebrafish
PhastCons genomic region containing each HAE were amplified by proofreading PCR (see primers list in supplementary table S4, Supplementary Material online) from human and chimpanzee genomic samples (chimpanzee samples were kindly donated by Adrian Sestelo) and cloned individually in the vector pXIG_cFos containing the minimal promoter cFos (Fisher, Grice, Vinton, Bessling, Urasaki, et al. 2006) that was kindly donated by Andy McCallion. Transgenic zebrafish were produced as described by Fisher, Grice, Vinton, Bessling, Urasaki, et al. (2006). Briefly, embryos were obtained by natural mating: the day before to the microinjection, two adult females and one adult male were placed in a breeding tank in which females were separated from the male by means of a division panel in the tank. The morning of the microinjections, the egg production started shortly after removing the division panel from the mating tank. Each HAE-cFos construct was coinjected with transposase mRNA in one- to two-cell zebrafish embryos. We performed two separate experiments for each construct, where a 1 nl mixture of DNA and RNA (5/25 ng/ml) was injected into approximately 200 embryos. Embryos were raised in E3 medium (5 mM NaCl, 0.17 mM, KCl, 0.33 mM CaCl2, 0.33 mM MgSO4) at 28.3 °C and were subsequently screened at 24 or 48 hpf for EGFP expression. When necessary, 0.1 mM of 1-phenyl-2-thiourea was added to the E3 medium to prevent pigment formation. Microscopy was carried out on tricaine-anaesthetized embryos mounted in 3% methyl cellulose. Whole-mount images were taken on an Olympus BX41 fluorescent microscope with an Olympus DP71 digital camera. EGFP expression patterns were observed in multiple embryos (on average 70 embryos for each element and stage) and we considered a positive result (an active enhancer) when at least 30% of embryos showed a consistent EGFP expression pattern (Fisher, Grice, Vinton, Bessling, Urasaki, et al. 2006). For those constructs that did not promote EGFP expression, we injected increasing amounts of DNA until reaching evident signs of toxicity and analyzed them at 24 hpf. For generation of stable transgenic lines, injected larvae were raised to adulthood and screened for stable germ-line insertion. Where indicated, patterns identified in mosaic animals were verified in at least three independent stable germ-line insertions.
Identification of TFBS Gain and Loss in HAEs
Comparative analysis of human and chimpanzee HAE sequences to detect gain and loss of TFBS was performed using the matrix-based program MatInspector (www.genomatix.de, last accessed February 28, 2013). MatInspector identifies TFBS in nucleotide sequences using a large library of weight matrices, which is designed to represent the best of current knowledge in terms of specificity and sensitivity regarding TFBS (Cartharius et al. 2005).
Coding Sequence Evolutionary Analysis
To analyze the evolution of the NPAS3 coding region, we obtained the coding sequences of the following species: human, chimp, orangutan, gibbon, macaque, marmoset, tree shrew, mouse, rat, guinea pig, rabbit, horse, cow, dolphin, pig, microbat, elephant, and opossum.
These sequences were first downloaded from Ensembl, using the orthologs tool and then compared with those stored in GenBank and UCSC’s Genome Browser. RefSeq sequences were prioritized when available. Missing data in the source sequences were manually curated using Genscan and N-Scan open reading frame predictors and NCBI’s traces-BLAST.
We aligned the sequences using ClustalW and calculated the % of identity (ID) using MEGA5 (Tamura et al. 2011). To carry out adaptive evolutionary analysis of NPAS3, the modified branch-site model A (test 2) of positive selection was carried out in NPAS3 mammalian sequences (Yang et al. 2005; Zhang et al. 2005) using PAML (Yang 2007). The focal branches in this test were the human and the primate lineages. Two nested hypotheses were tested for each focal branch. In the alternative hypothesis, three ω values were calculated: the ω0, ω1, and ω2 representing the codons under negative, null, and positive selection, respectively. However, in the null hypothesis, only two ω values (ω0 and ω1) were estimated and no positively selected sites are allowed. In the case where positive selection was found in our focal branch, particular sites with positive selection were detected by the Bayes empirical Bayes method (Yang et al. 2005; Zhang et al. 2005).
PhyloP Analysis of NPAS3 Genomic Region
With the aim of studying the genomic background in which NPAS3 in embedded, we selected the genomic portion that spans from the last base of AKAP6 to the first base of EGLN3, the 5′ and 3′ adjacent genes to NPAS3, respectively. That selection renders a genomic interval of 1,091,152 base pairs that covers from base 32,372,020 to base 33,463,171 of chromosome 14 in the human genome (assembly 2006: NCBI36/hg18). We processed this genomic interval using the open web-based platform Galaxy (https://main.g2.bx.psu.edu, last accessed February 28, 2013) to split it into a few partitions of different genomic characteristics. Our first step was cross this genomic interval with the RefSeq table using UCSC’s Table Browser to subtract the intervals corresponding to NPAS3 exons. Similarly, we subtracted the 17-way most conserved PhastCons elements, obtaining finally the intervals corresponding to the noncoding, nonconserved sections within the genomic interval studied. As coding sequences match the conditions to qualify as conserved elements, they are included in the PhastCons collection, but as we wanted to analyze them separately, we subtracted exon intervals from the PhastCons intervals. Likewise, we subtracted the HAEs from PhastCons to use them as our conserved sequences reference. Thus, we obtained four groups of genomic intervals representing each one of the partitions of interest, namely NPAS3 coding region in exon intervals, HAEs intervals, PhastCons intervals without HAEs and exons, and lastly the group of noncoding, nonconserved intervals. By applying the “Stitch MAF blocks,” using the 8-way multiZ MAF type to include orangutan in our alignments, and “Concatenate FASTA alignments” Galaxy tools, we obtained for each of these partitions a fasta alignment with four selected study species: human, chimpanzee, orangutan, and rhesus.
Using phyloP program, from the PHAST software package, we computed P values of conservation and acceleration for out test alignments under the SPH method (Siepel et al. 2006). The conservation and acceleration values were calculated in each case for a subtree holding only the human branch and a supertree made up of the remaining species: chimpanzee, orangutan, and rhesus. The neutral model of evolution with which to compare all the test alignments was derived from the primates.mod neutral model used in UCSC’s conservation track. We used the tree_doctor program, also from the PHAST package, to prune the original primate neutral model tree and keep only our four study species.
RT-PCR and In Situ Hybridization Assays
Total RNA was extracted from pools of 50 zebrafish embryos at 24 and 48 hpf or from a pool of two adult zebrafish brains. RNA isolation was carried out using TRIZOL reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s recommendations. For RT-PCR, 10 μg of total RNA was reverse transcribed at 37 °C for 60 min in a 20 μl final volume using SuperScript II Reverse Transcriptase (Invitrogen) and oligo-dT primers. PCR experiments were performed in 30 μl final volume, containing 3 μl of 1:20 reverse transcription sample as template. An initial denaturation step of 5 min at 94 °C was followed by amplification (30 s at 94 °C, 30 s at 64 °C, and 30 s at 72 °C) and final extension for 5 min at 72 °C. Primer pairs used to amplify NPAS3 cDNAs were as follows: NPAS3-Dr-E2-241-F (5′-CACCCAGGCTCTGAGGAAGGAGAA-3′), NPAN3-Dr-E2-241-R (5′CTTTGACGGAGGTGTTGG-3′), NPAS3-Dr-EGF-451-F (5′-CACCCTCCGGTTCCCTGCTCTAC-3′), NPAS3-Dr-EGF-451-R (5′-TGCGATCCAGAGTCTGTGTG-3′), and for β-actin we used the following primers BACT_Dr_F (5′-GCAGAAGGAGATCACATCCCTGGC-3′) and BACT_Dr_R (5′- CATTGCCGTCACCTTCACCGTTC-3′) obtained from the work of Casedei et al. (2011). The resulting NPAS3 PCR fragments were cloned into pENTR/D (Invitrogen), sequenced to confirm identity, and used as probes in our whole-mount in situ hybridization (WMISH) assay. Plasmids containing the probes were linearized using NotI, and transcribed using T7 RNA polymerase (Roche, Penzberg, Germany) to generate digoxigenin-labeled NPAS3 antisense riboprobe.
For whole-mount in situ hybridization, larvae were fixed overnight in 4% paraformaldehyde in phosphate-buffered saline (PBS), dehydrated in graded methanol concentrations (30%, 50%, 70%, and 100%) and stored at −20 °C. Then larvae were rehydrated to PBST (PBS with 0.2% Tween 20) and treated for 15 min (24 and 48 hpf) to 30 min (5 days postfertilization; dpf) with 10 µg/ml proteinase K in PBST, washed twice in PBST and postfixed for 20 min in 4% paraformaldehyde in PBS, washed in PBST, and then washed three times in a hybridization solution containing transfer RNA (tRNA) and heparin. Prehybridizations were performed for 2 h at 65 °C in a hybridization solution (50% formamide, 5× SSC, 500 µg/ml torula tRNA, 50 µg/ml heparin, 0.1% Tween 20, 5 mM citric acid). Hybridizations were performed overnight at 65 °C in a fresh hybridization solution containing 1 µg/ml antisense digoxygenin-labeled riboprobes. Larvae were then washed 5 min in 66% hybridization solution 33% 2× SSC at 65 °C, then washed 5 min in 33% hybridization solution 66% 2× SSC at 65 °C, and finally washed 5 min in SSC at 65 °C. Larvae were then washed 20 min in 0.2× SSC, 0.1% CHAPS and 20 min 0.1× SSC, 0.1% CHAPS. Finally, the samples were washed 5 min in 66% 0.2× SSC, 33% PBST, 5 min in 33% 0.2× SSC, 66% PBST and 5 min in PBST at room temperature. To perform anti-DIG antibody incubation, samples were first incubated in blocking solution (2% normal goat serum [NGS] and 2 mg/ml bovine serum albumin) for 1 h at room temperature. Anti-DIG antibody (Roche) was incubated at 1/5,000 overnight at 4 °C. The following day samples were washed five times 15 min each in PBST and then washed four times in a solution containing 100 mM NaCl, 50 mM MgCl2, 0.1% Tween 20, and 100 mM Tris–HCl (pH 9.5). Then samples were incubated with NBT/BCIP (Roche) in the abovementioned buffer overnight at room temperature and then stopped in PBS 1× and mounted in glycerol 70%.
For in situ hybridization (ISH) in cryosections, zebrafish embryos were dechorionated and then euthanized by submersion in ice water. The embryos were fixed by immersion in phosphate-buffered 4% paraformaldehyde solution (PB; 0.1 M, pH 7.4) at 4 °C overnight. The specimens were rinsed twice in PBS, cryoprotected in 10% sucrose solution (in PBS), and embedded in 10% gelatine with 10% sucrose in the same buffer. The blocks were frozen for 1 min in isopentane cooled to –70 °C in dry ice and were stored at –80 °C. Serial 14 -μm-thick (48 hpf) and 16 -μm-thick (72 hpf and 5 days) sections were cut in a cryostat (Leica 1510S, Germany), collected on Super-Frost Plus slides (FisherScientific, USA), and stored at –80 °C until use. EG and E2 NPAS3 probes were synthesized as mentioned previously and labeled using digoxigenin-UTP (Roche, Germany). We performed ISH on cryosections as previously described (Hidalgo-Sanchez et al. 2005). Briefly, the thawed sections were postfixed with 4% paraformaldehyde in PBS for 10 min and then washed with PBS for 15 min. Thereafter, prehybridization occurred at room temperature for 4 h in a solution containing 50% formamide, 10% dextran sulfate, 5× Denhardt’s solution, and 250 mg/ml tRNA. Hybridization was performed with 300 ng/ml of the probe in the same hybridization solution at 72 °C for 16 h. After hybridization, the sections were washed with 0.2% SSC at 72 °C for 1 h and then twice with a solution containing 100 mM NaCl, 0.1% Triton X-100, and 100 mM Tris–HCl (pH 7.5). After treatment with 10% NGS in the same solution for 4 h, the sections were incubated overnight with alkaline phosphatase-conjugated antidigoxigenin Fab fragments (Roche, Germany; 1:3,500). Then, the sections were treated and revealed as mentioned for our WMISH protocol. Both NPAS3 probes displayed indistinguishable results in our WMISH and ISH studies, whereas a scramble probe did not produce any pattern (data not shown). The sections were coverslipped with Mowiol (Polyvinyl alcohol, Glycerol in Tris 0.2 M, pH 8.5) and photographed as mentioned earlier. Images were processed using Panaview in cases where two images had to be used to generate a composite of one zebrafish embryo and images were size adjusted, cropped, contrast enhanced, and annotated in Adobe Photoshop.
Immunohistochemistry
Primary antibodies used were rabbit polyclonal anti-GFP (1/500; Abcam) and mouse monoclonal anti-HuC/D (1/20; Molecular Probes). Secondary antibodies used were goat anti-rabbit-AF488 (1/1,000; Molecular Probes, Invitrogen) and donkey anti-mouse-AF555 (1/1,000, Invitrogen). The specimens were fixed, cryoprotected, and frozen as mentioned previously for the ISH procedure. Cryostat sections (14 μm) were cut and washed three times for 10 min in 1× PBS to remove gelatin. Sections were then incubated for 2 h at room temperature in blocking solution (10% normal donkey serum, 0.1% triton X-100, and 1× PBS) followed by incubation with a mix of primary antibodies diluted in blocking solution overnight at 4 °C. Sections were washed three times in 1× PBS and then incubated sequentially for 1 h at room temperature with each secondary antibody. Finally, sections were washed three times in 1× PBS and mounted in Vectashield (Vector Laboratories Inc.). Images were acquired using a NIKON C1 Confocal Microscope using the EZ-C1 2.20 software and a PlanApo 40×/0.95 objective.
Supplementary Material
Supplementary tables S1–S4 and figure S1 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Drs. Sabina Domené and Nora Calcaterra for excellent technical advice in transgenic zebrafish production and handling. They thank Adrian Sestelo for chimpanzee samples. They express their gratitude to Marcelo Rubinstein for permanent support and valuable suggestions on experimental design, data interpretation, and critical comments on the manuscript. This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica grant PICT 1071, the Consejo Nacional de Investigaciones Científicas y Técnicas grant CONICET-Argentina PIP 0299 to L.F.F., and doctoral fellowships from the CONICET to F.P. and G.B.K.
References
- Alarcón M, Abrahams BS, Stone JL, et al. (13 co-authors) Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82:150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnby G, Abbott A, Sykes N, Morris A, Weeks DE, Mott R, Lamb J, Bailey AJ, Monaco AP. Candidate-gene screening and association analysis at the autism-susceptibility locus on chromosome 16p: evidence of association at GRIN2A and ABAT. Am J Hum Genet. 2005;76:950–966. doi: 10.1086/430454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill EW, Ehrman LA, Williams MT, Klanke J, Hammer D, Schaefer TL, Sah R, Dorn GW, 2nd, Potter SS, Vorhees CV. Abnormal neurodevelopment, neurosignaling and behaviour in Npas3-deficient mice. Eur J Neurosci. 2005;22:1265–1276. doi: 10.1111/j.1460-9568.2005.04291.x. [DOI] [PubMed] [Google Scholar]
- Brunskill EW, Witte DP, Shreiner AB, Potter SS. Characterization of npas3, a novel basic helix-loop-helix PAS gene expressed in the developing mouse nervous system. Mech Dev. 1999;88:237–241. doi: 10.1016/s0925-4773(99)00182-3. [DOI] [PubMed] [Google Scholar]
- Bush EC, Lahn BT. A genome-wide screen for noncoding elements important in primate evolution. BMC Evol Biol. 2008;8:17. doi: 10.1186/1471-2148-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Casadei R, Pelleri MC, Vitale L, Facchin F, Lenzi L, Canaider S, Strippoli P, Frabetti F. Identification of housekeeping genes suitable for gene expression analysis in the zebrafish. Gene Expr Patterns. 2011;11:271–276. doi: 10.1016/j.gep.2011.01.003. [DOI] [PubMed] [Google Scholar]
- Charrier C, Joshi K, Coutinho-Budd J, et al. (11 co-authors) Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YF, McGrath JA, Thornquist MH, Wolyniec PS, Nestadt G, Swartz KL, Lasseter VK, Liang KY, Pulver AE. Genetic heterogeneity in schizophrenia II: conditional analyses of affected schizophrenia sibling pairs provide evidence for an interaction between markers on chromosome 8p and 14q. Mol Psychiatry. 2002;7:658–664. doi: 10.1038/sj.mp.4001045. [DOI] [PubMed] [Google Scholar]
- Dennis MY, Nuttle X, Sudmant PH, et al. (16 co-authors) Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky RI, Sheldahl LC, Moon RT. A transgenic Lef1/beta-catenin-dependent reporter is expressed in spatially restricted domains throughout zebrafish development. Dev Biol. 2002;241:229–237. doi: 10.1006/dbio.2001.0515. [DOI] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Duret L, Semon M, Piganeau G, Mouchiroud D, Galtier N. Vanishing GC-rich isochores in mammalian genomes. Genetics. 2002;162:1837–1847. doi: 10.1093/genetics/162.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- Erbel-Sieler C, Dudley C, Zhou Y, Wu X, Estill SJ, Han T, Diaz-Arrastia R, Brunskill EW, Potter SS, McKnight SL. Behavioral and regulatory abnormalities in mice deficient in the NPAS1 and NPAS3 transcription factors. Proc Natl Acad Sci U S A. 2004;101:13648–13653. doi: 10.1073/pnas.0405310101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Choi SS, Lahn BT. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Hum Mol Genet. 2004;13:1139–1145. doi: 10.1093/hmg/ddh126. [DOI] [PubMed] [Google Scholar]
- Evans PD, Anderson JR, Vallender EJ, Gilbert SL, Malcom CM, Dorus S, Lahn BT. Adaptive evolution of ASPM, a major determinant of cerebral cortical size in humans. Hum Mol Genet. 2004;13:489–494. doi: 10.1093/hmg/ddh055. [DOI] [PubMed] [Google Scholar]
- Evans PD, Gilbert SL, Mekel-Bobrov N, Vallender EJ, Anderson JR, Vaez-Azizi LM, Tishkoff SA, Hudson RR, Lahn BT. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science. 2006;312:276–279. doi: 10.1126/science.1124070. [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, Urasaki A, Kawakami K, McCallion AS. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc. 2006;1:1297–1305. doi: 10.1038/nprot.2006.230. [DOI] [PubMed] [Google Scholar]
- Fortna A, Kim Y, MacLaren E, et al. (16 co-authors) Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JI, Vrijenhoek T, Markx S, et al. (17 co-authors) CNTNAP2 gene dosage variation is associated with schizophrenia and epilepsy. Mol Psychiatry. 2008;13:261–266. doi: 10.1038/sj.mp.4002049. [DOI] [PubMed] [Google Scholar]
- Galtier N, Duret L. Adaptation or biased gene conversion? Extending the null hypothesis of molecular evolution. Trends Genet. 2007;23:273–277. doi: 10.1016/j.tig.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Gould P, Kamnasaran D. Immunohistochemical analyses of NPAS3 expression in the developing human fetal brain. Anat Histol Embryol. 2011;40:196–203. doi: 10.1111/j.1439-0264.2010.01059.x. [DOI] [PubMed] [Google Scholar]
- Hidalgo-Sanchez M, Martinez-de-la-Torre M, Alvarado-Mallart RM, Puelles L. A distinct preisthmic histogenetic domain is defined by overlap of Otx2 and Pax2 gene expression in the avian caudal midbrain. J Comp Neurol. 2005;483:17–29. doi: 10.1002/cne.20402. [DOI] [PubMed] [Google Scholar]
- Huang J, Perlis RH, Lee PH, et al. (12 co-authors) Cross-disorder genomewide analysis of schizophrenia, bipolar disorder, and depression. Am J Psychiatry. 2010;167:1254–1263. doi: 10.1176/appi.ajp.2010.09091335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Li T, Pan Y, et al. (14 co-authors) CNTNAP2 is significantly associated with schizophrenia and major depression in the Han Chinese population. Psychiatry Res. 2012 doi: 10.1016/j.psychres.2012.09.024. pii:S0165-1781(12)00521-5. [DOI] [PubMed] [Google Scholar]
- Kamnasaran D, Muir WJ, Ferguson-Smith MA, Cox DW. Disruption of the neuronal PAS3 gene in a family affected with schizophrenia. J Med Genet. 2003;40:325–332. doi: 10.1136/jmg.40.5.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Kostka D, Hubisz MJ, Siepel A, Pollard KS. The role of GC-biased gene conversion in shaping the fastest evolving regions of the human genome. Mol Biol Evol. 2012;29:1047–1057. doi: 10.1093/molbev/msr279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (13 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Leamey CA, Glendining KA, Kreiman G, Kang ND, Wang KH, Fassler R, Sawatari A, Tonegawa S, Sur M. Differential gene expression between sensory neocortical areas: potential roles for Ten_m3 and Bcl6 in patterning visual and somatosensory pathways. Cereb Cortex. 2008;18:53–66. doi: 10.1093/cercor/bhm031. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, et al. (86 co-authors) A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham K, Schuurmans C, Weiss S. STAT5A/B activity is required in the developing forebrain and spinal cord. Mol Cell Neurosci. 2007;35:272–282. doi: 10.1016/j.mcn.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Martin CL, Duvall JA, Ilkin Y, et al. (12 co-authors) Cytogenetic and molecular characterization of A2BP1/FOX1 as a candidate gene for autism. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:869–876. doi: 10.1002/ajmg.b.30530. [DOI] [PubMed] [Google Scholar]
- Mekel-Bobrov N, Gilbert SL, Evans PD, Vallender EJ, Anderson JR, Hudson RR, Tishkoff SA, Lahn BT. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- Park HC, Hong SK, Kim HS, Kim SH, Yoon EJ, Kim CH, Miki N, Huh TL. Structural comparison of zebrafish Elav/Hu and their differential expressions during neurogenesis. Neurosci Lett. 2000;279:81–84. doi: 10.1016/s0304-3940(99)00940-4. [DOI] [PubMed] [Google Scholar]
- Peñagarikano O, Abrahams BS, Herman EI, et al. (14 co-authors) Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147:235–246. doi: 10.1016/j.cell.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñagarikano O, Geschwind DH. What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol Med. 2012;18:156–163. doi: 10.1016/j.molmed.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard BS, Christoforou A, Thomson PA, Fawkes A, Evans KL, Morris SW, Porteous DJ, Blackwood DH, Muir WJ. Interacting haplotypes at the NPAS3 locus alter risk of schizophrenia and bipolar disorder. Mol Psychiatry. 2008;14:874–884. doi: 10.1038/mp.2008.24. [DOI] [PubMed] [Google Scholar]
- Pickard BS, Malloy MP, Porteous DJ, Blackwood DH, Muir WJ. Disruption of a brain transcription factor, NPAS3, is associated with schizophrenia and learning disability. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:26–32. doi: 10.1002/ajmg.b.30204. [DOI] [PubMed] [Google Scholar]
- Pickard BS, Pieper AA, Porteous DJ, Blackwood DH, Muir WJ. The NPAS3 gene—emerging evidence for a role in psychiatric illness. Ann Med. 2006;38:439–448. doi: 10.1080/07853890600946500. [DOI] [PubMed] [Google Scholar]
- Pieper AA, Wu X, Han TW, et al. (11 co-authors) The neuronal PAS domain protein 3 transcription factor controls FGF-mediated adult hippocampal neurogenesis in mice. Proc Natl Acad Sci U S A. 2005;102:14052–14057. doi: 10.1073/pnas.0506713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, King B, et al. (13 co-authors) Forces shaping the fastest evolving regions in the human genome. PLoS Genet. 2006;2:e168. doi: 10.1371/journal.pgen.0020168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, et al. (16 co-authors) An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Prabhakar S, Noonan JP, Paabo S, Rubin EM. Accelerated evolution of conserved noncoding sequences in humans. Science. 2006;314:786. doi: 10.1126/science.1130738. [DOI] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, et al. (32 co-authors) Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Pollard KS, Haussler D. New methods for detecting lineage-specific selection. Proceedings of the 10th International Conference on Research in Computational Molecular Biology (RECOMB 2006), Venice, Italy; 2006. P. 190–205. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan GC, Doke TF, Ashburner J, Wood NW, Frackowiak RS. Normal variation in fronto-occipital circuitry and cerebellar structure with an autism-associated polymorphism of CNTNAP2. Neuroimage. 2012;53:1030–1042. doi: 10.1016/j.neuroimage.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Han JC, Morales A, Menzie CM, Williams K, Fan YS. Characterization of 11p14-p12 deletion in WAGR syndrome by array CGH for identifying genes contributing to mental retardation and autism. Cytogenet Genome Res. 2008;122:181–187. doi: 10.1159/000172086. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive selection. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.