Abstract
The molecular bases for the evolution of male–female sexual dimorphism are possible to study in volvocine algae because they encompass the entire range of reproductive morphologies from isogamy to oogamy. In 1978, Charlesworth suggested the model of a gamete size gene becoming linked to the sex-determining or mating type locus (MT) as a mechanism for the evolution of anisogamy. Here, we carried out the first comprehensive study of a candidate MT-linked oogamy gene, MAT3/RB, across the volvocine lineage. We found that evolution of anisogamy/oogamy predates the extremely high male–female divergence of MAT3 that characterizes the Volvox carteri lineage. These data demonstrate very little sex-linked sequence divergence of MAT3 between the two sexes in other volvocine groups, though linkage between MAT3 and the mating locus appears to be conserved. These data implicate genetic determinants other than or in addition to MAT3 in the evolution of anisogamy in volvocine algae.
Keywords: gender-based divergence, gene conversion, male–female sexual dimorphism, MAT3/RB, mating type locus, volvocine algae
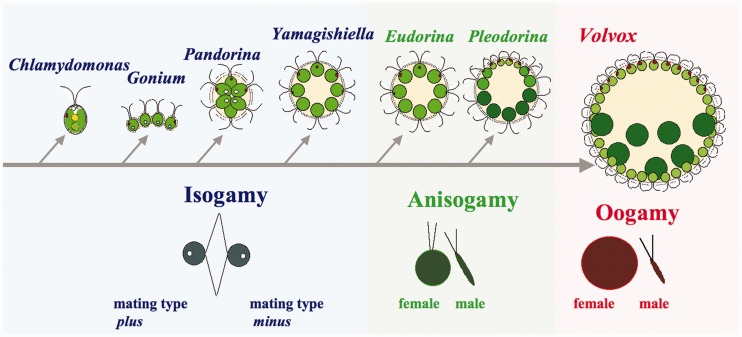
Sexual reproduction in the eukaryotes is classified according to gamete size and motility, with three major types: isogamy (equal sized gametes), anisogamy (large and small gametes), and oogamy (large immotile eggs and small motile sperm) (Bold and Wynne 1985). Anisogamous and oogamous organisms represent male–female sexual dimorphism that has arisen repeatedly in evolution, presumably from simpler isogamous mating systems in ancestral unicellular species (Parker et al. 1972; Kirk 2006). However, the molecular-evolutionary bases for the transitions from isogamy to anisogamy to oogamy are difficult or impossible to study in most extant lineages due to the ancient origins of oogamy (Kirk 2006; Nozaki et al. 2006). Volvocine algae or colonial Volvocales are exceptional because they are a relatively young group, yet they encompass the entire range of reproductive morphologies from isogamy to oogamy in an extant, phylogenetically coherent lineage (Nozaki 2003; Kirk 2006; Herron et al. 2009) (fig. 1).
Fig. 1.
Simplified diagram for stepwise evolution of colonial volvocine algae and their transition from isogamy to anisogamy and oogamy (Nozaki 2003; Herron et al. 2009).
In volvocine algae, sex is determined by a single mating type locus (MT) with two haplotypes that specify sexual differentiation. Although MT segregates as a single Mendelian trait, it is a complex genomic region that contains both shared and sex-limited genes that are rearranged with respect to each other and which do not undergo meiotic recombination (Umen 2011). A recent comparative study of MT from an oogamous volvocine species, Volvox carteri, with MT from an isogamous species, Chlamydomonas reinhardtii, revealed major differences in the size- and sex-based differentiation of MT genes (Ferris et al. 2010). Volvox MT was found to be about five times larger than Chlamydomonas MT, to contain more genes than Chlamydomonas MT, and to show a much higher degree of sex-based differentiation in its shared genes (those with an allele in both mating haplotypes and sexes). Shared genes that have become masculinized and feminized in sequence and/or expression as occurred in V. carteri are candidates for contributing to male–female sexual dimorphism. However, to date there has been no investigation of MT genes in the context of the isogamy to anisogamy/oogamy transition in volvocine algae besides the previously described comparison of C. reinhardtii and V. carteri (Charlesworth D and Charlesworth B 2010; Ferris et al. 2010).
One candidate regulator of gamete size is the mating locus gene MAT3. MAT3 encodes a homolog of the retinoblastoma (RB) tumor suppressor protein. In Chlamydomonas, MAT3 is tightly linked to MT and regulates cell size and cell cycle progression (Umen and Goodenough 2001; Fang et al. 2006). Based on its repertoire of cell cycle regulatory proteins that are orthologous to those in Chlamydomonas, a similar function is predicted for the Volvox MAT3/RB pathway (Prochnik et al. 2010). However, in contrast to Chlamydomonas where the minus and plus MAT3 alleles are nearly identical and function interchangeably (Umen and Goodenough 2001; Merchant et al. 2007), a high degree of male–female sequence differentiation and sex-regulated alternative splicing was observed for V. carteri MAT3 (Ferris et al. 2010). This observation led Ferris et al. (2010) to suggest that MAT3 homologs might be related to control of gamete size in colonial volvocine algae as predicted earlier by the gamete size regulator recruitment model for the evolution of anisogamy/oogamy from isogamous mating types (Charlesworth 1978). If this model applies to MAT3, then the degree of differentiation between MAT3 alleles from each mating type in isogamous species should be much lower than in males–females from anisogamous and oogamous species where MAT3 would have acquired sex-specific functions in gamete size control.
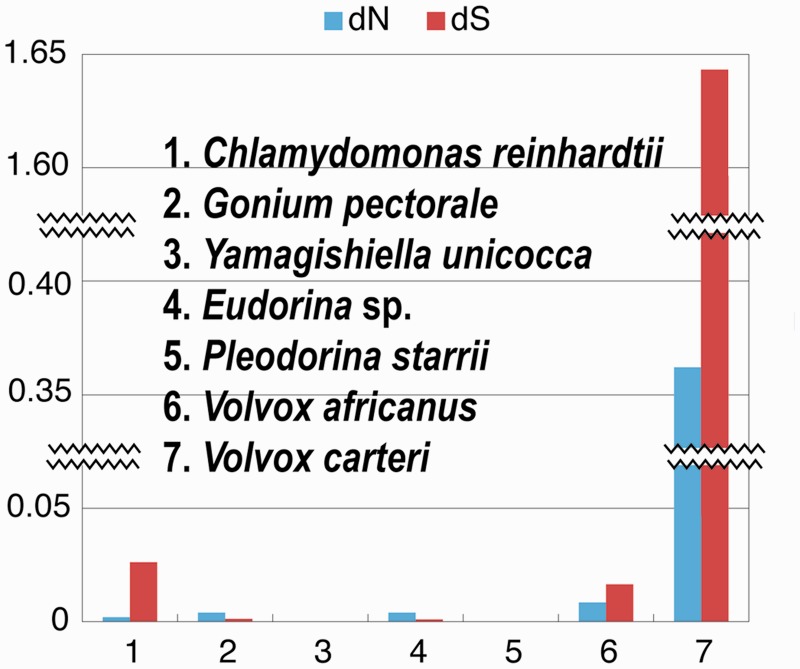
Here, we sequenced full-length coding regions of MAT3 from plus and minus mating types of isogamous Gonium pectorale and Yamagishiella unicocca, and from males and females of anisogamous Eudorina sp. and Pleodorina starrii, and from males and females of oogamous V. africanus (supplementary fig. S1 and table S1, Supplementary Material online). In contrast to V. carteri where the male and female MAT3 alleles show large differences in structure and sequence (Ferris et al. 2010), MAT3 homologs from the five colonial species examined here had almost identical nucleotide sequences between the two sexes (fig. 2).
Fig. 2.
Gender-based divergence of MAT3 genes from six species of the colonial Volvocales and Chlamydomonas reinhardtii (supplementary table S2, Supplementary Material online). Bar graph depicting dN (number of substitutions per nonsynonymous site) and dS (number of substitutions per synonymous site) between MAT3 alleles from each of the two mating types or sexes.
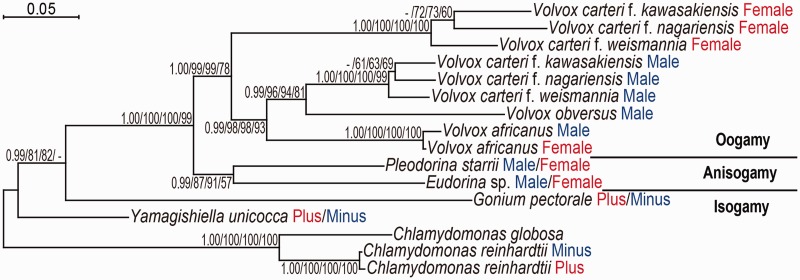
Our phylogenetic analysis of MAT3 sequences demonstrated that the extensive MAT3 divergence in the V. carteri lineage might have occurred recently in the ancestor of the three V. carteri forms after their divergence from the anisogamous lineage containing P. starrii and Eudorina sp. (fig. 3). Therefore, the extreme, gender-based MAT3 divergence observed in V. carteri species may not be directly related to the evolution of male and female dimorphism within the colonial Volvocales as a whole because it is not observed in their nearest anisogamous relatives (figs. 2 and 3). Moreover, in the oogamous species V. africanus MAT3 showed only modest levels of male–female divergence compared with V. carteri male and female MAT3 (fig. 2; supplementary table S2, Supplementary Material online).
Fig. 3.
Phylogeny of MAT3 proteins from the colonial Volvocales and Chlamydomonas. The analysis is based on 464 amino acid positions of slowly evolving regions of the alignment (supplementary fig. S1 and alignment, Supplementary Material online). Branch labels indicate from left to right: Posterior probabilities (≥0.90) from Bayesian inference/Bootstrap values (≥50%) obtained using 1,000 replicates with RAxML/Bootstrap values (≥50%) using 1,000 replicates with PhyML/Bootstrap values (≥50%) using 1,000 replicates with maximum parsimony. For details, see supplementary information, Supplementary Material online.
This study demonstrates that MAT3 genes from the two sexes have been undergoing some form of genetic exchange between mating types or sexes during most of their evolution within the colonial Volvocales (fig. 3) that serves to preserve homogeneity between alleles from each mating type. Gene conversion as proposed recently (Teshima and Innan 2004; Umen 2011) may contribute to sequence homogenization of MT-linked genes such as MAT3 alleles that cannot undergo crossover recombination. Gene conversion was also observed in a recent study of the Cryptococcus neoformans mating locus that is structured similarly to MT from volvocine algae with two rearranged haplotypes (Sun et al. 2012), but gene conversion has not been reported for volvocine algal MT genes. It can be speculated that complete loss of recombination and gene conversion of MT genes in V. carteri was a major contributor to male–female differentiation in this sublineage.
Sex-specific expression and/or alternative splicing of MAT3 (Ferris et al. 2010) that might contribute to male–female gamete size differences in anisogamous and oogamous volvocine algae were not detected in our reverse transcriptase polymerase chain reaction experiments for isogamous Gonium pectorale, anisogamous Eudorina sp. and oogamous V. africanus (supplementary fig. S2, Supplementary Material online). Although our data do not completely rule out a role for MAT3 in controlling gamete size in anisogamous and oogamous volvocine algae, they raise the intriguing question of what other MT genes are involved in specifying this trait. Future molecular genetic studies aimed at characterizing MT genes from anisogamous and oogamous volvocine species will help to answer this question.
Materials and Methods
Volvocalean strains and MAT3 sequences used in this study are listed in supplementary table S1, Supplementary Material online. Amino acid alignment of MAT3 proteins is available in supplementary alignment, Supplementary Material online. Other details of Materials and Methods are described in supplementary information, Supplementary Material online.
Supplementary Material
Supplementary information, alignment, figures S1 and S2, and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
Dr H. Innan (Graduate University for Advanced Studies, Hayama, Japan) kindly discussed the gene conversion. Shotgun sequencing was carried out by Kazusa DNA Research Institute (Kisarazu, Chiba, Japan). This research was mainly done at the University of Tokyo. The alignment of MAT3 is available (ID: S13735) via TreeBASE (http://www.treebase.org/treebase-web/home.html). This work was supported by the Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science KAKENHI grants 24112707 and 24247042 to H.N. and 22-40216 to H.K.-T. and the National Institutes of Health grant GM078376 to J.U.
References
- Bold HC, Wynne MJ. Introduction to the algae: structure and reproduction. 2nd ed. Englewood Cliffs (NJ): Prentice Hall; 1985. [Google Scholar]
- Charlesworth B. The population genetics of anisogamy. J Theor Biol. 1978;73:347–357. doi: 10.1016/0022-5193(78)90195-9. [DOI] [PubMed] [Google Scholar]
- Charlesworth D, Charlesworth B. Evolutionary biology: the origins of two sexes. Curr Biol. 2010;20:R519–R521. doi: 10.1016/j.cub.2010.05.015. [DOI] [PubMed] [Google Scholar]
- Fang S-C, de los Reyes C, Umen JG. Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet. 2006;2:e167. doi: 10.1371/journal.pgen.0020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P, Olson BJSC, De Hoff PL, et al. (15 co-authors) Evolution of an expanded sex-determining locus in Volvox. Science. 2010;328:351–354. doi: 10.1126/science.1186222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron MD, Hackett JD, Aylward FO, Michod RE. Triassic origin and early radiation of multicellular volvocine algae. Proc Natl Acad Sci U S A. 2009;106:3254–3258. doi: 10.1073/pnas.0811205106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk DL. Oogamy: inventing the sexes. Curr Biol. 2006;16:R1028–R1030. doi: 10.1016/j.cub.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, et al. (117 co-authors) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H. Origin and evolution of the genera Pleodorina and Volvox (Volvocales) Biologia. 2003;58:425–431. [Google Scholar]
- Nozaki H, Mori T, Misumi O, Matsunaga S, Kuroiwa T. Males evolved from the dominant isogametic mating type. Curr Biol. 2006;16:R1018–R1020. doi: 10.1016/j.cub.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Parker GA, Baker RR, Smith VGF. The origin and evolution of gamete dimorphism and the male-female phenomenon. J Theor Biol. 1972;36:529–553. doi: 10.1016/0022-5193(72)90007-0. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, et al. (28 co-authors) Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Hsueh Y-P, Heitman J. Gene conversion occurs within the mating-type locus of Cryptococcus neoformans during sexual reproduction. PLoS Genet. 2012;8:e1002810. doi: 10.1371/journal.pgen.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima KM, Innan H. The effect of gene conversion on the divergence between duplicated genes. Genetics. 2004;166:1553–1560. doi: 10.1534/genetics.166.3.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG. Evolution of sex and mating loci: an expanded view from volvocine algae. Curr Opin Microbiol. 2011;14:634–641. doi: 10.1016/j.mib.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW. Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 2001;15:1652–1661. doi: 10.1101/gad.892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.