Abstract
The genome is extensively transcribed into long intergenic noncoding RNAs (lincRNAs), many of which are implicated in gene silencing1,2. Potential roles of lincRNAs in gene activation are much less understood3,4,5. Development and homeostasis require coordinate regulation of neighboring genes through a process termed locus control6. Some locus control elements and enhancers transcribe lincRNAs7,8,9,10, hinting at possible roles in long range control. In vertebrates, 39 Hox genes, encoding homeodomain transcription factors critical for positional identity, are clustered in four chromosomal loci; the Hox genes are expressed in nested anterior-posterior and proximal-distal patterns co-linear with their genomic position from 3′ to 5′of the cluster11. Here we identify HOTTIP, a lincRNA transcribed from the 5′ tip of the HOXA locus that coordinates the activation of multiple 5′ HOXA genes in vivo. Chromosomal looping brings HOTTIP into close proximity to its target genes. HOTTIP directly binds the adaptor protein WDR5 and targets WDR5/MLL complexes across HOXA, driving histone H3 lysine 4 trimethylation and gene transcription. Induced proximity is necessary and sufficient for HOTTIP activation of its target genes. Thus, by serving as key intermediates that transmit information from higher order chromosomal looping into chromatin modifications, lincRNAs may organize chromatin domains to coordinate long-range gene activation.
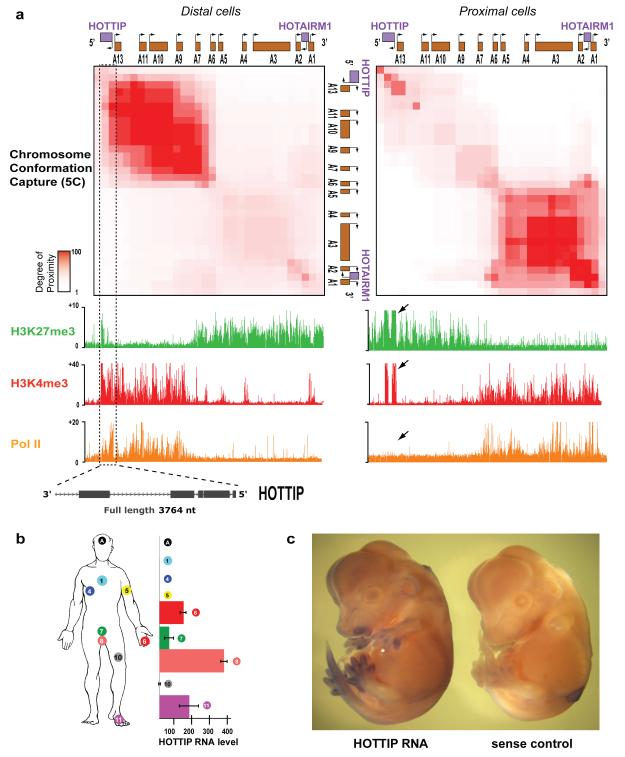
We examined chromosome structure and histone modifications in primary fibroblasts derived from several anatomic sites12, and found distinctive differences in the HOXA locus. High throughput chromosome conformation capture (5C)13 across HOXA revealed that its higher order structure is dependent on positional identity. In anatomically distal cells (e.g. foreskin and foot fibroblasts), we detected abundant chromatin interactions within the transcriptionally active 5′ HOXA locus (with reference to the directions of transcription of constituent Hox genes), pointing to a compact and looped conformation. In contrast, no long-range chromatin interactions are detected within the transcriptionally silent 3′ HOXA which appears largely linear (Fig. 1A). Strikingly, anatomically proximal cells (e.g. lung fibroblasts) have the diametrically opposite pattern. The ON and OFF states of Hox and other key developmental genes are maintained by the MLL/Trithorax (Trx) and Polycomb group (PcG) proteins, which mediate trimethylation of histone H3 lysine 4 (H3K4me3) to activate genes or lysine 27 (H3K27me3) to repress genes14. The portions of HOXA in tight physical interaction are marked by broad domains of H3K4me3, whereas H3K27me3 marks the physically extended and transcriptional silent regions (Fig. 1A).
Figure 1. HOTTIP is a lincRNA transcribed in distal anatomic sites.
(A) Chromatin state map of distal vs. proximal cells. Top panels: Chromosome Conformation Capture-Carbon Copy (5C) analysis of distal (foreskin) and proximal (lung) human fibroblasts. Heat map representations (generated by my5C30) of 5C data (bin size 30 kb, step size 3 kb) for HOXA in foreskin and lung fibroblasts. Red intensity of each pixel indicates relative interaction between the two points on the genomic coordinates. The diagonal represents frequent cis interactions between regions located in close proximity along the linear genome. 5C signals that are away from the diagonal represent long-range looping interactions. Bottom: Chromatin occupancy across HOXA. X-axis is genomic coordinate; Y-axis depicts occupancy of the indicated histone marks or protein (ChIP/input). Box and arrows highlight chromatin states of HOTTIP. (B) HOTTIP expression in primary human fibroblasts from 11 anatomic sites. Mean + S.E. is shown in all panels with error bars. (C) In situ hybridization of HOTTIP in E13.5 mouse embryo.
On the very 5′ and 3′ edges of the two respective interaction clusters are two lincRNA loci that exhibit distinct chromatin modifications. The 3′element has been previously identified as the myelopoiesis-associated lincRNA HOTAIRM115. The 5′ element, for which we suggest the name HOTTIP for “HOXA transcript at the distal tip”, exhibits bivalent H3K4me3 and H3K27me3, a histone modification pattern associated with poised regulatory sequences16. (Henceforth we refer to the DNA element with italics and the RNA without italics). Comparison with RNA polymerase II occupancy and RNA expression showed that the bivalent H3K4me3 and H3K27me3 modifications on HOTTIP do not require HOTTIP transcription, but transcription of HOTTIP is associated with increased H3K4me3 and decreased H3K27me3 at HOTTIP itself (Fig. 1A, left). ChIP analysis confirmed that HOTTIP is occupied by both PRC2 and MLL complexes, consistent with the bivalent histone marks (Fig. S1A).
HOTTIP transcription yields a 3764 nucleotide, spliced, and polyadenylated lincRNA that initiates ~330 bases upstream of HOXA13. Only the strand antisense to HOXA genes is transcribed (Fig. S1B). Genes near the 5′ end of each HOX cluster tend to be expressed in more posterior and/or distal anatomical locations. Consistent with its genomic location 5′ to HOXA13, HOTTIP is expressed in distal and/or posterior anatomic sites, (Fig. 1B). In situ hybridization of developing mouse and chick embryos confirmed that HOTTIP is expressed in posterior and distal sites in vivo, suggesting a conserved expression pattern from development to adulthood (Fig. 1C and Fig. S1C). Even in distal cells where HOTTIP is expressed, its RNA level is very low and estimated to be ~0.3 copies per cell (Fig. S2).
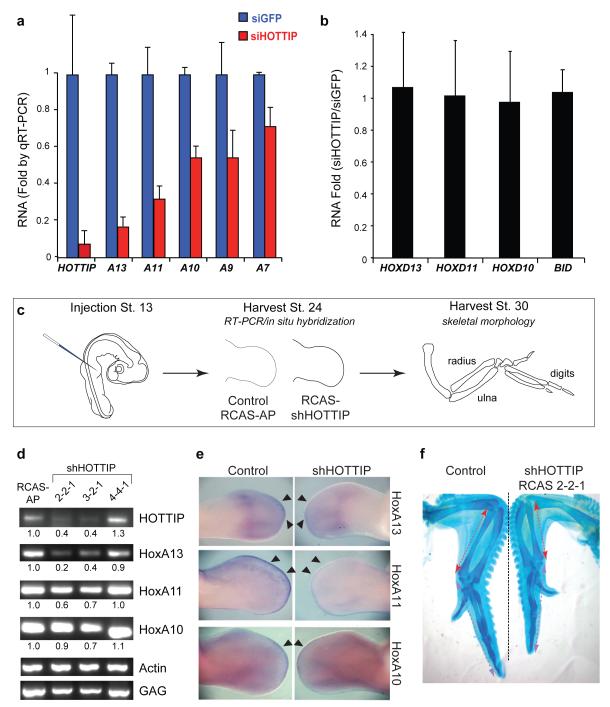
We employed small interfering RNAs (siRNAs) to knock down HOTTIP RNA in fibroblasts from a distal anatomic site (foreskin), and examined expression of 5′ HOXA genes by quantitative reverse transcription PCR. Notably, HOTTIP knockdown abrogated expression of distal HOXA genes across 40 kilobases with a trend dependent on the distance to HOTTIP. The strongest blockade was observed for HOXA13 and HOXA11, with progressively less severe effects on HOXA10, HOXA9, and HOXA7 (Fig. 2A). The effect on gene transcription appeared to be unidirectional, as there were no appreciable changes in the levels of EVX1, located ~40 kilobases 5′ of the HOXA cluster (data not shown). HOTTIP knockdown did not affect expression of the highly homologous HOXD genes, other control genes, nor induce antisense transcription at its own locus (Fig. 2B, Fig. S3A). Multiple independent siRNAs targeting HOTTIP yielded similar results (Fig. S3B). These results suggest that HOTTIP RNA is necessary to coordinate activation of 5′ HOXA genes.
Figure 2. HOTTIP is required for coordinate activation of 5′ HOXA genes.
(A) Knockdown of HOTTIP abrogates expression of 5′ HOXA genes in foreskin fibroblasts, but not HOXD genes or BID (B). Mean ± s.d. are shown. (C) Schematic of chick RNAi experiment. (D) HOTTIP is required for 5′ HoxA gene expression in vivo. RT-PCR of the indicated genes from control or HOTTIP-depleted distal limb bud is shown; quantitation and normalization by Actin signal is shown below each band. GAG signal confirms successful retroviral transduction in all cases. (E) In situ hybridization of 5′ HoxA genes in chick limb buds. Arrowheads highlight distal domains of high HoxA gene expression that are impacted by HOTTIP knockdown. (F) Shortening of distal bony elements in HOTTIP-depleted forelimbs. Alcian blue staining highlights the skeletal elements. Red and purple lines highlight radius and 3rd digit lengths, respectively.
We next addressed the function of HOTTIP RNA in vivo in the developing chick limb bud (Fig. 2C). While prior genetic studies of ncRNAs involved deletion or insertion into the gene locus17, we wished to distinguish the functions of HOTTIP RNA from its corresponding DNA element. HOTTIP can nucleate H3K4 and H3K27 methylation independent of transcription (Fig. 1A), and the precise genomic distance between upstream enhancer elements and Hox genes is critical for their proper co-linear activation17. Therefore, we employed RNAi in chick embryos, where replication-competent retroviruses can deliver short-hairpin RNAs (shRNAs) with high penetrance and precise spatiotemporal control18 (Fig. S4). In the limb bud, 5′ HoxA genes are transcribed in a nested pattern along the proximal-distal axis19. In this tissue, HoxA function is highly redundant with that of the HoxD locus, which allowed us to assess altered HoxA expression patterns without major changes in anatomic landmarks20. We injected retroviruses carrying shRNAs against chick HOTTIP into upper limb buds of stage 13 chicks; RT-PCR and in situ hybridization were performed on both control and knockdown samples after 2-4 days. Knockdown of HOTTIP by two independent shRNAs in limb buds decreased expression of HoxA13, HoxA11, and HoxA10—again with a graded impact depending on genomic proximity to HOTTIP. Vector control or an shRNA that fails to deplete HOTTIP had little effect on Hox gene expression (Fig. 2D). In situ hybridization on whole embryos (Fig. 2E) and sections (Fig. S5) revealed that HOTTIP most strongly affects HoxA gene expression at the distal edge of the developing limb bud, where the 5′ HoxA genes are most strongly expressed. By stage 36, limbs depleted of HOTTIP showed notable shortening and bending of distal bony elements, including the radius, ulna, and third digit (~20% length reduction for each compared to contralateral and stage-matched limbs treated with control virus, p < 0.05, Student′s t-test, Fig. 2F). This phenotype resembled some of the defects in mice lacking HoxA11 and HoxA1321,22,23. Together, these data indicate that HOTTIP RNA controls activation of distal Hox genes in vivo.
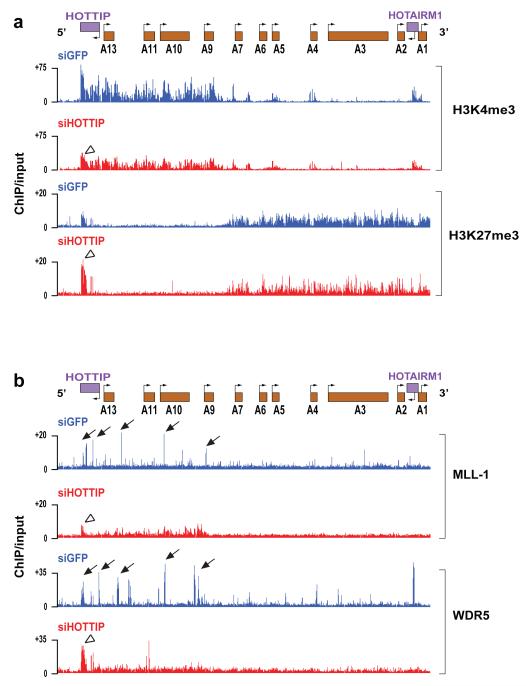
The broad impact of HOTTIP on gene activation across HOXA locus is reminiscent of the broad domains of chromatin modifications demarcating active and silent chromosomal domains12. 5C analysis of control and HOTTIP-depleted cells showed little change in higher order chromosomal structure, suggesting that the chromosomal looping is pre-configured and upstream of gene expression (Fig. S6A). In contrast, HOTTIP knockdown led to broad loss of H3K4me3 and H3K4me2 across the HOXA locus, mostly prominently over 5′ HOXA and HOTTIP itself (Fig. 3A, Figs. S6B and S7). HOTTIP knockdown also increased H3K27me3 focally over HOTTIP, but had little impact on H3K27me3 across HOXA. These results indicate that HOTTIP RNA is required for maintenance of H3K4me3 across the HOXA. These findings also imply that loss of 5′ HOXA gene transcription upon HOTTIP knockdown is likely due to loss of H3K4me3 (or other changes) rather than ectopic spread of H3K27me3.
Figure 3. HOTTIP is required for the active chromatin state of 5′ HOXA cluster.
(A) Knockdown of HOTTIP broadly decreases H3K4me3 across 5′ HOXA locus but focally affects H3K27me3 at HOTTIP itself. Display is as in Fig. 1A. (B) Knockdown of HOTTIP abrogates peaks of MLL1 and WDR5 occupancy near TSSs of 5′ HOXA genes and leads to accumulation of these proteins at HOTTIP itself. Arrows highlight peaks of MLL1 and WDR5 occupancy; open arrowheads highlight chromatin state of HOTTIP upon HOTTIP RNA knockdown.
H3K4 methylation of the HOX loci is carried out by the MLL family of complexes24. In mammals, at least six MLL family members of SET domain-containing lysine methyltransferases interact with a core complex of WDR5, ASH2, RBBP5, as well as with other proteins, for substrate recognition and genomic targeting24. Genetic analyses indicate that MLL1 and 2 are most essential for Hox gene expression in fibroblasts25, and MLL1 in particular is recruited to promoters of HOX genes to maintain their activation states26. In distally-derived cells, MLL1 and WDR5 densely occupied extended region of the 5′ HOXA cluster, coincident with the H3K4me3 domain, with specific “peaks” of occupancy near the transcriptional start sites (TSS) of multiple 5′ HOXA genes (Fig. 3B). Strikingly, HOTTIP knockdown abrogated the peaks of MLL1 and WDR5 occupancy near TSS, resulting in diffuse and less intense binding of MLL1 and WDR5 across HOXA cluster, most prominently over the 5′ HOXA domain. HOTTIP knockdown also led to increased accumulation of MLL1 and WDR5 on HOTTIP itself (Fig. S8). Thus, HOTTIP appears critical for maintaining a specific pattern of MLL complex occupancy across the HOXA locus to facilitate H3K4me3 and active transcription.
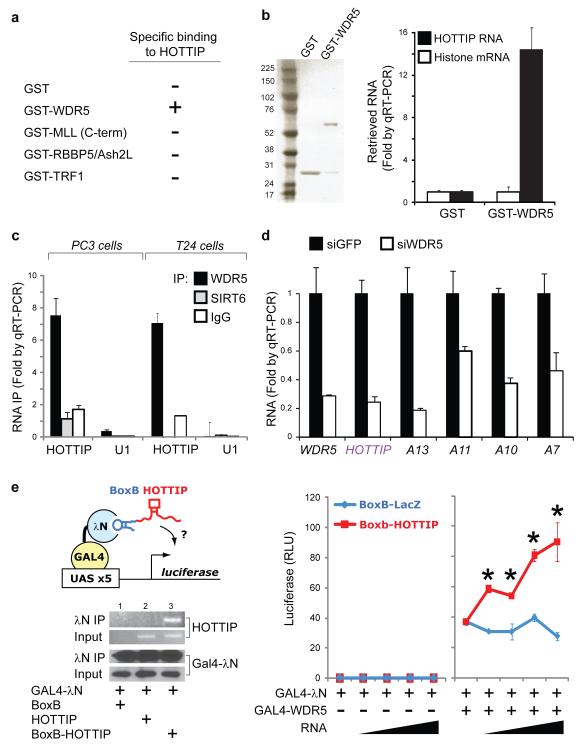
To define the molecular link between HOTTIP and MLL complex, we reasoned that HOTTIP RNA may physically interact with one or more subunits of the MLL complex. Purified, in vitro transcribed, full length HOTTIP RNA bound specifically to recombinant GST-WDR5, but not to GST, RBBP5, ASH2L, or the telomeric protein TRF1 (Fig. 4A, B). The C-terminus of MLL1, containing the SET domain, bound nonspecifically to all RNAs, consistent with previous studies27. Immunoprecipitation (IP) of endogenous WDR5 from two different cell lines each specifically retrieved endogenous HOTTIP RNA (Fig. 4C), indicating that WDR5 and HOTTIP interact in living cells. IP of an epitope-tagged WDR5 from a stable cell line that previously enabled stochiometric purification of WDR5 interacting proteins28 also specifically retrieved HOTTIP (Fig. S9). Knockdown of WDR5 broadly inhibited expression of 5′ HOXA genes, and also abrogated HOTTIP transcription, demonstrating mutual interdependence between HOTTIP and WDR5 (Fig. 4D).
Figure 4. HOTTIP programs active chromatin via WDR5.
(A) Summary of RNA-protein interaction studies. Each of the indicated recombinant protein was purified and used to retrieve purified HOTTIP or control histone RNA in vitro. Only GST-WDR5 specifically retrieved HOTTIP. (B) HOTTIP binds directly and specifically to WDR5. Left: Purified GST and GST-WDR5 are visualized by SDS-PAGE and Comassie Blue staining. Right: Retrieved RNAs are quantified by qRT-PCR. (C) HOTTIP binds specifically to WDR5 in cells. IP of endogenous WDR5 protein from PC3 (prostate) and T24 (bladder) carcinoma cells specifically retrieved HOTTIP, but not control IPs with IgG or chromatin binder SIRT6. (D) WDR5 is required for 5′ HOXA gene expression, including HOTTIP. (E) HOTTIP recruitment potentiates transcription. Left: The BoxB tethering system. BoxB-RNA specifically binds λN fused to GAL4 DNA binding domain, recruiting the complex to a UAS-luciferase reporter gene. After transient transfection, IP of GAL4- λN specifically retrieves BoxB-HOTTIP. Right: Luciferase activity after co-transfection of the indicated constructs (mean ± s.d., n=4 triplicates, * indciates p<0.05 Student’s t-test comparing BoxB-LacZ vs. BoxB-HOTTIP). Sloped triangle indicates increasing input of plasmids encoding ncRNAs.
HOTTIP appears to regulate genes in cis, due to its low copy number, distance dependence of HOXA target gene activation on endogenous HOTTIP, and the physical proximity of HOTTIP and its target genes as seen in 5C. Indeed, ectopic expression of HOTTIP by retroviral transduction of lung fibroblasts, which do not express HOTTIP, failed to activate expression of distal HOXA genes, nor change H3K4me3 and H3K27me3 patterns across HOXA (Fig S10). Moreover, in foreskin fibroblasts that express endogenous HOTTIP, ectopic HOTTIP expression did not induce 5′ HOXA genes, nor rescue the effects of depleting endogenous nascent HOTTIP (Fig. S11). The lack of response in foreskin fibroblasts is notable because endogenous HOTTIP is active in these cells, suggesting that the protein partners of HOTTIP are all present and target genes are receptive. Ectopically expressed HOTTIP, being transcribed from retroviral insertion sites scattered randomly in the genome, may not be able to find 5′HOXA genes. In contrast, endogenous HOTTIP is directly positioned near the 5′ HOXA genes by chromosomal looping, allowing interaction and control.
To test the requirement of an exogenous targeting mechanism, we engineered an allele of HOTTIP that can be artificially recruited to a reporter gene. Addition of five copies of the BoxB RNA element29 to HOTTIP allows the fusion transcript to be recruited to the λN RNA binding domain fused to a GAL4 DNA binding domain (Fig. 4E). Recruitment of HOTTIP to a silent GAL4 promoter is not sufficient to initiate transcription, but can significantly boost transcription if the promoter is also bound by WDR5 and transcriptionally active (Fig. 4E). By uncoupling the sites of HOTTIP transcription vs. HOTTIP RNA function, this experiment suggests that the proximity of HOTTIP RNA—rather than the act of transcription—maintains target gene expression. To further support the functionality HOTTIP RNA, deletion analysis identified a ~1 kb domain in the 5′ of HOTTIP (HOTTIPExons 1-2) that retains WDR5 binding activity (Fig. S12A). Enforced overexpression of HOTTIPExons 1-2 in foreskin fibroblasts inhibited 5′ HOXA gene expression in an apparently dominant negative manner (Fig. S12B).
In summary, HOTTIP is a key locus control element of HOXA genes and distal identity. Chromosomal looping brings HOTTIP in close proximity to the 5′ HOXA genes. HOTTIP transcription acts as a switch to produce HOTTIP lincRNA, which binds to and targets WDR5-MLL complexes to the 5′ HOXA locus, yielding a broad domain of H3K4me3 and transcription activation (Fig. S13). The mutual interdependence between HOTTIP and WDR5 creates a positive feedback loop that maintains the ON state of the locus. These findings provide an integrated view linking three dimensional genome organization to dynamic programming of chromatin states, and ultimately to developmental pattern formation.
H3K4 methylation is a feature of almost all transcribed genes, and MLL family proteins are involved in many cell fate decisions in development and disease24. Our findings suggest that additional lincRNAs, especially those associated with enhancers or enhancer-like activities8,9,10 may also be involved in gene activation by programming active chromatin states, and highlight WDR5 and other WD40 repeat proteins as candidate adaptors that link chromatin remodeling complexes to lincRNAs. Cis-restricted lincRNAs may be ideally suited to link chromosome structure and gene expression. Because such lincRNA can only act on its neighbors in space, information in higher order chromosomal looping can be faithfully transmitted to chromatin modification via RNA recruitment of enzymatic activities, and thus into gene expression.
Online-only Methods
Cells
Primary human fibroblasts derived from different anatomic sites were as described1,2,3,4,5,6,7. Primary human fibroblasts in culture retain their positional identity and have been used to examine chromatin states associated with positional memory, which have been confirmed in vivo3,4,8.
Chromatin immunoprecipitation followed by microarray analysis
ChIP-chip was performed using anti-H3K27me3 (Abcam, Cambridge, MA), anti-H3K4me3 (Abcam), anti-H3K4me2 (Abcam), anti-histone H3 (Abcam), anti-PolII (Abcam), anti-MLL1 (gift of R. Roeder, Rockefeller University), and anti-WDR59 antibodies as previously described5. Chromatin from each indicated cell type or RNAi treatment is split into multiple tubes and subject to ChIP with different antibodies in parallel. Retrieved DNA and input chromatin were competitively hybridized to custom tiling arrays interrogating human HOX loci at 5 bp resolution as previously described5.
5C analysis of the ENm010 HoxA1 region
5C primers were designed at HindIII restriction sites using 5C primer design tools previously developed10 and made available online at http://my5C.umassmed.edu11. Reverse primers were designed for fragments overlapping a known transcription start site from GENCODE transcripts12, or overlapping a start site as experimentally determined by CAGE Tag data of the ENCODE pilot project13. Forward primers were designed for all other HindIII restriction fragments. Primers were excluded if highly repetitive sequences prevented the design of a sufficiently unique 5C primer. Primers settings were: U-BLAST: 3; S-BLAST: 130: 15-MER: 1320; MIN_FSIZE: 40; MAX_FSIZE: 50000; OPT_TM: 65; OPT_PSIZE: 40. DNA sequence of the universal tails of forward primers was CCTCTCTATGGGCAGTCGGTGAT; DNA sequence for the universal tails of reverse primers was AGAGAATGAGGAACCCGGGGCAG. A 6 base barcode was included between the specific part of the primers and the universal tail. In total 17 reverse primers and 90 forward primers were designed in the 500 Kb HoxA1 locus (ENm010) and hence a total of 1530 cis interaction were interrogated in this region. Primer sequences are available separately (Supplementary Table 1).
3C was performed with HindIII as previously described14 separately for fetal lung and foreskin fibroblasts (FB) and also for the control and HOTTIP knockdown foreskin FBs. For the 5C reaction, a total of 107 forward and reverse primers of HoxA1 region were mixed with either the ENCODE random region (ENr) primer pool comprising of 2673 forward and 523 reverse primers (covering 30 additional ENCODE regions) or the ENr313 primer pool comprising of 57 forward and 58 reverse primers (covering 1 additional ENCODE region). 5C was then performed in 10 reactions each containing an amount of 3C library that represents 200,000 genome equivalents and 1 fmol of each primer. The 5C analysis of HoxA1 region was carried out in two biological replicates of fetal lung and foreskin FBs. 5C ligation products were amplified using a pair of universal primers that recognize the common tails of the 5C forward and reverse primers described above and pooled together. To facilitate paired end DNA sequence analysis on the Illumina GA2 platform, paired end adapter oligos were ligated to the 5C library using the Illumina PE protocol and PCR amplification of the library was carried out for 18 cycles with Illumina PCR primer PE 1.0 and 2.0. The 5C library was then sequenced on the Illumina GA2 platform generating 36 base paired end reads. For fetal lung FBs we obtained 7,625,276 and 10,947,424 mapped reads for two biological replicates of which 1,339,861 and 242,301 could be specifically mapped back to interactions within ENm010 using Novoalign (http://www.novocraft.com), respectively. For two biological replicates of foreskin FBs we obtained 7,311,386 and 5,731,107 mapped reads of which 2,752,789 and 66,769 could be mapped back to the ENm010 region, respectively. In the case of the knockdown study, control GFP knockdown foreskin FB 5C library yielded 4,909,482 mapped reads while HOTTIP knockdown foreskin FB had 5,565,389 mapped reads of which 39,168 and 38,950 could be mapped back to ENm010 for control GFP and HOTTIP knockdown, respectively. In the set with fetal lung and foreskin fibroblast samples, 5C for ENm010 was multiplexed for deep sequencing with 5C of one other region, ENr313; in the set containing the knockdown samples, ENm010 was multiplexed with 5C of 30 other genomic regions. The different extent of multiplexing resulted in different number of sequencing reads mapping back to ENm010. In all instances the mappable reads were proportional to the degree of multiplexing, indicating equivalent library quality despite different read numbers. Supplementary Table 2 outlines the library composition of each experiment. The heat maps are scaled as follows—for Figure 1A, distal (foreskin) FBs: 262-17467, proximal (lung) FBs: 7-5846; for Figure S6, siGFP: 1-100, siHOTTIP: 1-100. Raw data from the 5C experiments used to generate the binned heatmaps in Figures 1A and S6 can be found in Supplementary File 1. Raw data are available by request.
HOTTIP cloning, sequence, and expression analysis
We previously identified a portion of HOTTIP as a non protein-coding transcribed region named ncHOXA13-965. This region also overlaps EST clone AK093987 that was previously observed to be expressed in cancer cell lines derived from posterior anatomic sites15. 5′ and 3′ RACE (RLM Race kit, Applied Biosystems/Ambion, Foster City, CA) revealed full length HOTTIP to be 3764 nucleotides, extending the known transcribed region by more than 1400 bases. BLAST and BLAT confirmed that portions of HOTTIP are well conserved in mammals and even in avians but had no protein coding potential. Full length HOTTIP has been deposited at NCBI (accession #GU724873). qRT-PCR with SYBR Green was conducted as recommended by the manufacturer (Agilent Technologies, La Jolla, CA). Primer sequences specific for HOTTIP were CCTAAAGCCACGCTTCTTTG (HOTTIP-F) and TGCAGGCTGGAGATCCTACT (HOTTIP-R). For Fig. S11, endogenous nascent HOTTIP was distinguished from ectopic HOTTIP expressed from cDNA using primers that spanned intron-exon junctions.
Strand-specific RT-PCR
RNA extracted from primary foreskin fibroblasts was reverse transcribed (SuperScript III, Invitrogen) using combinations of the previously described HOTTIP-specific primers HOTTIP-F and/or HOTTIP-R as diagrammed in Supplementary Fig. 1B. Resulting cDNA was then PCR amplified using both HOTTIP-F and HOTTIP-R primers to visually determine strand specificity.
HOTTIP transcript count per cell
The level of HOTTIP transcript per cell was calculated from the level of HOTTIP in 500,000 cells. Full length HOTTIP in pcDNA3.1+ was assayed by qPCR using primers HOTTIP-F and HOTTIP-R at predetermined concentrations in triplicate to generate a linear amplification curve dependent on the moles of template DNA (Supplementary Fig 2). The qRT-PCR value from 500,000 foreskin fibroblasts was determined and plotted, and the corresponding total molecules of transcript was divided by 500,000 to determine the approximate number of transcripts per cell.
Single Molecule RNA Fluorescence in situ Hybridization (RNA-FISH)
Single molecule RNA-FISH was performed as described in Raj et al.16 with the following modifications: the amount of hybridization solution per chamber was doubled to allow for proper coating of the chamber and the amount of glucose-oxidase buffer was tripled to assist in image acquisition. Images were acquired using an Olympus FV1000 confocal microscope within 2 hours of the addition of the glucose-oxidase buffer.
RNA Interference
Primary foreskin fibroblasts were transfected with siRNAs targeting HOTTIP and WDR5 using Lipofectamine 2000 (Invitrogen, Carlsbad, California, United States) per manufacturer’s instructions. Total RNA was harvested 48-72 hours later using TRIzol (Invitrogen) and RNeasy Mini Kits (Qiagen, Valencia, California, United States) as previously described4. For the intronic HOTTIP knockdown experiment in Figure S11, a pool of 10 siRNAs (Supplementary Table 3) targeting intronic regions in HOTTIP were transfected into foreskin fibroblasts, and RNA isolated as above.
Generation of shRNAs against chicken HOTTIP
A reporter construct encoding a GFP-chicken HOTTIP fusion transcript was used in a small-scale screen to identify highly effective shRNA constructs. Eleven shRNAs targeting conserved regions of chicken HOTTIP were designed and inserted into the pSMP system (Thermo/Open Biosystems, Huntsvihotlle, AL). The reporter construct and shRNA constructs were cotransfected into Phoenix cells, and HOTTIP transcript levels were analyzed via reduced GFP fluorescence and by qRT-PCR. Three shRNAs that were effective in vitro were then cloned into RCAS vector for studies in chick embryos17.
Chick RNAi
RCAS HOTTIP hairpin and RCAS AP viruses were made by transfecting DF-1 cells with viral DNA. Transfected DF-1 cells were grown and passaged, after which the virus-containing supernatant was collected, concentrated, and titered. Fertilized chicken eggs were incubated in a humidified rotating incubator at 37°C until they reached Hamilton/ Hamburger stage 10. Eggs were then windowed to expose the embryos. After gently removing the vitelline membrane, chicken embryos were microinjected with RCAS-HOTTIP hairpins and RCAS-AP viruses at the prospective wing and leg buds. All viral stocks have titers of 1 × 108 IU/ml, and each limb was injected five times. The infected embryos were allowed to incubate at 37°C and were harvested 2 or 4 days after injection to detect viral infection by immunohistochemistry. Total RNA was extracted from injected forelimbs, and RT-PCR analysis was performed 4 days after injection. Chicken embryos were harvested 9 days post-injection to carry out whole mount alcian blue staining. A total of 50 animals were injected.
HOTTIP overexpression
Full length HOTTIP and a truncated transcript consisting of exons 1 and 2 (HOTTIP exons 1-2) were cloned into the LZRS vector (gift of P. Khavari, Stanford University), and then transfected into Phoenix cells (gift of G. Nolan, Stanford University) to generate amphotrophic retroviruses. Primary human fibroblasts were infected with either LZRS-full length HOTTIP (lung), LZRS-truncated HOTTIP (foreskin), or LZRS-GFP (both lung and foreskin), then passaged over 60 days, with periodic testing of HOXA and HOTTIP expression by qRT-PCR. These cells were used in the rescue experiments depicted in Figure S11.
GST Pulldown
Full length HOTTIP, truncated HOTTIP containing exons 1 and 2 (HOTTIPExons1-2), and histone H2B1 mRNA were transcribed in vitro using T7 polymerase according to manufacturer′s instructions (Promega, Madison, Wisconsin, United States), denatured, and refolded in folding buffer (100 mM KCl, 10 mM MgCl2, Tris pH 7.0). GST-tagged WDR5, C-terminal MLL1, RBBP5/Ash2L, and TRF1 were expressed in E. coli and purified as described18. Each GST-fusion protein was bound to glutathione beads (Amersham/GE Healthcare, Piscataway, NJ) and blocked with excess yeast total mRNA in PB100 buffer (20mM HEPES pH 7.6, 100 mM KCl, 0.05% NP40, 1 mM DTT, 0.5 mM PMSF) for one hour at room temperature. Beads were then incubated with either in vitro transcribed HOTTIP or histone H2B1 mRNA for 45 minutes at room temperature. After three washes in PB200 buffer (20 mM HEPES pH 7.6, 200 mM KCl, 0.05% NP40, 1 mM DTT, 0.5 mM PMSF), bound RNAs were extracted and analyzed by qRT-PCR, as previously described.
RNA Immunoprecipitation
HeLa-WDR5-FLAG cells: 48 hours after Lipofectamine 2000-mediated transfection of HOTTIP into HeLa WDR5-Flag cells (approximately 107), total protein was extracted as previously described, with modifications19. Briefly, cells were resuspended in Buffer A (10 mM Hepes pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 1.0 mM PMSF), lysed in 0.25% NP40, and fractionated by low speed centrifugation. The nuclear fraction was resuspended and lysed in Buffer C (20 mM Hepes pH 7.5, 10% glycerol, 0.42 M KCl, 4 mM MgCl2, 0.5 mM DTT, 1.0 mM PMSF). Combined nuclear and cytoplasmic fractions were immunoprecipitated with mouse anti-FLAG M2 monoclonal antibody (Sigma, St. Louis, MO) or mouse IgG affixed to agarose beads (Sigma) for three to four hours at 4C. Beads were washed four times with wash buffer (50 mM TrisCl pH 7.9, 10% glycerol, 100 mM KCl, 5 mM MgCl2, 10 mM betamercaptoethanol, 0.1% NP40). After elution using FLAG peptide (Sigma), coimmunoprecipitated RNA was extracted and analyzed by qRT-PCR.
Endogenous WDR5 and SIRT6 RIP: cellular fractions were isolated as above and incubated with the anti-WDR51 or anti-Sirt6 (ab62739, Abcam) antibodies overnight at 4°C. Samples were washed in wash buffer, and coimmunoprecipitated RNA was extracted and analyzed by qRT-PCR.
RNA chromatography
Full length in vitro transcribed HOTTIP RNA was conjugated to adipic acid dehydrzide agarose beads as described 20. The complexed beads were incubated with whole cell lysates from Hela WDR5-Flag cells, washed, and bound proteins visualized by Western blotting.
BoxB Tethering Assay
293T cells were grown to about 50% confluence in 6 well plates on the day of transfection. Using Lipofectamine 2000 (Invitrogen), a plasmid encoding a luciferase gene under the control of five tandem GAL4 UAS sites were co-transfected with plasmids encoding GAL4-WDR5, GAL4-lambdaN (the 22 amino acid RNA-binding domain of the lambda bacteriophage antiterminator protein N) peptide fused to a c-terminal GFP tag, BoxB (containing five repeats of the lambdaN-specific 19 nucleotide binding site), BoxB fused to full-length LacZ, or BoxB fused to full-length HOTTIP. Cells were lysed 48 hours after transfection, and luciferase assay kit (Promega) was used to determine relative levels of the luciferase gene product, following the manufacturer’s protocol.
Supplementary Material
Acknowledgements
We thank C. Tabin for chick Hox gene probes, M. Scott and members of our labs for input, and M. Lin for use of the confocal microscope and imaging expertise. Supported by grants from the California Institute for Regenerative Medicine (H.Y.C., J.W.), the National Institutes of Health (HG003143 to J.D.), and the Scleroderma Research Foundation (H.Y.C.). K.C.W. is a recipient of a Dermatology Foundation Career Development Award. J.D. is a recipient of the W. M. Keck Foundation Distinguished Young Scholar Award. H.Y.C. and M.L. are Early Career Scientists of the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions K.C.W., R.A.G., and H.Y.C. initiated the project; K.C.W. and H.Y.C. designed the experiments; K.C.W., Y.W.Y., B.L., A.S., R.C.Z., B.L., A.P., R.A.F., J.D., and J.A.H. conducted the experiments and analyzed the data; Y.C. and M.L. purified the recombinant proteins; J.W. provided antibodies and cell lines; K.C.W. and H.Y.C. prepared the manuscript with inputs from all co-authors.
Author Information: Sequence for human HOTTIP has been deposited with GenBank under the accession number GU724873. Reprints and permissions information is available at www.nature.com/nature/reprints. The authors declare no competing financial interests.
References and Notes
- 1.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- 4.Petruk S, et al. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Ashe HL, Monks J, Wijgerde M, Fraser P, Proudfoot NJ. Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev. 1997;11:2494–2509. doi: 10.1101/gad.11.19.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Santa F, et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang HY. Anatomic demarcation of cells: genes to patterns. Science. 2009;326:1206–1207. doi: 10.1126/science.1175686. [DOI] [PubMed] [Google Scholar]
- 12.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): A Massively Parallel Solution for Mapping Interactions between Genomic Elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, et al. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood. 2009;113:2526–2534. doi: 10.1182/blood-2008-06-162164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 17.Kmita M, Fraudeau N, Herault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420:145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- 18.Harpavat S, Cepko CL. RCAS-RNAi: a loss-of-function method for the developing chick retina. BMC Dev Biol. 2006;6:2. doi: 10.1186/1471-213X-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nelson CE, et al. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122:1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 20.Kmita M, et al. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005;435:1113–1116. doi: 10.1038/nature03648. [DOI] [PubMed] [Google Scholar]
- 21.Small KM, Potter SS. Homeotic transformations and limb defects in Hox A11 mutant mice. Genes Dev. 1993;7:2318–2328. doi: 10.1101/gad.7.12a.2318. [DOI] [PubMed] [Google Scholar]
- 22.Davis AP, Witte DP, Hsieh-Li HM, Potter SS, Capecchi MR. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature. 1995;375:791–795. doi: 10.1038/375791a0. [DOI] [PubMed] [Google Scholar]
- 23.Fromental-Ramain C, et al. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122:2997–3011. doi: 10.1242/dev.122.10.2997. [DOI] [PubMed] [Google Scholar]
- 24.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 25.Wang P, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29:6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guenther MG, et al. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krajewski WA, Nakamura T, Mazo A, Canaani E. A motif within SET-domain proteins binds single-stranded nucleic acids and transcribed and supercoiled DNAs and can interfere with assembly of nucleosomes. Mol Cell Biol. 2005;25:1891–1899. doi: 10.1128/MCB.25.5.1891-1899.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 29.Baron-Benhamou J, Gehring NH, Kulozik AE, Hentze MW. Using the lambdaN peptide to tether proteins to RNAs. Methods Mol Biol. 2004;257:135–154. doi: 10.1385/1-59259-750-5:135. [DOI] [PubMed] [Google Scholar]
- 30.Lajoie BR, van Berkum NL, Sanyal A, Dekker J. My5C: web tools for chromosome conformation capture studies. Nat Methods. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online-only Methods
- 1.Chang HY, et al. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc Natl Acad Sci U S A. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang HY, et al. Gene expression signature of fibroblast serum response predicts human cancer progression: Similarities between tumors and wounds. PLoS Biology. 2004;2:206–214. doi: 10.1371/journal.pbio.0020007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY. Anatomic demarcation by positional variation in fibroblast gene expression programs. PLoS Genet. 2006;2:e119. doi: 10.1371/journal.pgen.0020119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, et al. A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 2008;22:303–307. doi: 10.1101/gad.1610508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinn JL, et al. A systems biology approach to anatomic diversity of skin. J Invest Dermatol. 2008;128:776–782. doi: 10.1038/sj.jid.5700986. [DOI] [PubMed] [Google Scholar]
- 8.Soshnikova N, Duboule D. Epigenetic temporal control of mouse Hox genes in vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 9.Wysocka J, et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 10.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lajoie BR, van Berkum NL, Sanyal A, Dekker J. My5C: web tools for chromosome conformation capture studies. Nat Methods. 2009;6:690–691. doi: 10.1038/nmeth1009-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrow J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):S4, 1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 15.Sasaki YT, Sano M, Kin T, Asai K, Hirose T. Coordinated expression of ncRNAs and HOX mRNAs in the human HOXA locus. Biochem Biophys Res Commun. 2007;357:724–730. doi: 10.1016/j.bbrc.2007.03.200. [DOI] [PubMed] [Google Scholar]
- 16.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harpavat S, Cepko CL. RCAS-RNAi: a loss-of-function method for the developing chick retina. BMC Dev Biol. 2006;6:2. doi: 10.1186/1471-213X-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 19.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michlewski G, Caceres JF. RNase-assisted RNA chromatography. Rna. 2010;16:1673–1678. doi: 10.1261/rna.2136010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 22.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.