Abstract
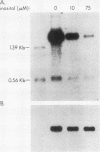
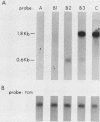
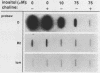
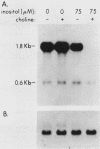
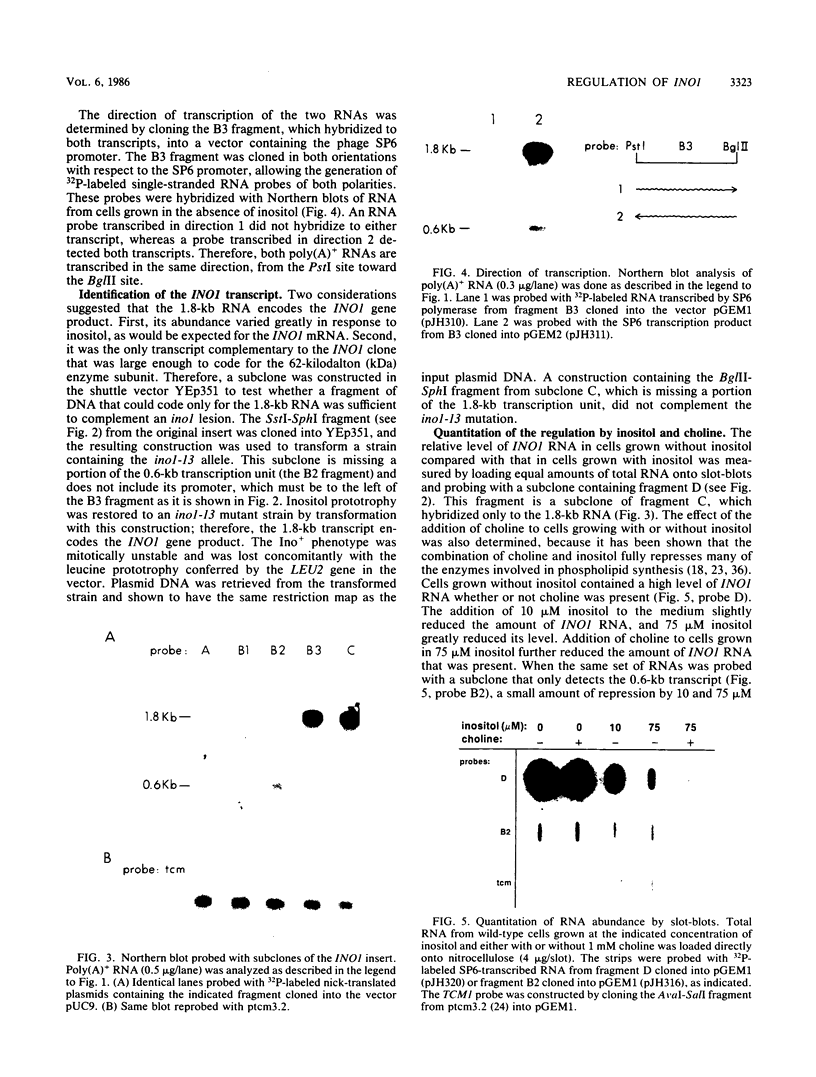
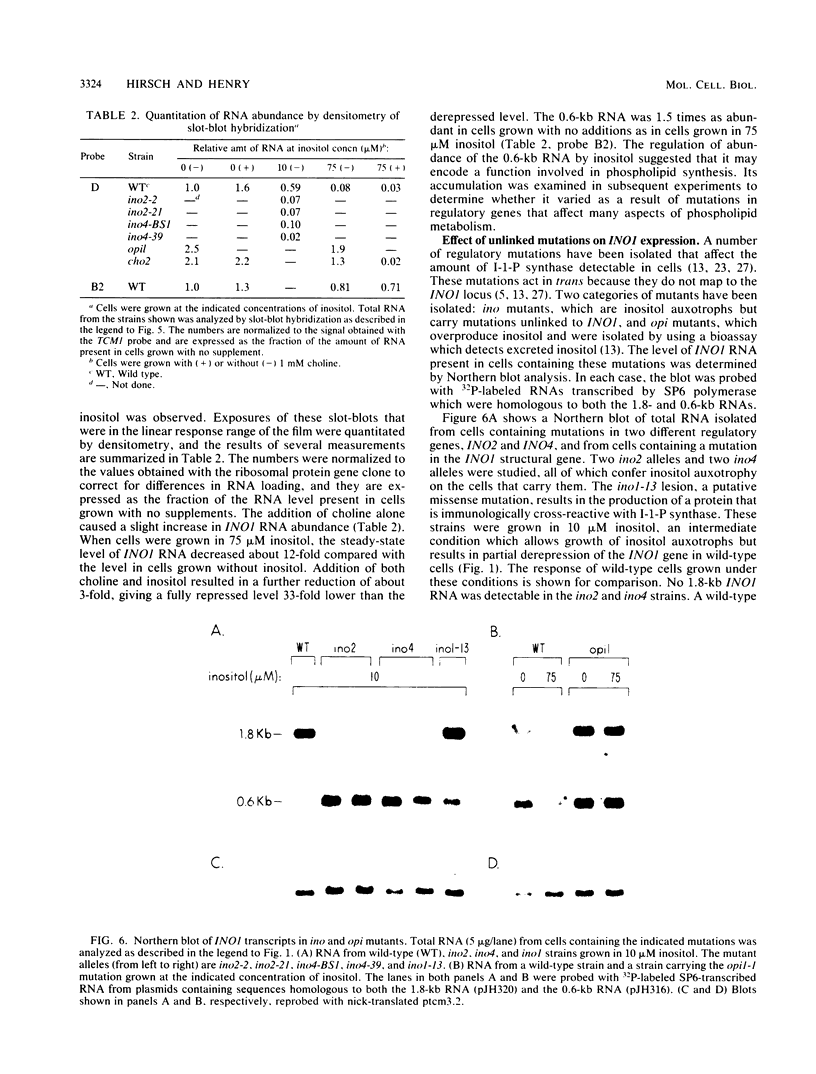
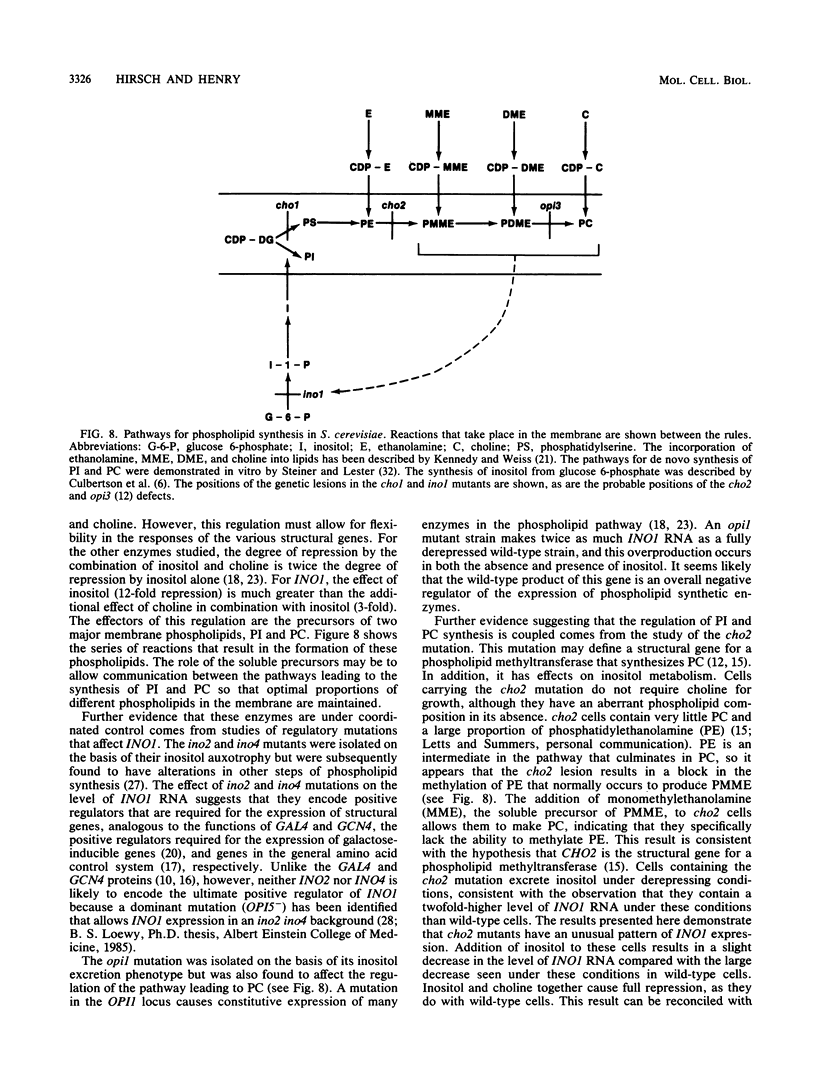
The INO1 gene of Saccharomyces cerevisiae encodes the regulated enzyme inositol-1-phosphate synthase, which catalyzes the first committed step in the synthesis of inositol-containing phospholipids. The expression of this gene was analyzed under conditions known to regulate phospholipid synthesis. RNA blot hybridization with a genomic clone for INO1 detected two RNA species of 1.8 and 0.6 kb. The abundance of the 1.8-kb RNA was greatly decreased when the cells were grown in the presence of the phospholipid precursor inositol, as was the enzyme activity of the synthase. Complementation analysis showed that this transcript encoded the INO1 gene product. The level of INO1 RNA was repressed 12-fold when the cells were grown in medium containing inositol, and it was repressed 33-fold when the cells were grown in the presence of inositol and choline together. The INO1 transcript was present at a very low level in cells containing mutations (ino2 and ino4) in regulatory genes unlinked to INO1 that result in inositol auxotrophy. The transcript was constitutively overproduced in cells containing a mutation (opi1) that causes constitutive expression of inositol-1-phosphate synthase and results in excretion of inositol. The expression of INO1 RNA was also examined in cells containing a mutation (cho2) affecting the synthesis of phosphatidylcholine. In contrast to what was observed in wild-type cells, growth of cho2 cells in medium containing inositol did not result in a significant decrease in INO1 RNA abundance. Inositol and choline together were required for repression of the INO1 transcript in these cells, providing evidence for a regulatory link between the synthesis of inositol- and choline-containing lipids. The level of the 0.6-kb RNA was affected, although to a lesser degree, by many of the same factors that influence INO1 expression.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Donahue T. F., Henry S. A. Control of inositol biosynthesis in Saccharomyces cerevisiae: properties of a repressible enzyme system in extracts of wild-type (Ino+) cells. J Bacteriol. 1976 Apr;126(1):232–242. doi: 10.1128/jb.126.1.232-242.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson M. R., Henry S. A. Inositol-requiring mutants of Saccharomyces cerevisiae. Genetics. 1975 May;80(1):23–40. doi: 10.1093/genetics/80.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Henry S. A. myo-Inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem. 1981 Jul 10;256(13):7077–7085. [PubMed] [Google Scholar]
- Elion E. A., Warner J. R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984 Dec;39(3 Pt 2):663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- Fried H. M., Warner J. R. Cloning of yeast gene for trichodermin resistance and ribosomal protein L3. Proc Natl Acad Sci U S A. 1981 Jan;78(1):238–242. doi: 10.1073/pnas.78.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Greenberg M. L., Goldwasser P., Henry S. A. Characterization of a yeast regulatory mutant constitutive for synthesis of inositol-1-phosphate synthase. Mol Gen Genet. 1982;186(2):157–163. doi: 10.1007/BF00331845. [DOI] [PubMed] [Google Scholar]
- Greenberg M. L., Klig L. S., Letts V. A., Loewy B. S., Henry S. A. Yeast mutant defective in phosphatidylcholine synthesis. J Bacteriol. 1983 Feb;153(2):791–799. doi: 10.1128/jb.153.2.791-799.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M. L., Reiner B., Henry S. A. Regulatory mutations of inositol biosynthesis in yeast: isolation of inositol-excreting mutants. Genetics. 1982 Jan;100(1):19–33. doi: 10.1093/genetics/100.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S. A., Klig L. S., Loewy B. S. The genetic regulation and coordination of biosynthetic pathways in yeast: amino acid and phospholipid synthesis. Annu Rev Genet. 1984;18:207–231. doi: 10.1146/annurev.ge.18.120184.001231. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Sep;5(9):2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann M. J., Henry S. A., Carman G. M. Regulation of CDP-diacylglycerol synthase activity in Saccharomyces cerevisiae. J Bacteriol. 1985 Sep;163(3):1265–1266. doi: 10.1128/jb.163.3.1265-1266.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston S. A., Hopper J. E. Isolation of the yeast regulatory gene GAL4 and analysis of its dosage effects on the galactose/melibiose regulon. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6971–6975. doi: 10.1073/pnas.79.22.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KENNEDY E. P., WEISS S. B. The function of cytidine coenzymes in the biosynthesis of phospholipides. J Biol Chem. 1956 Sep;222(1):193–214. [PubMed] [Google Scholar]
- Klig L. S., Henry S. A. Isolation of the yeast INO1 gene: located on an autonomously replicating plasmid, the gene is fully regulated. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3816–3820. doi: 10.1073/pnas.81.12.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klig L. S., Homann M. J., Carman G. M., Henry S. A. Coordinate regulation of phospholipid biosynthesis in Saccharomyces cerevisiae: pleiotropically constitutive opi1 mutant. J Bacteriol. 1985 Jun;162(3):1135–1141. doi: 10.1128/jb.162.3.1135-1141.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Letts V. A., Henry S. A. Regulation of phospholipid synthesis in phosphatidylserine synthase-deficient (chol) mutants of Saccharomyces cerevisiae. J Bacteriol. 1985 Aug;163(2):560–567. doi: 10.1128/jb.163.2.560-567.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letts V. A., Klig L. S., Bae-Lee M., Carman G. M., Henry S. A. Isolation of the yeast structural gene for the membrane-associated enzyme phosphatidylserine synthase. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7279–7283. doi: 10.1073/pnas.80.23.7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewy B. S., Henry S. A. The INO2 and INO4 loci of Saccharomyces cerevisiae are pleiotropic regulatory genes. Mol Cell Biol. 1984 Nov;4(11):2479–2485. doi: 10.1128/mcb.4.11.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton D. A., Krieg P. A., Rebagliati M. R., Maniatis T., Zinn K., Green M. R. Efficient in vitro synthesis of biologically active RNA and RNA hybridization probes from plasmids containing a bacteriophage SP6 promoter. Nucleic Acids Res. 1984 Sep 25;12(18):7035–7056. doi: 10.1093/nar/12.18.7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozek C. E., Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983 Jan;32(1):23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- St John T. P., Davis R. W. The organization and transcription of the galactose gene cluster of Saccharomyces. J Mol Biol. 1981 Oct 25;152(2):285–315. doi: 10.1016/0022-2836(81)90244-8. [DOI] [PubMed] [Google Scholar]
- Steiner M. R., Lester R. L. In vitro studies of phospholipid biosynthesis in Saccharomyces cerevisiae. Biochim Biophys Acta. 1972 Feb 21;260(2):222–243. doi: 10.1016/0005-2760(72)90035-5. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yamashita S., Oshima A., Nikawa J., Hosaka K. Regulation of the phosphatidylethanolamine methylation pathway in Saccharomyces cerevisiae. Eur J Biochem. 1982 Nov 15;128(2-3):589–595. doi: 10.1111/j.1432-1033.1982.tb07005.x. [DOI] [PubMed] [Google Scholar]