Abstract
EPF1-EPF2 and EPFL9/Stomagen act antagonistically in regulating leaf stomatal density. The aim of this study was to elucidate the evolutionary functional divergence of EPF/EPFL family genes. Phylogenetic analyses showed that AtEPFL9/Stomagen-like genes are conserved only in vascular plants and are closely related to AtEPF1/EPF2-like genes. Modeling showed that EPF/EPFL peptides share a common 3D structure that is constituted of a scaffold and loop. Molecular dynamics simulation suggested that AtEPF1/EPF2-like peptides form an additional disulfide bond in their loop regions and show greater flexibility in these regions than AtEPFL9/Stomagen-like peptides. This study uncovered the evolutionary relationship and the conformational divergence of proteins encoded by the EPF/EPFL family genes.
Introduction
Stomata are pores on the aerial surfaces of land plants that mediate gas exchange between plants and their environment. Stomata are found in mosses and hornworts in bryophytes and in vascular plants including lycophytes, ferns, gymnosperms and angiosperms [1]. In the evolution of land plants, stomatal density significantly shifted in response to fluctuations in the atmospheric environment such as CO2 concentration. Ancient land plants from the Silurian to the Devonian had a low stomatal density relative to extant land plants [2]. By the late Devonian/early Carboniferous, the density had increased by up to 100 times in response to a dramatic decline in the atmospheric CO2 concentration [3], which resulted in morphological advances such as planate leaves in vascular plants [4]. This evolutionary innovation in leaf tissues implies that early vascular plants had developed molecular systems elevating their stomatal density when they perceived the atmospheric fluctuation.
The EPF/EPFL (epidermal patterning factor/EPF-like) gene family encodes plant-specific secretory peptides, several of which play a role in controlling stomatal density and patterning in the plant epidermis [5]. Mature EPF/EPFL peptides have conserved six or eight cysteine residues that form intramolecular disulfide bonds [6]. The structure of Arabidopsis thaliana (At) EPFL9/Stomagen inferred by NMR indicates that the peptide is composed of two structural parts, a scaffold and a loop. The scaffold consists of a pair of ß-strands forming an anti-parallel ß-sheet supported by three disulfide bonds with a one-turn 310-helix, which is structurally required for the activity of the peptides. The loop domain connecting the two ß-strands contributes to the functional specificity of each EPF/EPFL peptide during stomatal development. In the model plant A. thaliana, AtEPF1 and AtEPF2 have a negative role in leaf stomatal development [7]–[9], in which an additional disulfide bond is formed between two cysteine residues in the loop region. Intriguingly, AtEPFL9/Stomagen lacking an additional disulfide bond acts as a positive regulator of leaf stomatal density and has an antagonistic action on AtEPF1 and AtEPF2 [6], [10].
The genes regulating stomatal density and patterning are well conserved from basal land plants. The small peptides AtEPF1 and AtEPF2 form ligand-receptor complexes with TOO MANY MOUTHS (TMM) and ERECTA family members in the A. thaliana [11]. TMM and the ERECTA family play roles as putative receptors for EPF peptides to initiate a signal cascade for stomatal development. The basal land plant Physcomitrella patens, which develops stomata around the base of its sporophytes [12], retains one copy of the EPF/EPFL gene, one copy of TMM and six copies of ERECTA family genes [13]–[15]. Intriguingly, an EPF/EPFL ortholog in P. patens has more similarity to negative regulators AtEPF1 and AtEPF2 than to positive regulator AtEPFL9/Stomagen, whereas in the genome of the lycophyte Selaginella moellendorffii, both AtEPFL9/Stomagen-like genes and AtEPF1/EPF2-like genes are conserved [15]. However, the evolutionary relationships between the negative and positive regulators are still unclear in the EPF/EPFL gene family.
In the present study, we examined the phylogenetic relationship of and structurally characterized the EPF/EPFL gene family to clarify evolutionary functional divergence between the negative and positive regulators. To this end, our extensive analysis reconstructed phylogenetic trees using EPF/EPFL genes in land plants and predicted the 3D structures of their products. The 3D structures were subjected to a computer simulation to analyze their molecular dynamical properties. This study revealed that the AtEPFL9/Stomagen-like genes conserved in vascular plants are closely related to AtEPF1/EPF2-like genes and that AtEPF1/EPF2-like peptides have greater flexibility in the loop region than AtEPFL9/Stomagen-like peptides. Our data allowed us to explore the evolutionary history of leaf stomatal density in early land plants.
Materials and Methods
Phylogenetic Analysis
EPF/EPFL genes were retrieved from genomic database for Arabidopsis thaliana (The Arabidopsis Information Resource, TAIR). To identify EPF/EPFL genes in Carica papaya (Cp), Medicago truncatula (Mt), Oryza sativa (Os), Picea glauca (Pg), Physcomitrella patens (Pp), Populus trichocarpa (Pt), Selaginella moellendorffii (Sm) and Sorghum bicolor (Sb), TBLASTN searches were performed against the genomic databases using amino acid sequences encoded by EPF/EPFL genes of A. thaliana (At) as queries: Rice Annotation Project Database for O. sativa, The Gene Index Project for P. glauca, and Phytozome v8.0 [16] for C. papaya, M. truncatula, P. trichocarpa, P. patens, S. bicolor and S. moellendorffii. Genes that retained the typical ß-sheet scaffold and the loop domain were retrieved from these genomic databases. Accession numbers or locus IDs of EPF/EPFLs are described in Table S1. For predicted genes lacking a conserved portion of the EPF/EPFL gene, the open reading frame of the gene was repredicted using an assembled EST sequence made by the PASA tool [17] in the Phytozome database. For unpredicted genes in the S. moellendorffii genomic database, the protein-coding region was predicted by the Fgenesh+ program [18]. Amino acid sequences for the C-terminal mature peptide region were aligned using the ClustalW program [19]. The number of amino acid substitutions between each pair of EPF/EPFL proteins was estimated by the Jones-Taylor-Thornton (JTT) model [20] with the complete-deletion option. From estimated numbers of amino acid substitutions, phylogenetic trees were reconstructed by the minimum evolution (ME) and neighbor-joining (NJ) methods. Bootstrap values were calculated with 1,000 replications. These procedures were performed using MEGA5 software [21].
Modeling
Three-dimensional structures of EPF/EPFL peptides were modeled with Modeller version 9.9 software [22]. The structure of AtEPFL9/Stomagen determined by NMR was employed as the template and SS-bonds were allowed to form during modeling. Structural figures were prepared using the MOLMOL graphics program [23].
Molecular Dynamics Simulation
The modeling structures were used as the starting structure for molecular dynamics (MD) simulation. The calculation was performed with the GROMACS program [24] using an AMBER99-SB force field [25]. Each structure was placed in the center of a 60 Å × 60 Å × 60 Å cubic box with periodic boundary conditions and solvated by SPC/E water molecules [26]. Na+ or Cl- counterions were added to satisfy the electroneutrality condition for each system. Berendsen temperature coupling and Parrinello-Rahman pressure coupling were used to keep the system in a stable environment (300 K, 1 bar), and the coupling constants were set to 0.1 ps. The particle mesh Ewald (PME) algorithm [27] was employed to calculate long-range electrostatic interactions with a cutoff value of 1.0 nm, and a cutoff of 1.0 nm was set for van der Waals interactions. The LINCS algorithm [28] for bond constraints was applied. Each system was energy-minimized with a steepest-descent algorithm for 1,000 steps; then the solvent, ions were equilibrated for 1 ns in NTP and NVT ensembles, respectively. Finally, all constraints were removed and a 10 ns MD simulation was performed for each system. All the trajectories were stored every 200 ps for further analysis.
Trajectories for each structure in aqueous solution were analyzed to obtain structural and dynamic properties using the GROMACS analysis tools package, including the root mean square fluctuation of the residues (RMSF), root mean square deviation (RMSD), and secondary structure prediction, which were performed using the STRIDE program [29].
Results
Phylogenetic Analysis of EPF/EPFL Gene Family
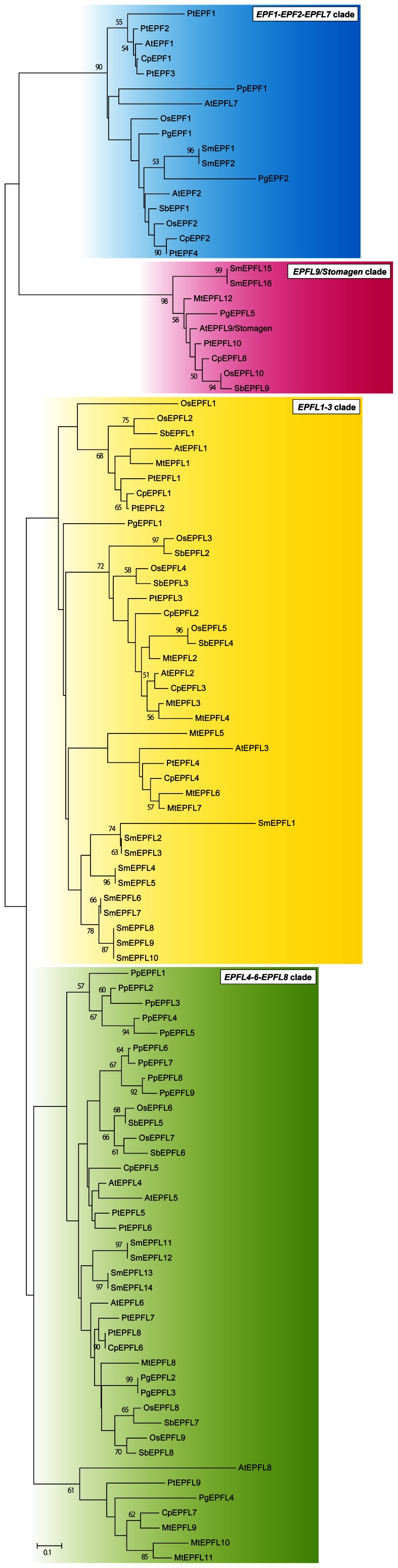
EPF/EPFL genes are conserved in land plants but not in algae such as Chlamydomonas, Ostreococcus and Cyanidioschyzon. Phylogenetic trees clearly showed that EPF/EPFL genes separated into four clades (EPF1-EPF2-EPFL7 clade, EPFL9/Stomagen clade, EPFL1-3 clade and EPFL4-6-EPFL8 clade; the clades were named after respective EPF/EPFLs in A. thaliana.) in land plants (Fig. 1, Fig. S1, Dataset S1). The EPF1-EPF2-EPFL7 clade and the EPFL4-6-EPFL8 clade contained genes from bryophyte and vascular plants including lycophytes, gymnosperms, monocots and eudicots. On the other hand, the EPFL9/Stomagen clade and the EPFL1-3 clade were constituted of genes only from vascular plants. The negative regulators such as AtEPF1 and AtEPF2 were found in the EPF1-EPF2-EPFL7 clade. AtEPFL9/Stomagen, the antagonist of AtEPF1 and AtEPF2, was a member of the EPFL9/Stomagen clade. The phylogenetic trees showed close relationships between the EPF1-EPF2-EPFL7 clade and the EPFL9/Stomagen clade and between the EPFL1-3 clade and the EPFL4-6-EPFL8 clade in the EPF/EPFL gene family.
Figure 1. Phylogenetic tree of EPF/EPFL genes.
Amino acid sequences for the mature peptide region were aligned using the ClustalW program. The phylogenetic tree was reconstructed by the ME method from the numbers of amino acid substitutions estimated by the JTT model. The numerals at the branch indicate bootstrap values calculated by the ME method with 1,000 replications. Bootstrap values >50% are shown.
Three-dimensional Structural Models of EPF/EPFL Peptides
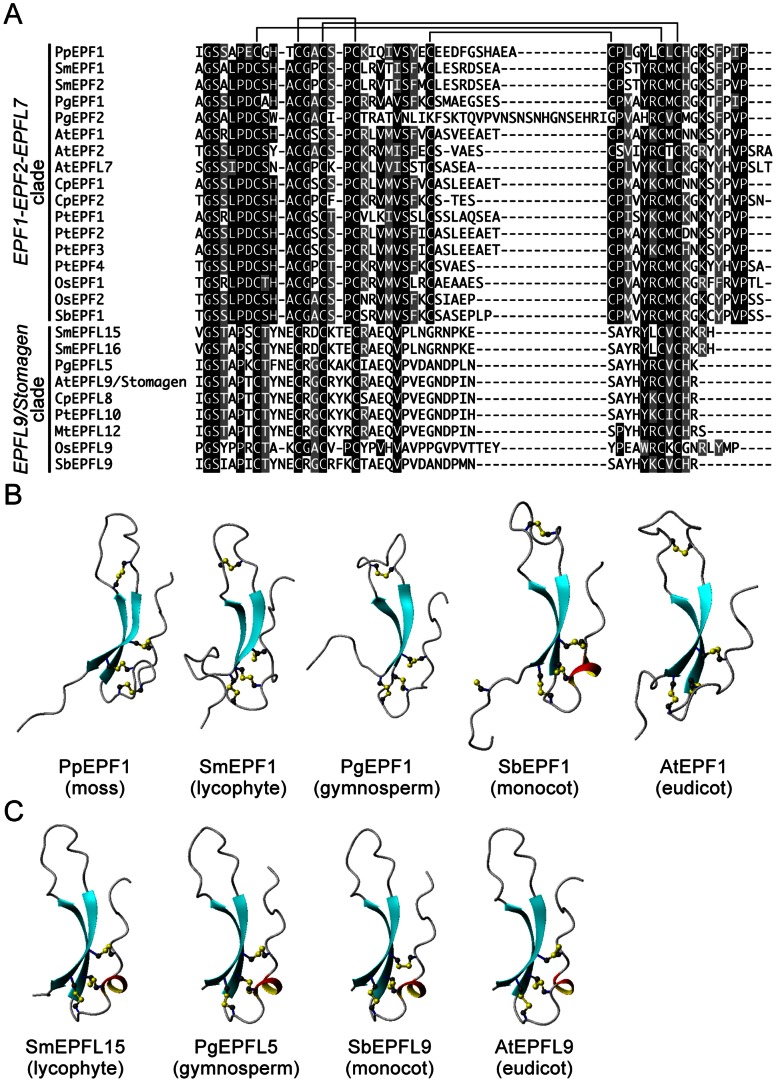
The phylogenetic analysis showed that the negative regulators AtEPF1 and AtEPF2 were closely related to the antagonist AtEPFL9/Stomagen. To characterize peptide sequences in the EPF1-EPF2-EPFL7 clade and the EPFL9/Stomagen clade, we analyzed their 1D structures. The prepropeptides for AtEPF1, AtEPF2 and AtEPFL9/Stomagen contain a signal peptide at their N-terminal regions [15]. A putative signal peptide was identified in genes from mosses, lycophytes, gymnosperms, monocots and eudicots using SignalP software [30] (Table S2). In the C-terminal mature peptide regions, six-cysteine residues located in a scaffold region were well conserved for all genes in both clades (Fig. 2A). Additional two-cysteine residues in the loop region were only conserved within the peptides in the EPF1-EPF2-EPFL7 clade except for PgEPF2.
Figure 2. Structures of AtEPF1/EPF2-like peptides and AtEPFL9/Stomagen-like peptides.
(A) Primary structures of AtEPF1/EPF2-like peptides and AtEPFL9/Stomagen-like peptides in land plants. Sequence alignment was generated by the ClustalW program. Pairs of cysteine residues forming disulfide bonds predicted for A. thaliana EPF/EPFL genes are indicated by lines. (B) and (C) Ribbon models of EPF/EPFL peptides. Structural models shown here were generated by homology modeling. The structure of AtEPFL9/Stomagen determined by NMR was used as the template for the homology modeling. A model of the disulfide bonds is shown as a ball-and-stick representation.
We next translated their 1D sequence information into 3D structural features. Homology modeling was performed to examine structural properties of the peptides in the EPF1-EPF2-EPFL7 clade and the EPFL9/Stomagen clade. Fig. 2B and 2C show 3D structures of the representative peptides from a moss (P. patens), a lycophyte (S. moellendorffii), a gymnosperm (P. glauca), a monocot (S. bicolor) and a eudicot (A. thaliana) in evolutionary lineages of plants. The 3D structure modeling clearly showed that the peptides shared an AtEPFL9/Stomagen-like structure that is composed of a consensus scaffold and a long loop. In the scaffold regions, three disulfide bonds were formed between an anti-parallel ß-strand and a one-turn 310-helix in SbEPF1, AtEPF1, SmEPFL15, PgEPFL5, SbEPFL9 and AtEPFL9. In PpEPF1, SmEPF1 and PgEPF1 peptides, one or two disulfide bonds were adopted between adjacent amino acids of the first ß-strand and the helix. The AtEPF1/EPF2-like peptides form an additional disulfide bond in their loop regions, although the AtEPFL9/Stomagen-like peptides do not. The PgEPF2, lacking any additional two-cysteine residues, does not retain a disulfide bond in its loop even though it is a member of the EPF1-EPF2-EPFL7 clade (Fig. S2A).
MD Simulation of EPF/EPFL Peptides
To clarify conformational divergence between AtEPF1/EPF2-like peptides and AtEPFL9/Stomagen-like peptides, 3D structural stability and flexibility of the EPF/EPFLs were examined by MD simulation. The 10 ns MD simulation was performed for six and four peptides, respectively, in the EPF1-EPF2-EPFL7 clade and EPFL9/Stomagen clade. The RMSD value of the AtEPF1 was higher than that of AtEPFL9/Stomagen (Fig. 3A). Likewise, the values for the AtEPF1/EPF2-like peptides were distinguishable from the AtEPFL9/Stomagen-like peptides. This indicates that the EPF/EPFLs in the EPF1-EPF2-EPFL7 clade are less conformationally stable than those in the EPFL9/Stomagen clade. For the RMSF values plotted through amino acid sequence, AtEPF1 showed a higher value in the loop region than AtEPFL9/Stomagen (Fig. 3B and 3C). Similarly to the A. thaliana EPF/EPFLs, the AtEPF1/EPF2-like peptides including moss EPF1 fluctuated more in the loop region than the AtEPFL9/Stomagen-like peptides (Fig. 3B and 3C, Fig. S2B). Taken together, these data suggest that the EPF/EPFLs in the EPF1-EPF2-EPFL7 clade have greater flexibility in their functional loop regions than those in the EPFL9/Stomagen clade.
Figure 3. Results of MD simulation of AtEPF1/EPF2-like peptides and AtEPFL9/Stomagen-like peptides.
(A) RMSD values (in nm) during a 10 ns molecular dynamic simulation. (B) and (C) RMSF value (in nm) through the amino acid sequence of each peptide. RMSD indicates conformational changes from the initial structure and RMSF indicates fluctuating area in the molecule. Blue and red bars indicate the ß-strand and loop, respectively.
Discussion
AtEPF1 and AtEPF2 have an antagonistic role with AtEPFL9/Stomagen in regulating stomatal density and patterning in leaf tissue. Although EPF/EPFL genes are widely conserved in land plants, their phylogenetic relationship remains to be determined. Here, we elucidated the evolutionary relationship of EPF/EPFL genes in land plants and examined their conformational divergence to deduce their emergence and acquisition of antagonistic function during land plant evolution.
In the phylogenetic trees, AtEPF1/EPF2-like genes and AtEPFL9/Stomagen-like genes showed a close relationship in EPF/EPFL family genes (Fig. 1, Fig. S1). AtEPF1/EPF2-like genes constitute the EPF1-EPF2-EPFL7 clade that contains AtEPF1 and AtEPF2, which negatively regulate stomatal development. AtEPFL9/Stomagen-like genes comprise the EPFL9/Stomagen clade that includes the positive regulator AtEPFL9/Stomagen. Although the EPF1-EPF2-EPFL7 clade is composed of genes from both bryophytes and vascular plants, the EPFL9/Stomagen clade does not conserve any gene from mosses. Collectively, these findings suggest that the EPF1-EPF2-EPFL7 clade and the EPFL9/Stomagen clade may share a common ancestral gene and the latter clade formed after the speciation of bryophytes and vascular plants.
AtEPF1/EPF2-like peptides and AtEPFL9/Stomagen-like peptides display different plasticity in their 3D structures, although they share a common conformation composed of a consensus scaffold and a loop (Figs. 2 and 3). In stomatal development of the model plant A. thaliana, both the ß-sheet scaffold and the loop domain are required for activity [6]. Moreover, the loop domain confers the functional specificity of EPF/EPFL peptides. The 3D structure modeling revealed differences in the loop region between AtEPF1/EPF2-like peptides and AtEPFL9/Stomagen-like peptides (Fig. 2). AtEPF1/EPF2-like peptides including the moss EPF1 form a disulfide bond in the loop region, whereas AtEPFL9/Stomagen-like peptides do not. Fig. 3 shows that the functional loop of the AtEPF1/EPF2-like peptides is more flexible than that of the AtEPFL9/Stomagen-like peptides. The additional disulfide bond is not always essential for loop flexibility because PgEPF2 exhibited a higher RMSF value in its loop (Fig. S2B). Amino acid composition and length of loops affect conformational stability, for example, longer inter-domain linkers show the greater flexibility [31]. The difference in the loop flexibility between AtEPF1/EPF2-like peptides and the AtEPFL9/Stomagen-like peptides would be defined by their loop length and additional disulfide bond that restricts the conformational degree of freedom in the loop domain [32]. The flexibility of the functional loop may contribute to the antagonistic function between the AtEPF1/EPF2-like peptides and the AtEPFL9/Stomagen-like peptides in stomatal development.
Our evolutionary and structural analyses revealed that the moss P. patens and vascular plants conserve the AtEPF1/EPF2-like gene in a structurally similar form to that of negative regulators and that vascular plants including lycophytes retain the AtEPFL9/Stomagen-like gene (Figs. 1, 2 and 3). AtEPF1 and AtEPF2 form ligand-receptor modules with TMM and ERECTA family members to initiate a signaling pathway in stomatal development [11]. Since TMM and the ERECTA family are found in P. patens, in which stomata are distributed around the sporophyte [12], it is strongly deduced that the ligand-receptor module was developed in early land plants. It is interesting that the dramatic increase in stomatal density that occurred in early vascular plants in the late Devonian [3] appears to coincide with the evolutionary acquisition of AtEPFL9/Stomagen-like peptide, which positively regulates stomatal development. Beerling et al. [4] hypothesize that the increased stomatal density on leaf surfaces was an essential driving force for evolving megaphyll leaves in vascular plants. The acquisition of an AtEPFL9/Stomagen-like gene would be potentially advantageous for this morphological innovation because the peptide hormone Stomagen is a potent inducer of leaf stomata [10]. Our findings provide new insights into functional divergence in the molecular evolution of the EPF/EPFL family genes and will facilitate future studies on the evolutionary development of stomata in land plants.
Supporting Information
Phylogenetic tree of EPF/EPFL genes. The phylogenetic tree was reconstructed by the NJ method from the numbers of amino acid substitutions estimated by the JTT model. The numerals at the branch indicate bootstrap values calculated by the NJ method with 1,000 replications. Bootstrap values >50% are shown.
(EPS)
Molecular properties of P. glauca EPF2. (A) Ribbon model and (B) MD simulation of PgEPF2.
(EPS)
EPF/EPFL genes used in the phylogenetic analyses.
(XLSX)
Putative signal peptide and cleavage site of the proteins encoded by genes in the EPF1-EPF2-EPFL7 clade and the EPFL9/Stomagen clade as predicted by the SignalP 4.0.
(XLSX)
Predicted EPF/EPFL genes in land plants. EPF/EPFL genes were retrieved from genomic databases for Arabidopsis thaliana (The Arabidopsis Information Resource, TAIR), Oryza sativa (Rice Annotation Project Database), Picea glauca (The Gene Index Project) and Carica papaya, Medicago truncatula, Populus trichocarpa, Physcomitrella patens, Sorghum bicolor and Selaginella moellendorffii (Phytozome v8.0).
(TXT)
Funding Statement
This work was principally supported by the JST Advanced Low Carbon Technology Research and Development Program (ALCA) from the Japan Science and Technology Agency (JST) (to NT, TT and MK). This work was supported in part by JST-SENTAN (to SO and MM). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vatén A, Bergmann DC (2012) Mechanisms of stomatal development: an evolutionary view. Evodevo 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Edwards D, Kerp H, Hass H (1998) Stomata in early land plants: an anatomical and ecophysiological approach. J Exp Bot 49: 255–278. [Google Scholar]
- 3. McElwain JC, Chaloner WG (1995) Stomatal density and index of fossil plants track atmospheric carbon dioxide in the Palaeozoic. Ann Bot 76: 389–395. [Google Scholar]
- 4. Beerling DJ, Osborne CP, Chaloner WG (2001) Evolution of leaf-form in land plants linked to atmospheric CO2 decline in the Late Palaeozoic. Nature 410: 352–354. [DOI] [PubMed] [Google Scholar]
- 5. Torii KU (2012) Mix-and-match: ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci 17: 711–719. [DOI] [PubMed] [Google Scholar]
- 6. Ohki S, Takeuchi M, Mori M (2011) The NMR structure of stomagen reveals the basis of stomatal density regulation by plant peptide hormones. Nat Commun 2: 512. [DOI] [PubMed] [Google Scholar]
- 7. Hara K, Kajita R, Torii KU, Bergmann DC, Kakimoto T (2007) The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev 21: 1720–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hara K, Yokoo T, Kajita R, Onishi T, Yahata S, et al. (2009) Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol 50: 1019–1031. [DOI] [PubMed] [Google Scholar]
- 9. Hunt L, Gray JE (2009) The signaling peptide EPF2 controls asymmetric cell divisions during stomatal development. Curr Biol 19: 864–869. [DOI] [PubMed] [Google Scholar]
- 10. Sugano SS, Shimada T, Imai Y, Okawa K, Tamai A, et al. (2010) Stomagen positively regulates stomatal density in Arabidopsis . Nature 463: 241–244. [DOI] [PubMed] [Google Scholar]
- 11. Lee JS, Kuroha T, Hnilova M, Khatayevich D, Kanaoka MM, et al. (2012) Direct interaction of ligand-receptor pairs specifying stomatal patterning. Genes Dev 26: 126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paton JA, Pearce JV (1957) The occurrence, structure and functions of the stomata in British bryophytes. Trans Brit Bryol Soc 3: 228–259. [Google Scholar]
- 13. Lehti-Shiu MD, Zou C, Hanada K, Shiu SH (2009) Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol 150: 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peterson KM, Rychel AL, Torii KU (2010) Out of the mouths of plants: the molecular basis of the evolution and diversity of stomatal development. Plant Cell 22: 296–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rychel AL, Peterson KM, Torii KU (2010) Plant twitter: ligands under 140 amino acids enforcing stomatal patterning. J Plant Res 123: 275–280. [DOI] [PubMed] [Google Scholar]
- 16. Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, et al. (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40: 1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, et al. (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Solovyev VV, Salamov AA, Lawrence CB (1994) Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res 22: 5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight-matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282. [DOI] [PubMed] [Google Scholar]
- 21. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fiser A, Sali A (2003) Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374: 461–491. [DOI] [PubMed] [Google Scholar]
- 23. Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. Mol Graph 14: 51–55. [DOI] [PubMed] [Google Scholar]
- 24. Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4: 435–447. [DOI] [PubMed] [Google Scholar]
- 25. Berensdsen HJC, Grigera JR, Straatma TP (1987) The missing term in effective pair potentials. J Phys Chem 91: 6269–6271. [Google Scholar]
- 26. Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65: 712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103: 8577–8593. [Google Scholar]
- 28. Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) LINCS: A linear constraint solver for molecular simulations. J Comp Chem 18: 1463–1472. [Google Scholar]
- 29. Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins 23: 566–579. [DOI] [PubMed] [Google Scholar]
- 30. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8: 785–786. [DOI] [PubMed] [Google Scholar]
- 31. Joo H, Chavan AG, Day R, Lennox KP, Sukhanov P, et al. (2011) Near-native protein loop sampling using nonparametric density estimation accommodating sparcity. PLoS Comput Biol 7: e1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Duché D, Izard J, González-Mañas JM, Parker MW, Crest M, et al. (1996) Membrane topology of the colicin A pore-forming domain analyzed by disulfide bond engineering. J Biol Chem 271: 15401–15406. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of EPF/EPFL genes. The phylogenetic tree was reconstructed by the NJ method from the numbers of amino acid substitutions estimated by the JTT model. The numerals at the branch indicate bootstrap values calculated by the NJ method with 1,000 replications. Bootstrap values >50% are shown.
(EPS)
Molecular properties of P. glauca EPF2. (A) Ribbon model and (B) MD simulation of PgEPF2.
(EPS)
EPF/EPFL genes used in the phylogenetic analyses.
(XLSX)
Putative signal peptide and cleavage site of the proteins encoded by genes in the EPF1-EPF2-EPFL7 clade and the EPFL9/Stomagen clade as predicted by the SignalP 4.0.
(XLSX)
Predicted EPF/EPFL genes in land plants. EPF/EPFL genes were retrieved from genomic databases for Arabidopsis thaliana (The Arabidopsis Information Resource, TAIR), Oryza sativa (Rice Annotation Project Database), Picea glauca (The Gene Index Project) and Carica papaya, Medicago truncatula, Populus trichocarpa, Physcomitrella patens, Sorghum bicolor and Selaginella moellendorffii (Phytozome v8.0).
(TXT)