Abstract
RtcB is an atypical RNA ligase that joins either 2′,3′-cyclic phosphate or 3′-phosphate termini to 5′-hydroxyl termini. In contrast to typical RNA ligases, which rely on ATP and Mg(II), catalysis by RtcB is dependent on GTP and Mn(II) with ligation proceeding through a covalent RtcB–histidine–GMP intermediate. Here, we present three structures of Pyrococcus horikoshii RtcB complexes that capture snapshots along the entire guanylylation pathway. These structures show that prior to binding GTP, a single manganese ion (Mn1) is bound to RtcB. To capture the step immediately preceding RtcB guanylylation, we solved a structure of RtcB in complex with Mn(II) and the unreactive GTP analogue guanosine 5′-(α-thio)-triphosphate (GTPαS). This structure shows that Mn1 is poised to stabilize the pentavalent transition state of guanylylation while a second manganese ion (Mn2) is coordinated to a nonbridging oxygen of the γ-phosphoryl group. The pyrophosphate leaving group of GTPαS is oriented apically to His404 with the ε nitrogen poised for in-line attack on the α phosphorus atom. The structure of RtcB in complex with GTPαS also reveals the network of hydrogen bonds that recognize GTP and illuminates the significant conformational changes that accompany the binding of this cofactor. Finally, a structure of the enzymic histidine–GMP intermediate depicts the end of the guanylylation pathway. The ensuing molecular description of the RtcB guanylylation pathway shows that RtcB and classical ATP/Mg(II)-dependent nucleic acid ligases have converged upon a similar two-metal mechanism for formation of the nucleotidylated enzyme intermediate.
RNA ligases catalyze the formation of a phosphodiester bond between RNA termini that are generated by specific endonucleases during tRNA splicing, the unfolded protein response and the antiphage response.1–4 These endonucleases generate 2′,3′-cyclic phosphate and 5′-OH termini upon cleavage.5, 6 Classical ATP-dependent RNA ligases in bacteria, fungi, and plants are components of multienzyme pathways that repair RNAs with 2′,3′-cyclic phosphate and 5′-OH ends.7, 8 Before ligation, the 2′,3′-cyclic phosphate is hydrolyzed to a 3′-OH by a phosphodiesterase and the 5′-OH is phosphorylated by a polynucleotide kinase to generate a 5′-phosphate (5′-P). Classical ligases then catalyze the ATP/Mg(II)-dependent joining of 5′-P and 3′-OH termini.1
The noncanonical RNA ligase RtcB catalyzes an unprecedented reaction: joining 2′,3′-cyclic phosphate and 5′-OH RNA termini.9–18 RtcB is an essential enzyme for the maturation of tRNAs in metazoa13 and possibly archaea,11 and shares no sequence or structural similarity19 with canonical nucleic acid ligases. In marked contrast to classical ligases, RtcB relies on GTP/Mn(II) for catalysis. Ligation proceeds through three nucleotidyl transfer steps, with 2′,3′-cyclic phosphate termini being hydrolyzed to 3′-P termini in a step that precedes 3′-P activation with GMP (Figure 1A).14, 16, 17 In the first nucleotidyl transfer step, RtcB reacts with GTP to form a covalent RtcB–histidine–GMP intermediate and release PPi; in the second step, the GMP moiety is transferred to the RNA 3′-P; in the third step, the 5′-OH from the other RNA strand attacks the activated 3′-P to form a phosphodiester bond and release GMP. Thus, a high-energy phosphoanhydride of GTP activates a 3′-P for intermolecular attack by a 5′-OH. Here, we provide insight into the chemical mechanism of an unusual nucleotidyl transfer reaction in this sequence—Mn(II)-dependent histidine guanylylation.
Figure 1.
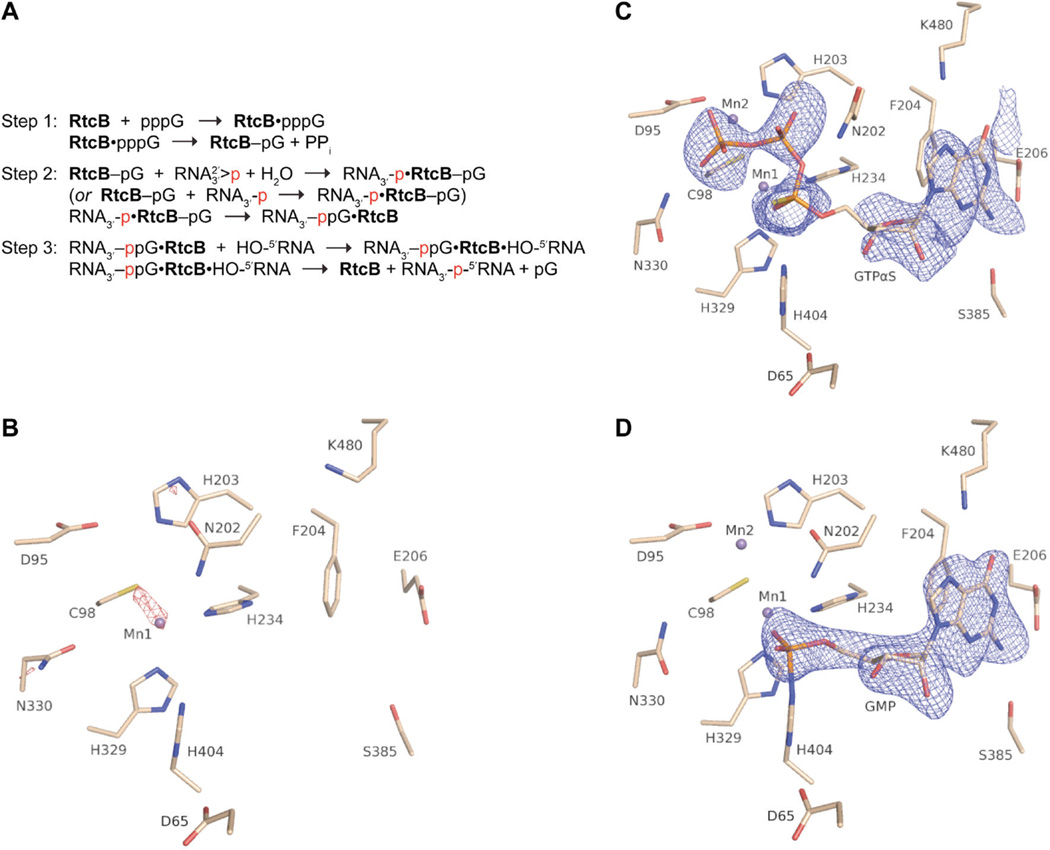
RtcB stepwise ligation reaction pathway and electron density maps of the RtcB active sites. (A) RtcB joins RNA in three nucleotidyl transfer steps, with 2′,3′-cyclic phosphate termini being hydrolyzed in a step preceding RNA 3′-P activation. The three nucleotidyl transfer steps are: (1) RtcB guanylylation, (2) RNA 3′-P activation, and (3) phosphodiester bond formation. The RNA 3′-P at the ligation junction is indicated in red to ease visualization of its incorporation into the newly generated 3′,5′-phosphodiester bond. (B) The RtcB/Mn(II) complex with red mesh representing a composite simulated annealing omit map contoured at 5.6σ to show the presence of one Mn(II) ion. (C) The RtcB/GTPαS/Mn(II) complex with blue mesh denoting an omit map (Fo – Fc) of the GTPαS density contoured at 2.6σ. (D) The RtcB–GMP/Mn(II) complex with blue mesh denoting an omit map (Fo – Fc) of the GMP density contoured at 3.5σ.
We sought to elucidate the entire pathway of RtcB guanylylation by determining the three-dimensional structures of key intermediates at atomic resolution. We present three structures of Pyrococcus horikoshii RtcB complexes: (i) a structure with bound Mn(II) represents the intermediate that precedes binding of GTP, (ii) a structure with bound Mn(II) and an unreactive GTP analogue, guanosine 5′-(α-thio)-triphosphate (GTPαS), captures the reaction step immediately preceding formation of the covalent enzyme intermediate, and (iii) a structure of the covalent RtcB–histidine–GMP intermediate depicts the end product of the guanylylation pathway. Our results show that RtcB coordinates a single Mn(II) ion prior to binding GTP and that GTP binds to RtcB in a complex with a second Mn(II) ion. This two-manganese mechanism of RtcB guanylylation is analogous to the two-magnesium mechanism of adenylylation used by canonical ATP-dependent nucleic acid ligases.20, 21
MATERIALS AND METHODS
RtcB Purification
A previously described plasmid expressing P. horikoshii RtcB was used except that the sequence encoding the hexa-histidine tag was removed via mutagenesis.15 Native P. horikoshii RtcB was expressed in BL21 cells by growing in Terrific Broth at 37 °C to an OD600 of 0.6, inducing with IPTG (0.5 mM) and continuing growth for 3 h. Cells were harvested by centrifugation and resuspended at 8 mL per gram of wet pellet in buffer A (50 mM MES– NaOH, pH 5.6, 45 mM NaCl and 1 mM Cleland’s reagent). Cells were lysed by passage through a cell disruptor (Constant Systems) at 20,000 psi and the lysate was clarified by centrifugation at 20,000g for 1h. Bacterial proteins were precipitated and removed by incubating the lysate at 70 °C for 25 min followed by centrifugation at 20,000g for 20 min. The clarified lysate was then loaded onto a 5 mL HiTrap HP SP cation-exchange column (GE Lifesciences). The column was washed with 25 mL buffer A, and RtcB was eluted with a NaCl gradient of buffer A (45 mM– 1.0 M) over 20 column volumes. Fractions containing RtcB were dialyzed against 4 L of buffer A overnight at 4 °C. Dialyzed RtcB was then loaded onto a 5 mL HiTrap heparin column (GE Lifesciences), and purified RtcB was eluted as described for the cation-exchange chromatography step. Purified RtcB was dialyzed against 4 L of buffer (10 mM HEPES–NaOH, pH 7.5, 200 mM NaCl) overnight at 4 °C.
14C-labeled GTP Binding Assays
Binding assays were performed in 250 µL of 50 mM HEPES buffer, pH 7.5, containing NaCl (200 mM), P. horikoshii RtcB (100 µM), various concentrations of MnCl2, and [8–14C]GTP (Moravek Biochemicals, Brea, CA).22 After incubation, free GTP was removed by applying the reaction to three 5-mL HiTrap desalting columns (GE Lifesciences) connected in series. The desalting columns were equilibrated with elution buffer (50 mM HEPES, pH 7.5, 200 mM NaCl), and protein was eluted in 0.5-mL fractions. Absorbance readings at A260 and A280 were obtained for each fraction. The protein fractions have high A280 readings, whereas the fractions with free GTP have higher A260 readings. In fractions containing protein, the RtcB concentrations were calculated from the A280 reading using an extinction coefficient of 62,340 M−1cm−1 (ExPASy). The concentration of [8–14C]GTP in the protein fractions was determined by liquid scintillation counting. Each 0.5-mL fraction was mixed with 3.5 mL of Ultima Gold MV liquid scintillation cocktail (Perkin Elmer) in a 4-mL vial, and counts were read on a MicroBeta TriLux liquid scintillation counter (Perkin Elmer). The concentration of GTP in each fraction was determined by comparing the counts per minute (cpm) in these samples to the cpm values obtained from standards of known concentration. Optimal formation of the RtcB–GMP complex was found to occur in reaction mixtures that included 1 mM GTP and 2 mM MnCl2. The optimal incubation conditions were found to be at 70 °C for 45 min. Under these conditions, the GTP:RtcB molar ratio was determined to be (0.76 ± 0.02):1. No binding of GTP to RtcB was detected in the absence of Mn(II).
RtcB Crystallization
RtcB was concentrated to 200 µM (11 mg/mL) by ultrafiltration using a spin concentrator (5,000 MWCO, Amicon) and passed through a 0.2-µm filter. To prepare the RtcB/Mn(II) complex, MnCl2 (1 mM) was added to the concentrated protein. For preparation of the RtcB/GTPαS/Mn(II) complex, MnCl2 (2 mM) and a 1:1 mixture of RP and SP diastereomers of GTPαS (1 mM) was added to the concentrated protein, and the resulting solution was incubated at 70 °C for 15 min. For preparation of the RtcB–GMP/Mn(II) complex, the covalent intermediate was formed as described above, and the solution was subjected to gel-filtration chromatography on a Superdex 16/60 column (GE Lifesciences) to remove PPi and excess MnCl2 and GTP. Each of the protein complexes was flash-frozen in liquid nitrogen and stored at −80 °C. Protein samples were crystallized using the hanging drop vapor diffusion method. Crystals were grown by mixing 1 µL of sample solution with 1 µL of reservoir solution. The RtcB/Mn(II) and RtcB/GTPαS/Mn(II) complexes were crystallized using identical reservoir solutions consisting of Bis–Tris (0.1 M, pH 5.5) and ammonium sulfate (2.1 M), the RtcB– GMP/Mn(II) complex used HEPES–NaOH (0.1 M, pH 7) and ammonium sulfate (2 M). Trays were incubated at 20 °C and crystals appeared within one week. Crystals were harvested and cryoprotected in reservoir solution containing sucrose (20% w/v) and cryopreserved in liquid nitrogen.
Data Collection, Structure Determination and Refinement
X-ray diffraction data were collected at 100 K at the Life Science Collaborative Access Team at the Advanced Photon Source at Argonne National Laboratory. Datasets were indexed and scaled using HKL2000.23 The apo-RtcB structure19 was used as a starting model and the structures were completed using alternating rounds of manual model building using COOT24 and refinement with phenix.refine.25 Structure quality was assessed by MolProbity26 and figures were generated using PyMOL.27 The GMP in the RtcB–GMP structure was fitted into the difference density and refined using phosphoramidate bond distance and angle values derived from the small-molecule X-ray crystal structure of 1-carboxymethyl-2-imino-3-phosphonoimidazolidine.28 Omit maps were calculated using Phenix.
RESULTS
A Structure with Mn(II) Represents the Intermediate that Precedes GTP Binding
For crystallization of the RtcB/Mn(II) complex, MnCl2 (1 mM) was added to the concentrated protein solution (200 µM) before crystallization. Crystals of this complex diffracted to a resolution of 2.34 Å, and the apo-RtcB structure19 was used as a starting model for refinement (Table S1 of the Supporting Information). The omit density map shows a single Mn(II) ion (Mn1) in a tetrahedral coordination complex with three amino acid residues (Cys98, His234, and His329) and a water molecule (Figure S1A of the Supporting Information and Figures 1B and 2A). Sulfate ions present in the structure substitute for putative RNA phosphoryl group-binding sites.
Figure 2.
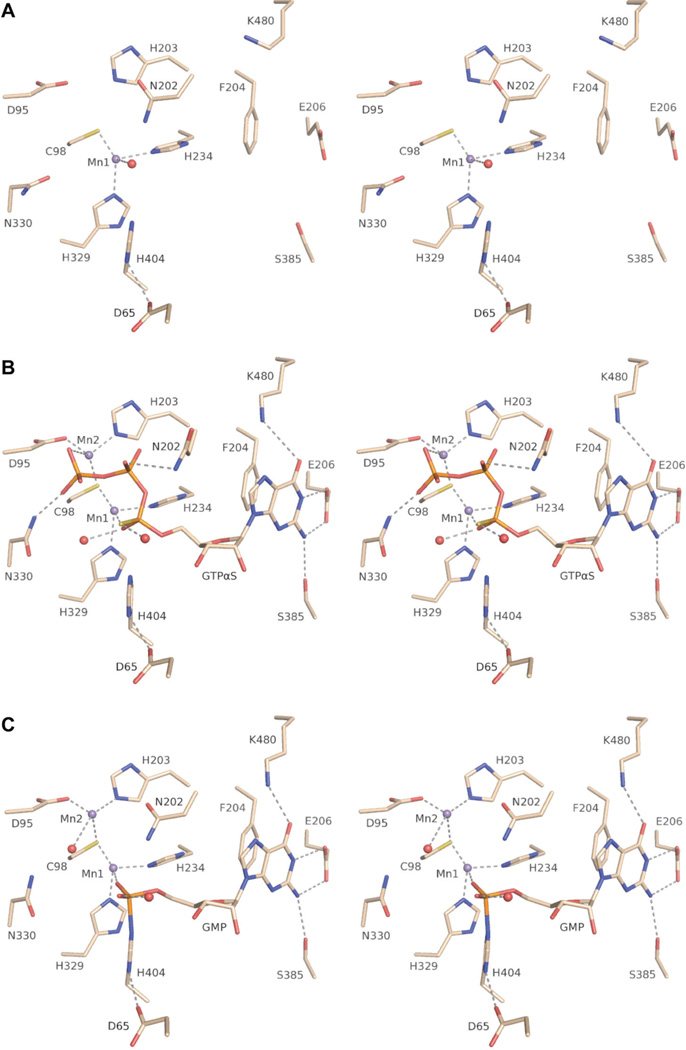
Wall-eyed stereo views of the active sites of RtcB complexes (subunit A). (A) The RtcB/Mn(II) complex (PDB ID 4isj). (B) The RtcB/GTPαS/Mn(II) complex (PDB ID 4isz). GTPαS is modeled as the RP diastereomer. (C) The RtcB–GMP/Mn(II) covalent intermediate (PDB ID 4it0). Hydrogen bonding to the guanosine ribose 2′-OH and 3′-OH with the backbone amides of Ala406 and Gly407, respectively, is not shown.
An earlier crystal structure of RtcB indicated the presence of two manganese ions in the active site, the second manganese ion being coordinated to three amino acid ligands (Asp95, Cys98 and His203) and inorganic sulfate.18 The intracellular concentration of Mn(II) is low (~0.1 mM),29 and its inclusion at 1 mM in our study (5× the RtcB concentration) should saturate the relevant coordination sites within the enzyme in the absence of GTP.
A Structure with GTPαS Captures the Step Immediately Preceding RtcB Guanylylation
We reported previously that RtcB is unreactive when the GTP analogues guanosine 5′-(α-thio)-triphosphate (GTPαS) and guanosine 5′-(α,β-methyleno)-triphosphate (GpCpp) are used as cofactors in ligation reactions, as the bond between the α- and β-phosphoryl groups is recalcitrant to cleavage by RtcB.15 We chose to pursue structural studies with GTPαS because the modification of a nonbridging oxygen causes minimal perturbation to the phosphoryl groups. To obtain the RtcB/GTPαS/Mn(II) complex, MnCl2 (2 mM) and GTPαS (1 mM) were added to a concentrated solution of protein (200 µM), and the resulting solution was incubated at 70 °C for 15 min to facilitate any conformational changes in the hyperthermophilic enzyme that are necessary for cofactor binding. The RtcB/GTPαS/Mn(II) complex was subsequently maintained at temperatures which disallow formation of the covalent intermediate. Crystals of this complex diffracted to an effective resolution of 2.45 Å (I/σI = 2); however, all data to 2.3 Å was used in refinement (Table S1 of the Supporting Information).
The omit density map of the RtcB/GTPαS/Mn(II) complex indicated the presence of GTPαS and two manganese ions in the RtcB active site (Figure S1B of the Supporting Information and Figures 1C and 2B). Mn1 remains in tetrahedral coordination geometry with ligands that include the same three amino acid residues as the structure with manganese only; however, the water molecule has been replaced with the nonbridging α-thiophosphate oxygen of GTPαS. A second manganese ion (Mn2) is in tetrahedral coordination geometry with ligands that include a nonbridging oxygen of the γ-phosphoryl group of GTPαS, as well as three amino acid residues (Asp95, Cys98, and His203). The β-phosphoryl group of GTPαS is oriented apically to His404, and its Nε is poised for in-line attack on the α-phosphoryl group.30 Furthermore, H–Nδ of His404 forms a hydrogen bond with Oδ1 of Asp65, which is strictly conserved and appears to orient and activate Nε for attack. A main-chain H–N forms a hydrogen bond with Oδ2 of Asp65, stabilizing an anti orientation of the carboxylate in the His⋯Asp dyad.
The presence of a cysteine residue bridging two manganese ions in the RtcB active site is unique. The Mn1⋯S and Mn2⋯S coordination distances in the RtcB/GTPαS/Mn(II) complex are 2.3 Å and 2.4 Å, respectively, and the Mn1⋯S⋯Mn2 angle is 100°. Thus, the two Mn(II) ions are separated by only 3.6 Å. This distance is similar to the 3.3 Å distance separating the Mn(II) ions of the renowned binuclear manganese cluster in the active site of arginase, where the Mn(II) ions are bridged by two aspartate residues.31 The manganese ion coordination distances in the RtcB/GTPαS/Mn(II) complex are listed in Table 1.
Table 1.
Manganese coordination distance in the RtcB/GTP?S/Mn(II) complex.
| Mn⋯ligand | r (Å) |
|---|---|
| Mn1⋯Sγ (Cys98) | 2.3 |
| Mn1⋯Nε (His234) | 2.4 |
| Mn1⋯Nε (His329) | 2.4 |
| Mn1⋯OPα (GTP?S) | 2.2 |
| Mn2⋯Oδ2 (Asp95) | 2.1 |
| Mn2⋯Sγ (Cys98) | 2.4 |
| Mn2⋯Nε (His203) | 2.3 |
| Mn2⋯OPγ (GTP?S) | 2.8 |
RtcB is the only known enzyme catalyzing nucleotidyl transfer that requires a NTP/Mn(II) complex rather than a NTP/Mg(II) complex as a cofactor. Indeed, RtcB is not active with Mg(II),12, 14 which is much more abundant than Mn(II) in both cells and the environment. The structure of the RtcB/GTPαS/Mn(II) complex provides an explanation for this unusual requirement. First, the ligands of the two bound Mn(II) ions have a tetrahedral geometry, which is disfavored by Mg(II). Second, the side chain of a cysteine residue interacts with both Mn(II) ions, which are more thiophilic than Mg(II) ions. This essential cysteine residue is strictly conserved throughout evolution and likely serves as a gatekeeper that selects for Mn(II) in each metal binding site. Third, Coulombic repulsion deters the close placement of two Mg(II) ions, which have a high charge density. Indeed, the two Mg(II) ions used by T4 RNA ligase are separated by 7.4 Å,21 a distance that is twofold greater than that of the Mn(II) ions in RtcB. The two Mg(II) ions in the active site of xylose isomerase, which are bridged by a glutamate carboxylate, have a shorter internuclear distance of 5.1 Å.32 More polarizable Mn(II) ions, however, can be accommodated in even closer proximity.
An intricate array of hydrogen bonds explains the specificity and high affinity for GTP. The triphosphate moiety forms hydrogen bonds with H–Nδ of two asparagine residues. Asn202 has now adopted a different conformation, and its H–Nδ forms a hydrogen bond with the β-phosphoryl group. Likewise, H–Nδ of Asn330 forms a hydrogen bond with the γ-phosphoryl group. The guanosine nucleoside is bound in an anti conformation with the guanine base stacked on Phe204 and with Tyr451 forming an edge of the guanine-binding pocket. Each carboxylate oxygen of Glu206 forms a hydrogen bond with guanine, one with H–N1 and the other with H– N2; Ser385 also interacts with the H–N2, while H–Nε of Lys480 forms a hydrogen bond with O6. The guanosine ribose 2′- and 3′-oxygens form hydrogen bonds with the main-chain H–N of Ala406 and Gly407, respectively.
The binding of GTPαS elicits significant conformational changes in the RtcB active site (Figure 3A). The loop that is displaced by the guanine base has a maximal Cα displacement of 2.5 Å at Ser380. In addition, the loop containing Ala406 and Gly407 changes conformation around the ribose 2′-OH and 3′-OH with a maximal Cα displacement of 1.4 Å at Ala406.
Figure 3.
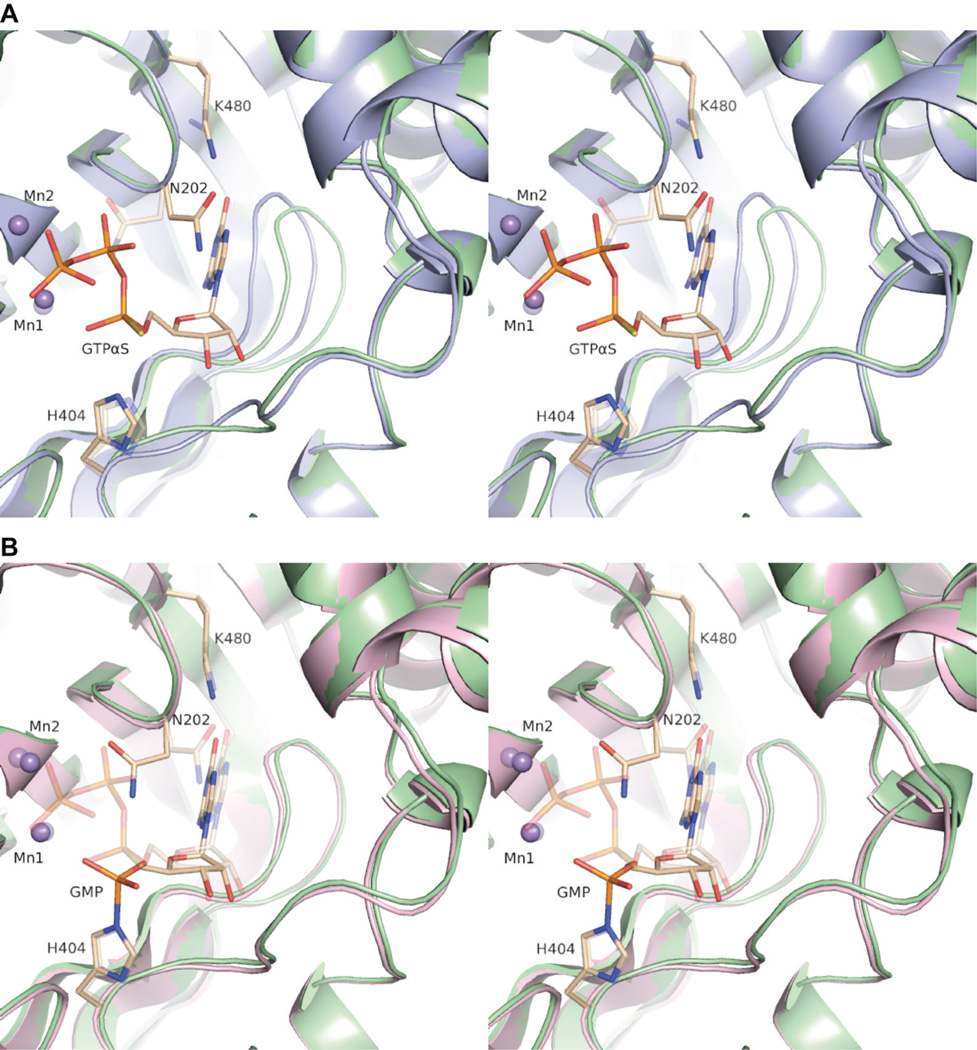
Wall-eyed stereo views of conformational changes in the RtcB active site along the guanylylation pathway. (A) Pairwise superpositions of the RtcB/Mn(II) (light blue) and RtcB/GTPαS/Mn(II) (light green) complexes; (B) pairwise superpositions of the RtcB/GTPαS/Mn(II) (light green) and RtcB–GMP/Mn(II) (light pink) complexes. Upon binding of GTPαS, two active-site loops make significant conformational rearrangements, Lys480 changes conformation to form hydrogen bonds with the guanine C6 carbonyl, and Asn202 changes conformation to accommodate the triphosphate moiety. After formation of the covalent intermediate and release of PPi, Asn202 again changes conformation to a position directly above the nascent phosphoramidate linkage. The amino acids, Mn(II) ions, and nucleotide from the antecedent step in each depiction are rendered transparent.
Structure of the RtcB–Histidine–GMP Covalent Intermediate
To determine the optimal reaction conditions that enable formation of the RtcB–GMP covalent intermediate, 14C-labeled GTP binding studies were performed.22 The optimal reaction conditions were found to contain purified RtcB (100 µM), GTP (1 mM), and MnCl2 (2 mM), with incubation at 70 °C for 45 min. Under these conditions, the maximal GMP:RtcB molar ratio was determined to be (0.76 ± 0.02):1. No binding of GTP to RtcB was detected in the absence of Mn(II). Using these reaction conditions, we formed the RtcB–GMP intermediate and removed unbound Mn(II), GTP, and PPi by gel-filtration chromatography. The protein was concentrated to 200 µM, and crystals of this complex diffracted to a resolution of 2.4 Å (Table S1 of the Supporting Information).
The omit density map indicated the presence of a covalent histidine–GMP and two manganese ions in the RtcB subunit A active site (Figure S1C of the Supporting Information and Figure 1D and 2C). The GMP density present in subunit B was too weak to model confidently. The guanine and ribose interactions are essentially identical to those in the RtcB/GTPαS/Mn(II) complex. Asn202, however, has shifted to a position near the nascent phosphoramidate bond (Figure 3B). The labile histidine–GMP33 is stabilized by coordination of one nonbridging oxygen of the GMP to Mn1 and the formation of a hydrogen bond of the other nonbridging oxygen with a water molecule. The phosphoimidazolium form of His404 is stabilized by a hydrogen bond from its H–Nδ to the carboxylate side chain of Asp65. Mn2 remains bound in tetrahedral coordination geometry; however, the metal contact to the γ-phosphoryl group has been replaced with a water molecule. Alanine-scanning mutagenesis of GMP-interacting residues revealed their importance for RNA-ligation activity, consistent with a previous report18 (Figures S2 and S3 of the Supporting Information).
DISCUSSION
RtcB Guanylylation Mechanism
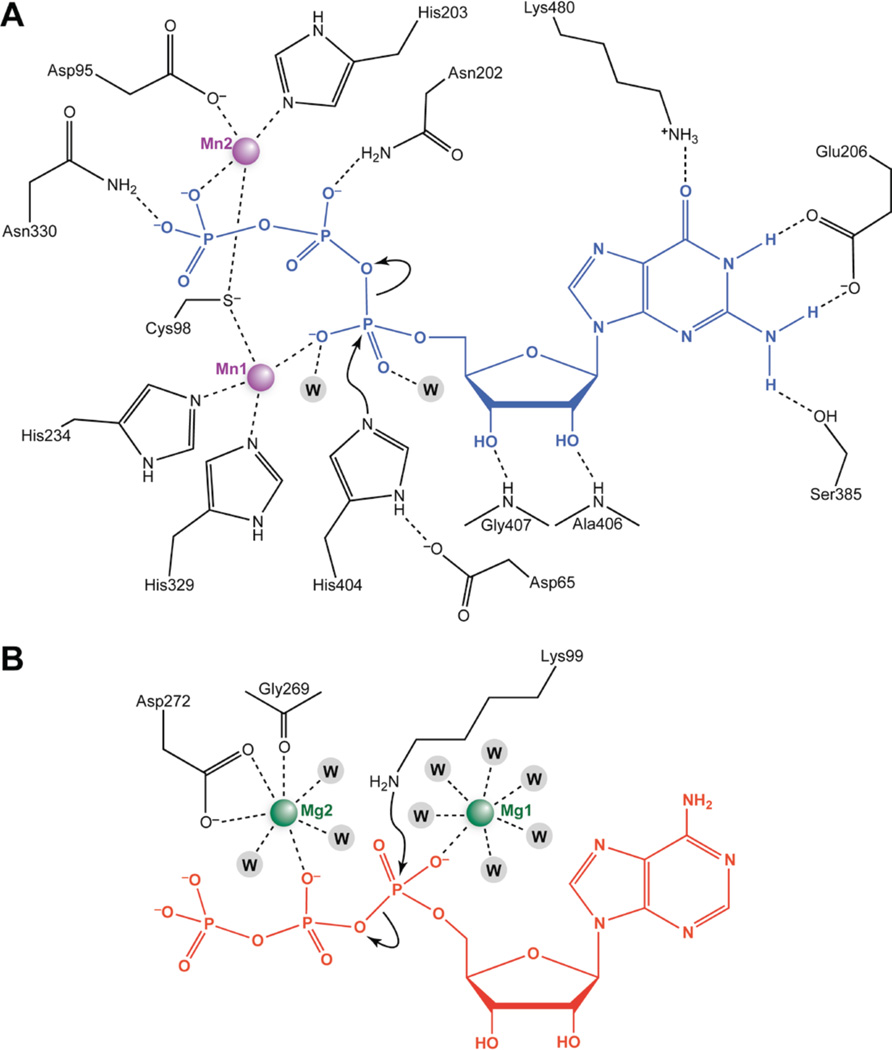
Histidine guanylylation is expected to proceed through an associative mechanism with the accumulation of negative charge on the nonbridging oxygens of the α-phosphoryl group in the pentavalent transition state.30 In the RtcB active site, guanylylation is promoted by neutralization of this negative charge by coordination to Mn1 and hydrogen bonds with water molecules. The PPi leaving group of GTP is oriented apically to Nε of His404 by coordination to Mn2. This orientation allows for in-line attack by Nε. The formation of a hydrogen bond between H–Nδ of His404 and the side-chain carboxylate of Asp65 orients Nε for attack on the α-phosphorus atom of GTP and stabilizes the phosphoimidazolium group in the ensuing intermediate. Similar hydrogen bonds are common features of other enzymes that are known to proceed through a phosphorylated/nucleotidylated histidine intermediate.34–38 In RtcB, the H–Nδ proton also serves to make the side chain of His404 into a much better leaving group during the subsequent step in which the GMP moiety is transferred to an RNA 3′-P (Figure 1A).
Our structural characterizations show that RtcB and classical ATP-dependent nucleic acid ligases share a similar metal-assisted mechanism for formation of the nucleotidylated enzyme intermediate (Figure 4), despite using a different metal ion. A structure of T4 RNA ligase bound to the ATP analogue adenosine 5′-(α,β-methyleno)-triphosphate (ApCpp) is consistent with a mechanism analogous to the one put forth here for RtcB guanylylation.21 In the T4 RNA ligase structure, a calcium ion is bound in place of one magnesium ion (Mg1) and coordinates to a nonbridging oxygen of the ApCpp α-phosphonate and a magnesium ion (Mg2) coordinates to the β-phosphonate group. Mg1 in the T4 ligase structure and Mn1 in the RtcB structure both promote enzyme nucleotidylation by neutralizing the negative charge on the α-phosphoryl group in the pentavalent transition state. The second metal ion observed in both T4 ligase and RtcB enters the active site in a coordination complex with the NTP cofactor and promotes catalysis by orienting the PPi leaving group and neutralizing the charge on the phosphoryl groups. Despite the absence of sequence or structural similarity between RtcB and classical ATP-dependent nucleic acid ligases, Nature has converged on analogous two-metal dependent nucleotidylation mechanisms.
Figure 4.
Proposed mechanism of RtcB guanylylation and comparison to the adenylylation mechanism of classical ATP-dependent ligases. (A) RtcB active site. Potential hydrogen bonds and metal contacts are indicated with dashed lines. Structure–function analyses suggest a mechanism in which nucleophilic attack by Nε of His404 on the α-phosphorus of GTP is promoted by: (i) orientation and stabilization of His404 by a hydrogen bond with H–Nδ of Asp65; (ii) orientation of the PPi leaving group apical to Nε of His404 by Mn2; and (iii) charge neutralization of the pentavalent transition state by Mn1 and two water molecules. (B) The two-metal mechanism of adenylylation used by T4 RNA ligase based on a structure in complex with magnesium ions and the non-hydrolyzable ATP analogue ApCpp (PDB ID 2c5u). Hydrogen bonds with ApCpp are not shown, and only the inner sphere ligands of Mg1 and Mg2 are shown. In the crystal structure, the Mg1 site was occupied by a Ca(II) ion, which was present at high concentration in the crystallization solution. Mg1 and Mg2 serve essentially identical mechanistic roles to Mn1 and Mn2, respectively, in the RtcB guanylylation mechanism.
The structures presented here resolve key issues about RtcB guanylylation. In particular, the structure of the RtcB/GTPαS/Mn(II) complex has revealed the orientation of the bound triphosphate cofactor and the role of Mn2. The consequent molecular description of the two-metal RtcB guanylylation mechanism contrasts with one put forth previously,18 which depicted an incorrect orientation for its β- and γ-phosphoryl groups.
All known nucleotidyl transferases require complexation of a nucleotide triphosphate cofactor to a metal ion. The roles of the metal ion in the NTP/metal complex include orientating the phosphoryl groups, neutralizing their charge, and enhancing their reactivity.39 Accordingly, many enzymes that catalyze the cleavage of a NTP α–β phosphoanhydride bond employ two Mg(II) ions, one that coordinates to the high-affinity site between the β- and γ-phosphoryl groups and a second that coordinates to the α-phosphoryl group.39–43 We have revealed how an enzyme can, instead, employ two Mn(II) ions in analogous roles to catalyze nucleotidyl transfer.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Prof. Aaron A. Hoskins for helpful discussions.
Funding
This work was supported in part by National Institutes of Health Grants F32 GM100681 (to K.K.D); Protein Structure Initiative grants U01 GM098248 (to G.N.P.) and U54 GM074901 (Center for Eukaryotic Structural Genomics); and R01 CA073808 (to R.T.R). The Life Sciences Collaborative Access Team has been supported by Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under contract W-31-102-ENG-38.
Footnotes
Supporting Information
Active site electron density (2Fo – Fc) of refined models (Figure S1); structure-guided mutagenesis of the guanylate-binding pocket (Figures S2 and S3); and crystallographic data collection and refinement statistics (Table S1). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Shuman S, Lima CD. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Abelson J, Trotta CR, Li H. tRNA splicing. J. Biol. Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 3.Levitz R, Chapman D, Amitsur M, Green R, Snyder L, Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J. 1990;9:1383–1389. doi: 10.1002/j.1460-2075.1990.tb08253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 5.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell. Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cuchillo CM, Nogués MV, Raines RT. Bovine pancreatic ribonuclease: Fifty years of the first enzymatic reaction mechanism. Biochemistry. 2011;33:7408–7414. doi: 10.1021/bi201075b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amitsur M, Levitz R, Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwer B, Sawaya R, Ho CK, Shuman S. Portability and fidelity of RNA-repair systems. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins KK, Furneaux H, Hurwitz J. Isolation and characterization of an RNA ligase from HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 1985;82:684–688. doi: 10.1073/pnas.82.3.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Filipowicz W, Shatkin AJ. Origin of splice junction phosphate in tRNAs processed by HeLa cell extract. Cell. 1983;32:547–557. doi: 10.1016/0092-8674(83)90474-9. [DOI] [PubMed] [Google Scholar]
- 11.Englert M, Sheppard K, Aslanian A, Yates JR, III, and Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka N, Shuman S. RtcB is the RNA ligase component of an Escherichia coli, RNA repair operon. J. Biol. Chem. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lührmann R, Söll D, Martinez J. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2010;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka N, Chakravarty AK, Maughan B, Shuman S. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011;286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai KK, Raines RT. tRNA ligase catalyzes the GTP-dependent ligation of RNA with 3′-phosphate and 5′-hydroxyl termini. Biochemistry. 2012;51:1333–1335. doi: 10.1021/bi201921a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakravarty AK, Subbotin R, Chait BT, Shuman S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakravarty AK, Shuman S. The sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTPdependent. Nucleic Acids Res. 2012;40:8558–85567. doi: 10.1093/nar/gks558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Englert M, Xia S, Okada C, Nakamura A, Tanavde V, Yao M, Eom SH, Konigsberg WH, Söll D, Wang J. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 2012;109:15235–15240. doi: 10.1073/pnas.1213795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada C, Maegawa Y, Yao M, Tanaka I. Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii, reveals a novel fold. Proteins. 2006;63:1119–1122. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- 20.Cherepanov AV, de Vries S. Kinetic mechanism of the Mg2+-dependent nucleotidyl transfer catalyzed by T4 DNA and RNA ligases. J. Biol. Chem. 2002;277:1695–1704. doi: 10.1074/jbc.M109616200. [DOI] [PubMed] [Google Scholar]
- 21.El Omari K, Ren J, Bird LE, Bona MK, Klarmann G, LeGrice SF, Stammers DK. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 22.Morar M, Anand R, Hoskins AA, Stubbe J, Ealick SE. Complexed structures of formylglycinamide ribonucleotide amidotransferase from Thermotoga maritima, describe a novel ATP binding protein superfamily. Biochemistry. 2006;45:14880–14895. doi: 10.1021/bi061591u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 28.Phillips GNJr, Thomas JWJr, Annesley TM, Quiocho FA. Stereospecificity of creatine kinase. Crystal structure of 1-carboxymethyl-2-imino-3-phosphonoimidazolidine. J. Am. Chem. Soc. 1979;101:7120–7121. [Google Scholar]
- 29.Finney LA, O’Halloran TV. Transition metal speciation in the cell: Insights from the chemistry of metal ion receptors. Science. 2003;300:931–936. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 30.Lassila JK, Zalatan JG, Herschlag D. Biological phosphoryl-transfer reactions: Understanding mechanism and catalysis. Annu. Rev. Biochem. 2011;80:669–702. doi: 10.1146/annurev-biochem-060409-092741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanyo ZF, Scolnick CR, Ash DE, Christianson DW. Structure of a unique binuclear manganese cluster in arginase. Nature. 1996;383:554–557. doi: 10.1038/383554a0. [DOI] [PubMed] [Google Scholar]
- 32.Lavie A, Allen KN, Petsko GA, Ringe D. X-ray crystallographic structures of D-xylose isomerase-substrate complexes position the substrate and provide evidence for metal movement during catalysis. Biochemistry. 1994;33:5469–5480. doi: 10.1021/bi00184a016. [DOI] [PubMed] [Google Scholar]
- 33.Hultquist DE. The preparation and characterization of phosphorylated derivatives of histidine. Biochim. Biophys. Acta. 1968;153:329–340. doi: 10.1016/0005-2728(68)90078-9. [DOI] [PubMed] [Google Scholar]
- 34.Puttick J, Baker EN, Delbaere LTJ. Histidine phosphorylation in biological systems. Biochim. Biophys. Acta. 2008;1784:100–105. doi: 10.1016/j.bbapap.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Wedekind JE, Frey PA, Rayment I. The structure of nucleotidylated histidine-166 of galactose-1-uridylyltransferase provides insight into phosphoryl group transfer. Biochemistry. 1996;35:11560–11569. doi: 10.1021/bi9612677. [DOI] [PubMed] [Google Scholar]
- 36.Moréra S, Chiadmi M, LeBras G, Lascu I, Janin J. Mechanism of phosphate transfer by nucleoside diphosphatekinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila, and Dictyostelium . Biochemistry. 1995;34:11062–11070. [PubMed] [Google Scholar]
- 37.Lima CD, Klein MG, Hendrickson WA. Structure-based analysis of catalysis and substrate definition in the HIT protein family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 38.Fraser ME, James MNG, Bridger WA, Wolodko WT. A detailed structural description of Escherichia coli, succinyl-CoA-synthetase. J. Mol. Biol. 1999;285:1633–1653. doi: 10.1006/jmbi.1998.2324. [DOI] [PubMed] [Google Scholar]
- 39.Cleland WW, Hengge AC. Enzymatic mechanisms of phosphate and sulfate transfer. Chem. Rev. 2006;106:3252–3278. doi: 10.1021/cr050287o. [DOI] [PubMed] [Google Scholar]
- 40.Tesmer JJ, Sunahara RK, Johnson RA, Gosselin G, Gilman AG, Sprang SR. Two-metal-ion catalysis in adenylyl cyclase. Science. 1999;285:756–760. doi: 10.1126/science.285.5428.756. [DOI] [PubMed] [Google Scholar]
- 41.Zheng J, Knighton DR, ten Eyck LF, Karlsson R, Xuong N, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 42.Steitz TA. DNA polymerases: structural diversity and common mechanisms. J. Biol. Chem. 1999;274:17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 43.Hyde SJ, Eckenroth BE, Smith BA, Eberley WA, Heintz NH, Jackman JE, Doublié S. tRNAHis guanylyltransferase (THG1), a unique 3′-5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′-3′ DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20305–20310. doi: 10.1073/pnas.1010436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.