Abstract
OBJECTIVE
The increased risk of thrombosis in systemic lupus erythematosus (SLE) may be partially explained by interrelated genetic pathways for thrombosis and SLE. In a case-control analysis, we investigated whether 33 established and novel single nucleotide polymorphisms (SNP) in 20 genes involved in hemostasis pathways that have been associated with deep venous thrombosis in the general population were risk factors for SLE development among Asians.
METHODS
Patients in the discovery cohort were enrolled in one of two North American SLE cohorts. Patients in the replication cohort were enrolled in one of four Asian or two North American cohorts. SLE cases met American College of Rheumatology classification criteria. We first genotyped 263 Asian SLE and 357 healthy Asian control individuals for 33 SNPs using Luminex multiplex technology in the discovery phase, and then used Taqman and Immunochip assays to examine 5 SNPs in up to an additional 1496 cases and 993 controls in the Replication phase. SLE patients were compared to healthy controls for association with minor alleles in allelic models. Principal components analysis was used to control for intra-Asian ancestry in an analysis of the replication cohort.
RESULTS
Two genetic variants in the gene VKORC1, rs9934438 and rs9923231, were highly significant in both the discovery and replication cohorts: OR(disc) = 2.45 (p=2×10−9), OR(rep) = 1.53 (p=5×10−6) and OR(disc) = 2.40 (p=6×10−9), OR(rep) = 1.53 (p=5×10−6), respectively. These associations were significant in the replication cohort after adjustment for intra-Asian ancestry: rs9934438 OR(adj) = 1.34 (p=0.0029) and rs9923231 OR(adj) = 1.34 (p=0.0032).
CONCLUSION
Genetic variants in VKORC1, involved in vitamin K reduction and associated with DVT, are associated with SLE development in Asians. These results suggest intersecting genetic pathways for the development of SLE and thrombosis.
Keywords: systemic lupus erythematosus, single nucleotide polymorphisms, genetic risk factors
Systemic lupus erythematosus (SLE) is characterized by autoantibody production, immune complex deposition and systemic inflammation. Manifestations vary from life-threatening nephritis to rash, oral ulcers, and arthritis. Individuals of Asian ancestry have been reported to have a higher prevalence of SLE, more severe disease, and higher cumulative organ damage compared to European Americans [1], yet are understudied compared to subjects of European descent.
Genome-wide association studies (GWAS) in Asians have confirmed some common genes as SLE risk factors in European Americans and Asians. While certain SLE-associated genes have similar risk allele frequencies in various ethnic groups (e.g. STAT4), others demonstrate large allele frequency differences across ancestral and population groups (e.g. PTPN22).
Given that genes may predispose to SLE across ethnicities [2], and the evidence for interrelated pathways between SLE and thrombosis [3, 4], we investigated 33 SNPs implicated in deep venous thrombosis (DVT) risk in the general European American population [5] to see if they confer risk for the development of SLE in Asians.
PATIENTS AND METHODS
Discovery cohort
Discovery cohort SLE subjects (n=263) were derived from the UCSF Lupus Genetics Project (n=230) and the PROFILE cohort (n=33). The UCSF Genetics Project is a multiethnic cohort of patients with SLE. The protocol was approved by the Institutional Review Board (IRB) at the University of California, San Francisco Subjects fulfill American College of Rheumatology (ACR) classification criteria for SLE, complete an extensive questionnaire, give permission for medical record review, and provide a DNA sample. Individuals are recruited from diverse sources including tertiary care and community hospitals and clinics.
Additional SLE patients in the Discovery Cohort were obtained from the PROFILE cohort [6]. Patients meet ACR criteria, are at least 16 years of age, and have disease duration of ≤ 10 years at cohort entry. Phenotype data included age, sex, and ethnicity. Healthy controls (n=357) were derived from UCSF, Celera and the University of Alabama at Birmingham.
Replication cohort
Significant results from the Discovery cohort were tested in our Replication Cohort, which consisted of 1496 SLE cases and 993 controls (except one SNP, rs1801690, typed on 398 cases and 427 controls) of self-reported Asian ancestry, including Chinese, Koreans, and Singaporean-Chinese. Samples were recruited from multiple centers under the approval of the IRB. All SLE cases meet ACR criteria and are at least 16 years of age at diagnosis.
Genotyping and Quality Control Methods
Primary predictors for this case-control study included 33 SNPs suggested to be associated with DVT [5] (Table 2 of [7] and Supplemental Table 1). Established and suggested genetic risk factors for thrombosis in the general population and in SLE in the literature (e.g. MBL) were also genotyped.
Table 2.
VKORC1 in replication cohort with adjustment for ancestry and stratification by ethnic subgroup
| Analysis | SNP | Case/Control | Case | Control | P | OR (95%CI) |
|---|---|---|---|---|---|---|
| All subjects adjusted for PC1, PC2, PC3 | rs9934438 | 1496/993 | 0.136 | 0.093 | 0.0029† | 1.34 (1.11–1.62) |
| rs9923231 | 1496/993 | 0.135 | 0.093 | 0.0032† | 1.34 (1.10–1.62) | |
| Chinese subgroup | rs9934438 | 1022/809 | 0.123 | 0.099 | 0.020‡ | 1.28 (1.04–1.58) |
| rs9923231 | 1022/809 | 0.122 | 0.099 | 0.028‡ | 1.27 (1.03–1.56) | |
| Korean/Japanese subgroup | rs9934438 | 260/67 | 0.102 | 0.052 | 0.076‡ | 2.06 (0.91–4.64) |
| rs9923231 | 260/67 | 0.102 | 0.045 | 0.043‡ | 2.38 (1.00–5.67) | |
| Southeast Asian subgroup | rs9934438 | 112/7 | 0.290 | 0.071 | 0.076‡ | 5.31(0.68–41.46) |
| rs9923231 | 112/7 | 0.290 | 0.071 | 0.076‡ | 5.31 (0.68–41.46) |
By logistic regression with first three principal components derived from Immunochip markers as covariates.
By chi-square test.
For rs6048 and rs2289252, genotyping in the discovery cohort was done by allele-specific real-time PCR using assays designed and validated at Celera [8]. For other SNPs, genotyping was performed using the Luminex multiplex technology in which genotypes were determined automatically by passing the raw Luminex L-100 signal data through an unsupervised classification algorithm. Approximately 96 to 98% of all genotypes were autocalled. A final manual review of the data was performed to assess each assay's technical performance to rescue any aberrant genotype autocalls. Call graphs for our 2 significant SNPs are given in Supplemental Figure 1.
Among the discovery cohort, nine SLE patients were dropped from genotyping due to failure to amplify; 11 patients were dropped because their self-report sex did not match sex typing; and 10 patients and 4 controls were dropped from analysis for missing more than 10% genotyping. Post-QC call rate per SNP was ≥ 98%. No additional subjects were dropped among the PROFILE patients. Deviations from Hardy-Weinberg Equilibrium (HWE) were assessed using an exact test (Supplemental Table 2).
In the replication cohort, SNPs rs9923231, rs9934438, rs1801131 and rs1613662 were typed on the Illumina Immunochip custom platform. A predesigned Taqman SNP assay (Applied Biosystems) was used to genotype rs1801690 in available replication samples. Post-QC call rates and HWE for these 5 SNPs are given in Supplemental Table 2. Outliers (n=95) had been previously removed from the Immunochip samples based on principal components analysis (PCA) performed with 1000 Genomes Asian (CHB, CHD, JPT) samples.
Statistical analysis
SNPs were first tested for SLE association in bivariate allelic analyses (p<0.01), adjusting for cohort. Heterogeneity of odd ratios (OR) was tested across Discovery and Replication cohorts.
To account for potential intra-Asian population stratification among the VKORC1 SNPs, we used approximately 9,000 autosomal SNPs with low LD (r2 < 0.2) in Asians and not associated with SLE (p>0.001) from the Immunochip platform to perform a principal components analysis (PCA). Data for the replication cohort were reanalyzed (Table 2) with PC1, PC2 and PC3 included as covariates to adjust for intra-Asian ancestry. The VKORC1 SNPs were also re-analyzed within reported ethnic subgroups.
Statistical analyses were performed using STATA SE v11, HAPLOVIEW, EIGENSOFT v4.2, and PLINK.
RESULTS
263 SLE cases and 357 controls were studied in the Discovery Cohort. Ninety percent of cases were female, the mean age at diagnosis was 27.5 years (SD 14), and the average disease duration was 9.2 years (SD 7.3). For the Replication Cohort, SLE patients were at least 16 years of age at diagnosis and 50.2% were females. The controls were well-matched for age, gender (61.4% were female), and ethnicity. After QC, 253 cases and 353 controls were analyzed.
Results of case-control analyses are shown in Table 1 for SNPs having p < 0.01 in the discovery cohort. Two of these were confirmed in our replication cohort, in VKORC1 (rs9923231, OR = 1.53, p=5.1×10−6 and rs9934438, OR = 1.54, p=4.3×10−6). These two SNPs were in high LD (r2=0.98). Heterogeneity between the discovery and replication associations was significant (p-values ranging from 0.008 to 0.015 for the 5 SNPs), probably reflecting different intra-Asian ancestry in the cohorts (see Table 2 subgroups); thus we did not combine these results with a meta-analysis.
Table 1.
Genetic Association Results for Asian SLE Patients vs. Control Individuals
| Discovery | Replication | ||||
|---|---|---|---|---|---|
| SNP (minor allele) | Gene | MAF* cases/controls n=253/353 |
SLE Association OR (p) |
MAF* cases/controls n=1496/993† |
SLE Association OR (p) |
| rs9923231 (C) | VKORC1 | 0.25/0.12 | 2.40 (6.1×10−9) | 0.14/0.092 | 1.53 (5.1×10−6) |
| rs9934438 (G) | VKORC1 | 0.26/0.12 | 2.45 (2.4×10−9) | 0.14/0.093 | 1.54 (4.3×10−6) |
| rs1801690 (G) | APOH | 0.036/0.073 | 0.47 (0.0053) | 0.104/0.083 | 1.27 (0.18) |
| rs1801131 (C) | MTHFR | 0.30/0.21 | 1.64 (0.00024) | 0.22/0.21 | 1.06 (0.46) |
| rs1613662 (G) | GP6/RDH13 | 0.055/0.008 | 6.85 (1.07×10−6) | 0.023/0.015 | 1.57 (0.043) |
minor allele frequency
For SNP rs1801690, case/control n=398/427; different subset of replication cohort was used due to sample availability
P-values were determined by chi-square test
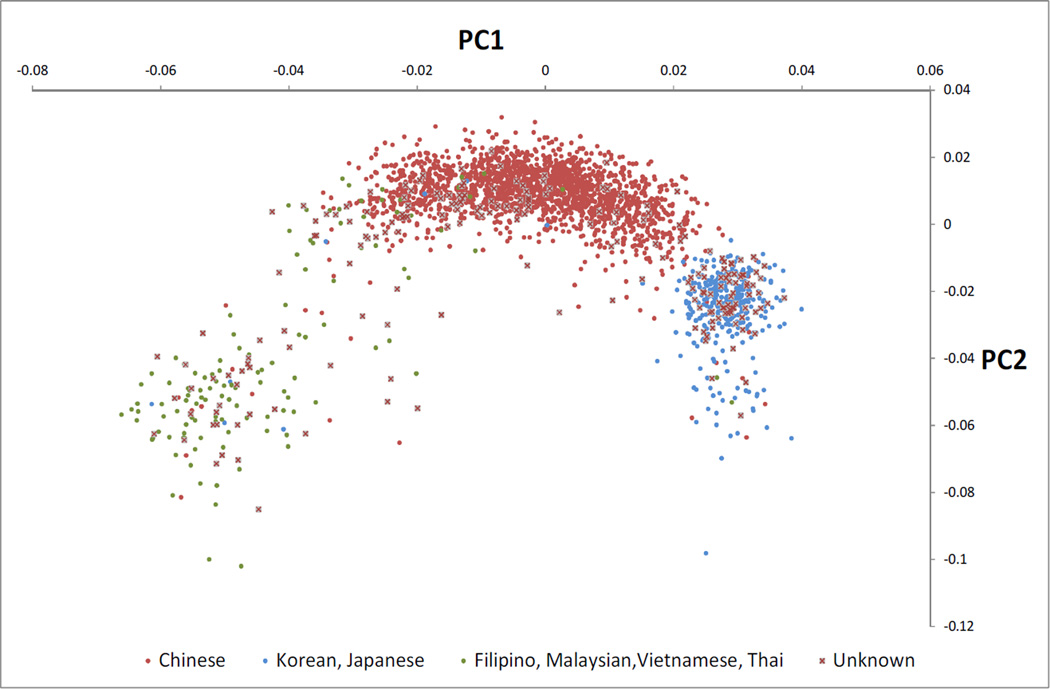
The first 2 PCs from our PCA of the replication cohort are shown in Figure 1, along with annotation for subjects with reported ancestry, demonstrating that these indeed reflect intra-Asian ancestry. Replication results adjusted for the top 3 PCs confirm the VKORC1 associations (Table 2): SNPs rs9934438 and rs9923231 continue to be associated with SLE after adjustment (rs9923231, OR 1.34, p=0.0032 and rs9934438, OR 1.34, p=0.0029). In sensitivity analyses we also tested the VKORC1 associations in reported ethnic subgroups of the replication set (Table 2). Although these analyses have low power, they suggest that the magnitude of association may vary by Asian subpopulation, and may explain the heterogeneity between the discovery and replication cohorts seen in Table 1.
Figure 1. First two principal components of replication cohort typed on the Immunochip*.
* Using approximately 9000 SNPs from Immunochip with r2 < 0.2 in Asians and SLE p > 0.001.
DISCUSSION
These results demonstrate that a genetic risk factor implicated in thrombosis is also a risk factor for SLE susceptibility in Asians. The vitamin K epoxide reductase enzyme complex (VKOR) is involved in the synthesis of vitamin K-dependent coagulation factors, is the target enzyme for warfarin, and is encoded by the gene VKORC1. SNPs in VKORC1 have been linked to reduced efficacy of vitamin K recycling resulting from lower VKOR activity. VKORC1 polymorphisms have primarily been studied in the context of warfarin dosing and a GWAS found that VKORC1 variants are one of the principal genetic determinants of warfarin dose [9]. Recent studies investigating their role in thrombosis in the non-SLE population have not found associations with venous thrombosis, although such an association was observed in one study [10]. However, the majority of these studies were performed in European Americans.
The SNPs in VKORC1 have not been implicated in published GWAS for either Europeans or Asians. We observed in results from one European GWAS [11] and in additional data for our subjects that they were not significantly associated with SLE (unpublished data). This adds to growing evidence that the genetics of SLE varies by ethnicity [2]. These SNPs were interrogated in the 610-Quad used in two prior Asian GWAS, Han et al [12] and Yang et al [13]. In the Han et al study (unpublished data), these SNPs have OR=1.32 (p=0.011), supporting our conclusion. They were not significant in Yang et al (unpublished data) although the effect is in the same direction (OR=1.11, p=0.30); this could be due to heterogeneity of clinical manifestations and related underlying genetic differences, as these SNPs are significant in Yang et al for multiple subphenotypes within Asian SLE subjects. For example, oral ulcers positive/negative has OR=1.80 (p=0.0023) for these SNPs, but is only present in 20.3% of Yang et al subjects versus 24.6% of our discovery subjects and 31.3% of Han et al subjects. Similarly, hematologic disorder positive/negative has OR=1.48 (p=0.026) for these SNPs, but is only present in 52.8% of Yang et al subjects versus 77.0% among our discovery subjects and 78.7% of Han et al subjects. Such heterogeneity may be due in part to ascertainment or sample selection bias in different studies.
Our results provide further evidence for common genetic pathways for SLE development and the complication of thrombosis and may help explain ethnic differences in SLE outcomes. Recent evidence suggests that the antiphospholipid syndrome (APS) and SLE are diseases along the same spectrum that may have common genetic underpinnings. Indeed, this hypothesis has been supported by a recent study involving 341 SLE and 90 APS patients which demonstrated that BANK1 (rs10516487), BLK (rs13277113), and rs10798269 in 1q25.1 were associated with both APS and SLE [3]. Other recent studies found that SLE predisposing SNPs in STAT4 (rs7574865) and BLK (rs2736340) [4] were associated with primary APS.
Asians have the lowest prevalence of venous thrombotic events in the general population [14] and in our SLE cohort (17% vs. 21–24%) compared to other ethnic groups. While Asians have a lower prevalence of thrombosis in general, Chinese patients with SLE are 12 times more prone to thrombosis than in the general Asian population, emphasizing the importance of this complication in this ethnic group [15]. Among Asian American SLE patients in the UCSF cohort, association of DVT risk allele carriers – including VKORC1 – with thrombosis subtypes were inconclusive due to low power in the Asian subgroups [7].
This is the largest study investigating SNPs implicated in thrombosis for risk of SLE and identifies a new genetic risk locus inVKORC1 for SLE susceptibility among Asians, an understudied ethnic subgroup.
Supplementary Material
Acknowledgments
SUPPORT These studies were performed in part in the General Clinical Research Center, Moffitt Hospital, University of California, San Francisco, California USA with funds provided by the National Center for Research Resources, 5 M01 RR-00079, US Public Health Service. This research was supported by: Arthritis Foundation Postdoctoral Fellow Award, an American College of Rheumatology Investigator Award, an Alliance for Lupus Research Target Identification in Lupus award, a Kirkland Scholar Award, the UCSF Rosalind Russell Medical Research Center for Arthritis, and the following NIH grants: P60 AR053308, R01 AR44804 and K24 AR02175. For the PROFILE collaborators, the Hopkins cohort is supported by NIH AR 43727, the Northwestern cohort is supported by K24 AR 002138, P60 2 AR 30692, PO1 AR 49084, and UL 1 RR 025741, the UAB cohort supported by P60 AR048095, P01 AR049084, and 5UL1 RR025777. Dr. Tsao’s work was supported by RO1 AR043814. Dr. Song’s work was supported by the R&D Program of MKE/KEIT (10035615). Dr. Lau and Dr. Yang are supported by Hong Kong General Research Fund (781709, 784611, and 770411).
Footnotes
DISCLOSURES Celera authors (YL, MC, and JC) declare their employment in the company. Other authors have nothing to disclose.
REFERENCES
- 1.Mok MY, Li WL. Do Asian patients have worse lupus? Lupus. 2010;19(12):1384–1390. doi: 10.1177/0961203310375832. [DOI] [PubMed] [Google Scholar]
- 2.Kim I, Kim YJ, Kim K, Kang C, Choi CB, Sung YK, et al. Genetic studies of systemic lupus erythematosus in Asia: where are we now? Genes and immunity. 2009 Jul;10(5):421–432. doi: 10.1038/gene.2009.24. [DOI] [PubMed] [Google Scholar]
- 3.Horita T, Nakagawa H, Oku K, Kataoka H, Yasuda S, Atsumi T, et al. Lupus Susceptible Gene Polymorphisms in patients wtih Antiphospholipid Syndrome. Arthritis and rheumatism. 2009 Oct;60(10) 2009. [Google Scholar]
- 4.Yin H, Borghi MO, Delgado-Vega AM, Tincani A, Meroni PL, Alarcon-Riquelme ME. Association of STAT4 and BLK, but not BANK1 or IRF5, with primary antiphospholipid syndrome. Arthritis and rheumatism. 2009 Aug;60(8):2468–2471. doi: 10.1002/art.24701. [DOI] [PubMed] [Google Scholar]
- 5.Bezemer ID, Bare LA, Doggen CJ, Arellano AR, Tong C, Rowland CM, et al. Gene variants associated with deep vein thrombosis. Jama. 2008 Mar 19;299(11):1306–1314. doi: 10.1001/jama.299.11.1306. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon GS, McGwin G, Jr, Petri M, Reveille JD, Ramsey-Goldman R, Kimberly RP. Baseline characteristics of a multiethnic lupus cohort: PROFILE. Lupus. 2002;11(2):95–101. doi: 10.1191/0961203302lu155oa. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser R, Li Y, Chang M, Catanese J, Begovich AB, Brown EE, et al. Genetic Risk Factors for Thrombosis in Systemic Lupus Erythematosus. The Journal of rheumatology. 2012 Jun 15; doi: 10.3899/jrheum.111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y, Nowotny P, Holmans P, Smemo S, Kauwe JS, Hinrichs AL, et al. Association of late-onset Alzheimer's disease with genetic variation in multiple members of the GAPD gene family. Proceedings of the National Academy of Sciences of the United States of America. 2004 Nov 2;101(44):15688–15693. doi: 10.1073/pnas.0403535101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS genetics. 2009 Mar;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lacut K, Larramendy-Gozalo C, Le Gal G, Duchemin J, Mercier B, Gourhant L, et al. Vitamin K epoxide reductase genetic polymorphism is associated with venous thromboembolism: results from the EDITH Study. J Thromb Haemost. 2007 Oct;5(10):2020–2024. doi: 10.1111/j.1538-7836.2007.02706.x. [DOI] [PubMed] [Google Scholar]
- 11.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. The New England journal of medicine. 2008 Feb 28;358(9):900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 12.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nature genetics. 2009 Nov;41(11):1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 13.Yang W, Shen N, Ye DQ, Liu Q, Zhang Y, Qian XX, et al. Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS genetics. 2010 Feb;6(2):e1000841. doi: 10.1371/journal.pgen.1000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klatsky AL, Armstrong MA, Poggi J. Risk of pulmonary embolism and/or deep venous thrombosis in Asian-Americans. The American journal of cardiology. 2000 Jun 1;85(11):1334–1337. doi: 10.1016/s0002-9149(00)00766-9. [DOI] [PubMed] [Google Scholar]
- 15.Mok CC, Ho LY, Yu KL, To CH. Venous thromboembolism in southern Chinese patients with systemic lupus erythematosus. Clinical rheumatology. Jun;29(6):599–604. doi: 10.1007/s10067-009-1364-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.