Abstract
Cell cycle proteins regulate processes as diverse as cell division and cell death. Recently their role in neuronal death has been reported in several models of neurodegeneration. We have reported previously that two key regulators of the cell cycle, the retinoblastoma susceptibility gene product (pRb) and transcription factor E2F1, exhibit altered immunostaining patterns in simian immunodeficiency virus encephalitis (SIVE). Here we show that E2F1 and the inactivated, hyperphosphorylated form of pRb (ppRb) also exhibit altered immunostaining patterns in human immunodeficiency virus encephalitis (HIVE). Quantification of E2F1 and ppRb staining by immunofluorescent confocal microscopy confirms a significant increase in E2F1 and ppRb in both HIVE and the simian model. This increase in E2F1 and ppRb staining correlates with an increase in the presence of activated macrophages, suggesting a link between changes in cell cycle proteins and the presence of activated macrophages. Changes in ppRb and E2F1 staining in SIVE also correlate with alterations in E2F/DNA binding complexes present in the nuclear and cytoplasmic fractions from both midfrontal cortex and basal ganglia. These findings suggest that changes in cell cycle proteins occur in both HIVE and the simian model and that these changes have functional implications for gene expression in neural cells under encephalitic conditions mediated by macrophage activation or infiltration.
Keywords: HIV, SIV, transcription, cell cycle, neurodegeneration, brain, encephalitis, neuron, phosphorylation, retinoblastoma, E2F
At autopsy, ∼25% of human immunodeficiency virus (HIV)-infected individuals exhibit brain macrophage infiltrates and multinucleated giant cells, pathologic hallmarks of HIV encephalitis (HIVE) (Bacellar et al., 1994; Maehlen et al., 1995; Davies et al., 1997; Masliah et al., 2000). A variable percentage of SIV-infected macaques also develop encephalitis exhibiting similar pathologic changes in the CNS (Sharer et al., 1991; Baskin et al., 1992; Joag et al., 1995; Czub et al., 1996;Mankowski et al., 1997; Zink et al., 1997; Strelow et al., 1998;Westmoreland et al., 1998). In both of these lentiviral encephalitides, severe neuronal damage occurs despite an absence of significant neuronal infection (Budka, 1991; Budka et al., 1991; Sharer et al., 1991; Baskin et al., 1992; Czub et al., 1996). The pathogenesis of neurodegeneration in HIV and simian immunodeficiency virus encephalitis (SIVE) is not completely understood but has been associated with abundant lentiviral-infected and activated brain macrophages (Achim and Wiley, 1992, 1996; Baskin et al., 1992; Masliah et al., 1992; Power et al., 1993; Tyor et al., 1993; Achim et al., 1994; Wiley and Achim, 1994; Glass et al., 1995; Mankowski et al., 1997; Zink et al., 1997;Wiley et al., 1998, 1999). Activated macrophages have been reported to secrete molecules that induce synaptic damage and/or neuronal death directly or indirectly through stimulation of surrounding glia (Price et al., 1988; Lipton et al., 1991; Lo et al., 1992; Brouwers et al., 1993; Gelbard et al., 1994; Pulliam et al., 1994, 1996; Crowe, 1995;Giulian et al., 1996; Heyes et al., 1998; Power et al., 1998).
The presence of activated and lentiviral-infected CNS macrophages has been tightly associated with the presence of aberrant cell-signaling molecules (e.g., cytokines, chemokines, neurotrophic factors) (Price et al., 1988; Lipton et al., 1991; Lo et al., 1992; Brouwers et al., 1993;Gelbard et al., 1994; Pulliam et al., 1994, 1996; Crowe, 1995; Giulian et al., 1996; Heyes et al., 1998; Power et al., 1998). Because some of these molecules are capable of initiating cell cycle in non-neuronal cells, we hypothesized that the aberrant CNS milieu may lead to inappropriate expression of cell cycle proteins in terminally differentiated neuronal elements resulting in their chronic damage and eventual death.
Signaling molecules secreted by HIV-infected macrophages can stimulate various responses, including proliferation, differentiation, or death. These processes are at least partially controlled by the activities of cell cycle regulatory proteins. Two important regulators of cell cycle that have been implicated in neuronal death are the retinoblastoma susceptibility gene product (pRb) and E2F1 (for review, see LaThangue, 1994; Adams and Kaelin, 1995; Kouzarides, 1995; Ross, 1996; Whyte, 1996; Scherr, 1998; Black and Azizkhan-Clifford, 1999). The E2F1 transcription factor is regulated by direct interaction with pRb (Chellappan et al., 1991, 1992). Hyperphosphorylation of pRb abolishes this interaction. The pRb-liberated E2F/DNA complex activates expression of genes needed for entry into S-phase of the cell cycle (Nevins, 1992; Farnham et al., 1993; LaThangue, 1994; Scherr, 1998). E2F1 and pRb have also been shown to participate in several models of neuronal cell death (Park et al., 1997, 2000; Giovanni et al., 1999,2000). Treatment of cultured rat cortical neurons with β amyloid causes neuronal death requiring phosphorylation of pRb (Giovanni et al., 1999). Excess E2F1 can bypass the need for phosphorylation of pRb, indicating that it is also necessary for neuronal death (Giovanni et al., 2000). In vivo, deletion of E2F1 is necessary and sufficient to rescue severe neuronal loss induced by pRb deletion (Pan et al., 1998; Tsai et al., 1998). These data support a role for pRb-liberated E2F1 in neuronal death.
Because cell cycle proteins participate in neuronal death and respond to stimuli present in HIVE, we proposed to test the hypothesis that cell cycle proteins are activated in the CNS during this disease. Our previous evidence suggested that E2F1 and hyperphosphorylated pRb (ppRb) exhibited altered subcellular localization in neuronal cultures treated with signaling molecules and in SIVE (Jordan-Sciutto et al., 2000, 2001). Here we show that both E2F1 and hyperphosphorylated pRb exhibit altered staining patterns in HIVE as compared with controls. Returning to the SIVE model, we find that the abundance of E2F1 and ppRb immunostaining correlates with the presence of activated macrophages. DNA binding assays using protein extracts from SIV and SIVE tissue demonstrate alterations in E2F DNA binding activity in SIVE compared with control extracts. These findings support the hypothesis that neural cells respond to infected and activated macrophages by altering cell cycle protein localization and activity.
MATERIALS AND METHODS
Human subjects. Brain tissue was obtained from HIV-infected patients at autopsy from the University of California San Diego brain bank. HIV encephalitis was defined by the presence of histopathologic hallmarks of the disease and high viral load in the brain as determined by branched DNA quantification for HIV viral production and the presence of viral proteins (Wiley et al., 1998). Using these criteria, seven cases were defined as HIVE and six as HIV without encephalitis. From these cases, tissues from three brain regions, the midfrontal cortex, the hippocampus, and the basal ganglia, were stained. In the hippocampus, special attention was paid to the CA1 region because these neurons are particularly vulnerable to injury.
Animal model. Rhesus macaques were housed and maintained according to strict Association for Assessment and Accreditation of Laboratory Animal Care standards. Macaques were derived from vaccine trials, infected with SIVdeltaB670, and killed at variable times after infection. SIVE was empirically defined as the presence of abundant perivascular mononuclear infiltrate and microglial nodules. Multinucleated giant cells were present in some lesions of all SIVE cases. A difference between the simian model of disease and the human disease is that the simian brains are harvested before end-stage disease either by immune deficiency, as in the human cases in the HIV group, or by encephalitis, as in the human cases in the HIVE group.
Immunohistochemistry and immunofluorescence. Paraffin-embedded sections were heated to 50°C for 20 min, and deparaffinized in Histoclear (15 min, three times) (National Diagnostics, Atlanta, GA). Sections were rehydrated as follows: 100% alcohol for 10 min, two times; 95% alcohol for 10 min; 90% alcohol for 10 min; 70% alcohol for 10 min; H2O for 5 min. Endogenous peroxidase activity was inactivated by immersing in 3% H2O2 for 30 min. Antigen unmasking was performed by placing slides in target retrieval solution (Dako, Carpinteria, CA) at 95°C for 1 hr. After gradual cooling to room temperature, tissue sections were blocked with 10% normal goat serum in PBS. Antibodies to E2F1, pRb (Santa Cruz Biotechnology, Santa Cruz, CA), and ppRb (New England Biolabs, Beverly, MA) previously characterized for immunohistochemistry (Silverstein et al., 1995; Guy et al., 1996; Riley et al., 1996) were used at commercially recommended and empirically defined dilutions (Table1) for detection by the tyramide amplification system (New England Biolabs) (Jordan-Sciutto et al., 2000). For immunofluorescent studies, antibodies to MAP2, GFAP, HAM-56, human leukocyte antigen (HLA)-DR, HIV p24, and SIV gp110 (Table1) were used without amplification. DNA staining was visualized by propidium iodide staining. RNA was removed by 30 min incubation with RNase A at 10 μg/ml. Propidium iodide was used at 10 μg/ml and incubated on the slides for 10 min. This abbreviated time was used to minimize “bleed through” of the propidium iodide staining into the FITC and Cy5 channels. Immunofluorescent slides were mounted in gelvatol (Ausubel et al., 1994) and analyzed by laser confocal microscopy (Molecular Dynamics, Sunnyvale, CA), as described previously (Soontornniyomkij et al., 1998). All images shown in the figures were captured with uniform threshold settings.
Table 1.
Antibody dilutions used for immunohistochemistry and immunoblot
| Antigen | Antibody type | IHC dilution | TSA | Source |
|---|---|---|---|---|
| E2F1 | Mouse monoclonal | 1:100 | Yes | Santa Cruz Biotechnology (Santa Cruz, CA) |
| pRb | Mouse monoclonal | 1:100 | Yes | Santa Cruz Biotechnology |
| p53 | Mouse monoclonal | 1:100 | Yes | Santa Cruz Biotechnology |
| ppRb | Rabbit polyclonal | 1:100 | Yes | New England Biolabs, Inc. (Beverly, MA) |
| GFAP | Rabbit polyclonal | 1:500 | No | Dako (Carpinteria, CA) |
| HAM-56 | Mouse monoclonal | 1:100 | No | Dako |
| MAP-2 | Mouse monoclonal | 1:500 | No | Sternberger Monoclonals Inc. (Lutherville, MD) |
| HIV p24 | Mouse monoclonal | 1:10 | No | Dako |
| HLA-DR | Mouse monoclonal | 1:1000 | No | Dako |
| SIV gp110 | Rabbit polyclonal | 1:236 | No | Gift from Kelly Stefano Cole (University of Pittsburgh, Pittsburgh, PA) |
Information on the antibodies used for immunohistochemistry (IHC) is listed. Also indicated are the type and source of each antibody used and whether tyramide amplification (TSA) was used.
Immunocytochemical staining was performed using an amino ethyl carabazole detection system (Biogenex, San Ramon, CA). Nucleic acids were labeled with hematoxylin as a counter stain, and slides were mounted in crystal mount.
Quantification by laser confocal microscopy. Consecutive sections were stained with hematoxylin and eosin (H and E) or immunohistochemically stained as described above. A dissecting microscope was used to identify anatomical regions on the H and E-stained section. Three regions of the SIV-infected macaque brains containing putamen, caudate, and midfrontal cortical gray matter were identified and marked on the H and E-stained slides. For HIVE, midfrontal cortical gray matter, midfrontal cortical white matter, putamen, and hippocampus were delineated on the H and E slides. The marked H and E tissue section was matched with the consecutive, immunostained tissue section, and the marked regions were traced on the immunofluorescent section. Immunohistochemically stained sections containing regions of interest were analyzed by laser confocal microscopy (Molecular Dynamics). This instrument is equipped with a Nikon inverted microscope with Plan-Apo 20× 0.75 numerical aperture (NA) (air), 40× 1.00 NA (oil), and 60× 1.40 NA (oil) objective lenses. The illumination is provided by an argon/krypton laser with 488, 568, and 647 nm primary emission lines. Each image was scanned along the z-axis, and the middle sectional plane was found (262,144 pixels per plane, 1 pixel = 0.25 μm2). Images were collected on a Silicon Graphics Inc. computer (Operating System Release 5.3, Farmington, MI) and analyzed using the Image Space software (Version 3.2, Molecular Dynamics).
Each brain region from every macaque was randomly scanned in 10 microscopic areas. The specimen was first scanned for FITC signals. Subsequently, the specimen was rescanned for Cy5 signal in the same sectional plane. All specimens were scanned at the same photomultiplier tube settings for each wavelength. A threshold was used to reduce contribution of autofluorescence to the pixel counts. For each channel, the number of pixels per image with an intensity that exceeded the threshold was counted using the same collection parameters. Each area that was scanned encompasses an area of 67,600 μm2. For quantification of E2F1, pRb, ppRb, SIV-gp110, or HAM56, the pixel count was obtained for 10 areas within a brain region for three macaques with encephalitis and three macaques without encephalitis. For HIV and HIVE cases, E2F1, ppRb, HIV p24, or HLA-DR pixels were quantified in 10 areas for each brain region of five cases.
To determine differences in total fluorescence, positive pixel areas were multiplied by the average pixel fluorescence to give a measurement of total fluorescence. When this analysis was done, the results were comparable to the area measurement, and therefore we present the data as positive pixel area per image.
Statistical analysis. Pixel counts for pRb, ppRb, or E2F1 were correlated either with the pixel number for a macrophage marker or a viral marker relevant to the species being used. Correlations are represented by Pearson's correlation coefficient, r. As a correlation becomes stronger, r approaches the absolute value of 1. We defined a strong correlation as r ≥ 0.8 and a moderate correlation as r ≥ 0.6. The statistical significance (p value) of the correlation coefficient was determined for each value using the correlation coefficient and the number of observations (α value). This value represents the probability that we would obtain the observed correlation coefficient by chance based on the number of observations.
Protein extracts. Protein extracts were prepared from basal ganglia and frontal cortex of three uninfected control monkeys, two SIV-infected monkeys, and three SIV-infected, encephalitic monkeys as described previously (Haas et al., 1993). Tissues were homogenized on ice in PBS with protease inhibitors (5 mm PMSF, 2 μg/ml pepstatin A, and 1 μg/ml leupeptin) until there were no large chunks. Separated cells were collected by centrifugation at 3000 rpm for 5 min at 4°C. Supernatants were removed to a separate tube, and pellets were resuspended in 4 vol of hypotonic buffer (20 mm HEPES, pH 7.9, 1.5 mmMgCl2, 10 mm KCl, 0.5 mm DTT, 0.5 mm PMSF, 2 μg/ml pepstatin A, and 1 μg/ml leupeptin). The suspension was homogenized for 10 sec and incubated on ice for 15 min. The cells were collected by centrifugation at 13,000 × g for 30 min. The supernatant was labeled “S1,” and the pellet was further extracted with high salt buffer (0.42 m NaCl, 20 mm HEPES, pH 7.9, 1.5 mmMgCl2, 0.2 mm EDTA, 0.5 mm DTT, 25% glycerol, 0.5 mm PMSF, 2 μg/ml pepstatin A, and 1 μg/ml leupeptin) on ice for 20 min. Residual insoluble material was removed by centrifugation at 14,000 × g for 30 min. The supernatant fraction was collected and termed “S2.” Protein concentrations for each fraction were determined by Bio-Rad protein assay.
Electrophoretic mobility shift assay. Oligonucleotides containing the E2F DNA consensus site were end labeled with32P. Protein extracted from frozen macaque brain tissue (24.5 μg) was incubated with 0.5 ng of32P-labeled E2F probe in 20 μl of electrophoretic mobility shift assay (EMSA) buffer (20 mm HEPES, pH 7.9, 20% glycerol, 200 mm KCl, 0.2 mm EDTA, 0.2 mm PMSF, 0.5 mm DTT) for 20 min at room temperature (Ausubel et al., 1994; Jordan et al., 1994). Included in the reaction was 0.5 μg of salmon sperm DNA as a nonspecific competitor before addition of labeled probe to reduce nonspecific DNA/protein interactions. For competition reactions, unlabeled competitive molecules were preincubated with the protein for 5 min on ice before addition of labeled probe. The reaction mixture was loaded onto a pre-run 4% nondenaturing polyacrylamide gel (Ausubel et al., 1994; Jordan et al., 1994) and electrophoresed at 100 V in a low ionic-strength buffer (6.8 mm Tris, pH 8.0, 1 mm EDTA, pH 8.0, and 3.3 mmNaOAc). The buffer was recirculated every 45 min. After 1.5–2 hr, the polyacrylamide gel was removed from the apparatus, dried, and exposed to autoradiography film. Exposure time varied depending on the abundance of E2F protein complexes present in the extract. The double-stranded oligonucleotides containing an E2F site were taken from the c-myc promoter (Hall, 1990). These sites were as follows: 5′-AATTCGCTTGGCGGGAAAAC-3′ and 3′-GCGAACCGCCCTTTTGTTAA-5′.
RESULTS
E2F1 and ppRb exhibited altered staining patterns in HIV encephalitis
To determine whether our observations in SIVE reflected similar processes occurring in HIVE, we compared staining patterns for E2F1 and ppRb in six patients diagnosed with HIVE and six HIV-infected patients with no encephalitic pathology as controls. In both sets of patients, we surveyed immunohistochemical staining patterns in three brain regions: the basal ganglia, the hippocampus, and the midfrontal cortex. Of the six HIV-positive control patients, only one exhibited any positive staining for E2F1 or ppRb (Table2). Most of the HIVE cases exhibited abundant staining for E2F1 and ppRb in all three brain regions examined (Table 2). These results suggest that there is increased staining for both E2F1 and ppRb in HIVE consistent with our observations in the simian model. The HIV-infected, non-encephalitic autopsy case that stained for E2F1 and ppRb also exhibited abundant type II Alzheimer's astrocytes, a condition associated with elevated serum ammonia levels. For this study, such a case may not be appropriate for use as a control.
Table 2.
E2F1 and ppRb immunostaining was abundant in HIVE brains but not in non-encephalitic brains
| Midfrontal cortex | Basal ganglia | Hippocampus | ||||
|---|---|---|---|---|---|---|
| HIV | HIVE | HIV | HIVE | HIV | HIVE | |
| E2F1 | 1/6 | 6/7 | 1/6 | 4/6 | 1/6 | 5/6 |
| ppRb | 1/6 | 6/7 | 1/6 | 5/6 | 1/6 | 6/6 |
Tissue sections from midfrontal cortex, basal ganglia, and hippocampus of six to seven human autopsy cases were immunostained for E2F1 or ppRb. Each column contains the number of E2F1 and ppRb immunopositive sections of the total number of sections analyzed.
In HIV encephalitis, increased E2F1 staining was observed predominantly in neuronal cytoplasm and not in astrocytes
We wanted to define the cell types exhibiting increased E2F1 staining in HIVE cases. Using triple-label immunofluorescent confocal microscopy, we assessed E2F1 localization in astrocytes (GFAP) and neurons (MAP2). Distribution to the nucleus or cytoplasm was determined by colocalization with nuclei stained with propidium iodide, a DNA intercalating agent. In HIV control cases, the neurons, astrocytes, and oligodendrocytes did not stain appreciably for E2F1 (Fig.1A–C,G–I). In the basal ganglia of HIVE autopsy tissues, some E2F1 (green) localized to the nuclei (red) of cells that did not colabel with GFAP (blue) (Fig.1D) but did label with MAP2 and thus were identified as neurons (Fig. 1J). In all three brain regions examined, E2F1 was observed in neuronal cytoplasm of HIVE cases (Fig.1D–F,J–L). In the white matter of the midfrontal cortex, E2F1 localized to the nuclei of oligodendrocytes in HIVE but not HIV cases (data not shown). Although there was some variability between different brain regions, overall E2F1 staining was more abundant in HIVE and localized predominantly to neurons in gray matter and to oligodendrocytes in white matter.
Fig. 1.

In HIVE, increased E2F1 staining was observed predominantly in neuronal cytoplasm and not astrocytes.A–F, Triple-label immunofluorescent confocal microscopy for E2F1 (green), nuclear DNA (propidium idodide; red), and astrocytes labeled with GFAP (blue). In HIV autopsies without encephalitis (A–C), minimal E2F1 staining is observed. In HIV autopsies with encephalitis (D–F), abundant E2F1 staining is observed but does not localize to astrocytes.G–L, Triple-label immunofluorescent confocal microscopy for E2F1 (green), nuclear DNA (propidium iodide; red), and MAP2 (blue). In HIV autopsies without encephalitis (G–I), minimal E2F1 staining is observed. In HIV autopsies with encephalitis (J–L), increased E2F1 staining is observed in neuronal cytoplasm [colocalization of blue (MAP2) andgreen (E2F1) staining appears aqua]. Thecolumns contain images from the following brain regions:Basal Ganglia, Hippocampus, and midfrontal cortex (Frontal Cortex). DNA staining may not be seen in all nuclei because of the staining protocol used. In these cases, MAP2 and GFAP, as cytoskeletal components, are used to delineate the cytoplasm. All micrographs are the same magnification. Scale bar, 20 μm (for all images).
ppRb localized predominantly to nuclei of neurons in HIV encephalitis
Specific cell types containing ppRb in HIVE were also identified using triple-label immunofluorescent laser confocal microscopy. In all three brain regions of HIV-positive control cases that were investigated, there was little or no detectable ppRb staining (Fig.2A–C,G–I). In the basal ganglia of encephalitic patients, ppRb localized to the nuclei of MAP2-positive neurons but not GFAP-positive astrocytes (Fig.2J,D). Staining for ppRb in HIVE hippocampus was both nuclear and cytoplasmic in neurons and nuclear in occasional astrocytes (Fig. 2K,E). Although occasional astrocytic nuclei were also positive for ppRb staining in the gray matter of HIVE midfrontal cortex, staining for ppRb was observed predominantly in neuronal nuclei (Fig.2F,L). In the white matter from midfrontal cortex, ppRb localized to the nuclei of oligodendrocytes (data not shown). Overall, ppRb staining was more abundant in HIVE and localized predominantly to the nuclei of neurons in gray matter.
Fig. 2.

ppRb localized predominantly to nuclei of neurons in HIVE. A–F, Triple-label immunofluorescent confocal microscopy for ppRb (green), nuclear DNA (propidium idodide;red), and astrocytes labeled with GFAP (blue). In HIV autopsies without encephalitis (A–C), minimal ppRb staining was observed. In HIV autopsies with encephalitis (D–F), abundant ppRb staining was observed but only occasionally localized to GFAP-positive astrocytes. G–L, Triple-label immunofluorescent confocal microscopy for ppRb (green), nuclear DNA (propidium iodide;red), and MAP2 (blue). In HIV autopsies without encephalitis (G–I), minimal ppRb staining was observed. In HIV autopsies with encephalitis (J–L), increased ppRb staining was observed in neuronal cytoplasm and nuclei [colocalization of blue(MAP2) and green (E2F1) staining appearsaqua, and colocalization between green(E2F1) and red (DNA) appears yellow]. Columns contain images from the following brain regions: Basal Ganglia, Hippocampus, and midfrontal cortex (Frontal Cortex). DNA staining may not be seen in all nuclei because of the staining protocol used. In these cases, MAP2 and GFAP, as cytoskeletal components, are used to delineate the cytoplasm. All micrographs are the same magnification. Scale bar, 20 μm (for all images).
E2F1 and ppRb exhibited increased staining in both SIV and HIV encephalitis, whereas pRb exhibited increased staining in SIV encephalitis only
In a previous study we demonstrated an increase in E2F1 protein by immunoblot analysis in the basal ganglia, but not midfrontal cortex, of macaques that developed SIVE (Jordan-Sciutto et al., 2000); however, we did see changes in E2F1 staining in midfrontal cortex. Similar to the simian model, we observed differences in E2F1 and ppRb staining in HIVE. Vagaries of postmortem autolysis prohibited reliable performance of immunoblot analysis using the human tissues. To quantify the differences in the staining present in encephalitic cases compared with non-encephalitic control cases, we quantified both the area and total fluorescent staining for each of the cell cycle proteins using immunofluorescent confocal microscopy in both the simian model and human autopsy tissue. Positive fluorescent area and total fluorescence gave comparable results. In the SIVE model, three SIVE macaques and three SIV-infected macaques without encephalitis were assessed for E2F1, pRb, or ppRb immunostaining levels. Paraffin-embedded sections containing basal ganglia and midfrontal cortex were labeled by immunofluorescence for E2F1, pRb, or ppRb. The basal ganglia and gray matter of midfrontal cortex were defined anatomically and marked on the slides. Within the identified regions, five random 67,600 μm2 fields were chosen by an individual blinded to the experimental design and imaged using confocal microscopy. The top of the stained tissue section was defined, and 10 sections at 0.34 μm intervals were collected to account for the three-dimensional nature of the cells in the tissue. The staining area for each marker was then determined by counting the number of pixels present in each image. The total fluorescence was obtained by multiplying the area by the average fluorescence intensity. Quantification of E2F1, pRb, and ppRb staining confirmed a statistically significant increase in both staining area and volume for each protein in midfrontal cortex (CTX), caudate (CAD), and putamen (PUT) of SIVE cases over non-encephalitic, SIV-infected controls (Fig. 3) (p < 0.001; data not shown.
Fig. 3.
E2F1, pRb, and ppRb staining was increased in SIVE. Three SIV and three SIVE tissue sections containing the basal ganglia and midfrontal cortex were stained for E2F1, pRb, or ppRb. Anatomic features were identified and marked on the slide as cortical gray matter (CTX), caudate (CAD), and putamen (PUT). Five random 67,600 μm2 fields were chosen from each region and imaged using confocal microscopy as described in Results. The number of pixels present in the captured image was quantified as described in Materials and Methods. Averages of the pixels per field for each brain region are indicated for the three SIV (stippled) and three SIVE (diagonal striped) cases for E2F1, pRb, andppRb. SDs are indicated by error bars. (∗ indicates significant difference between staining in SIV and SIVE cases for the indicated brain region; p < 0.001).
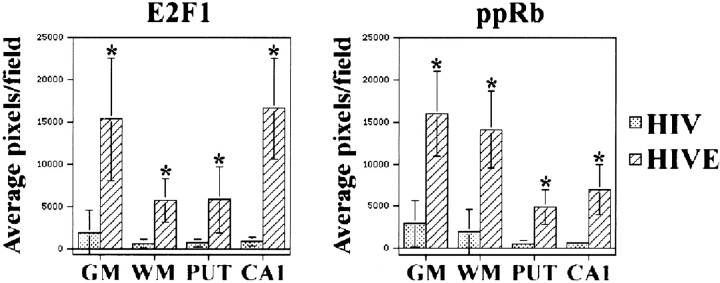
In human autopsy tissue, we quantified the area of positive E2F1 and ppRb staining present in basal ganglia, midfrontal cortex, and hippocampus from six HIVE and six HIV cases. Because our staining protocol for pRb did not differentiate between HIV, HIVE, and non-infected individuals, these data were not subjected to statistical analysis at this time. Neuro-anatomic regions of the brain were marked on the slide (midfrontal cortical gray matter, midfrontal cortical white matter, CA1 region of the hippocampus, and putamen), five random sites within the marked regions were imaged, and the average positive staining area and total fluorescence were calculated. As in the simian model, both E2F1 and ppRb exhibited statistically significant increases in staining in HIVE as compared with non-encephalitic controls (Fig.4) (p < 0.001). These data suggest that the number or availability of E2F1 and ppRb epitopes increased in both SIV and HIV encephalitis.
Fig. 4.
E2F1 and ppRb staining was increased in HIVE. Five HIVE and five HIV tissue sections containing the midfrontal cortex, basal ganglia, and hippocampus were stained for E2F1 or ppRb. Neuro-anatomic features were identified and marked on the slides corresponding to midfrontal cortical gray matter (GM), midfrontal cortical white matter (WM), putamen (PUT), and hippocampal CA1 region (CA1). In each case, five random 67,600 μm2 fields were chosen and imaged for each brain region. The amount of staining for a given field was quantified by counting the number of pixels present. Shown are the average number of pixels per field for each brain region within the five HIV (stippled) and five HIVE (diagonal striped) cases. SDs are indicated by error bars; ∗ indicates a significant difference between the average pixels per field in HIVE as compared with HIV (p < 0.001).
E2F1 staining correlated with macrophage activation in SIV encephalitis, whereas Rb and ppRb staining correlated with the presence of both SIV and activated macrophages
Because we observed alterations in the staining patterns and levels of E2F1 and ppRb in neurons of both SIV and HIV encephalitic individuals, we wanted to determine whether changes in these two proteins were spatially related to the presence of infected or activated macrophages. To decipher this relationship, we correlated the expression of our cell cycle protein of interest in the vicinity of macrophages or lentiviral infection.
In the SIV model, paraffin-embedded brain sections containing the basal ganglia and midfrontal cortex were labeled by immunofluorescence for E2F1, pRb or ppRb, and HAM56, a marker expressed abundantly in macrophages, or gp110, a viral coat protein from SIV. The basal ganglia and gray matter of midfrontal cortex were identified grossly and marked on the slides. Within the marked regions, five random 67,600 μm2 fields were imaged using double-label confocal microscopy by an individual blinded to the experimental design and to the cell cycle protein staining. As above, 10 planes at 0.34 μm intervals were captured, and the central image was subjected to analysis because we determined that there was no difference between using the value for this single image or the average of the 10 fields. In this field, the total number of pixels containing staining for the cell cycle protein and the cell phenotype marker were enumerated to give us a staining area. The staining area for the cell cycle protein being tested within each frame was then correlated with the staining area for macrophages or virus. There was little or no detectable staining for macrophages [HAM56(red)] or infected [gp110(red)] macrophages in either midfrontal cortex or the basal ganglia of SIV-infected macaques without encephalitis (Fig.5A–C,G–I) (data not shown). Brains from macaques infected with SIV without encephalitis also demonstrated minimal staining for E2F1, pRb, and ppRb (green) (Fig. 5, A and G,B and H, C and I, respectively), whereas staining for the cell cycle proteins was abundant in macaques with encephalitis (green) (Fig.5D–F,J–L). A strong correlation was observed between the area of E2F1 staining and the area of HAM56 staining present in the 67,000 μm2 fields in all three brain regions examined for SIVE (Table 3) (r > 0.78). The E2F1 staining area did not correlate with the area of viral staining except in putamen (Table 3) (r = 0.7601). Similar results were obtained when total E2F1 fluorescence was compared with total fluorescence of macrophages or virus (Table 3, E2F1, T). E2F1 staining did not colocalize with the markers for either macrophages or SIV, and only minimal E2F1 staining was observed in HAM56- or gp110-positive cells, indicating that the observed correlations were not caused by E2F1-positive infiltrating cells but by increased E2F1 staining in adjacent and nearby cells (Fig. 5D,J). These results show that overall, the change in E2F1 staining observed in SIVE correlated with the presence of macrophages.
Fig. 5.
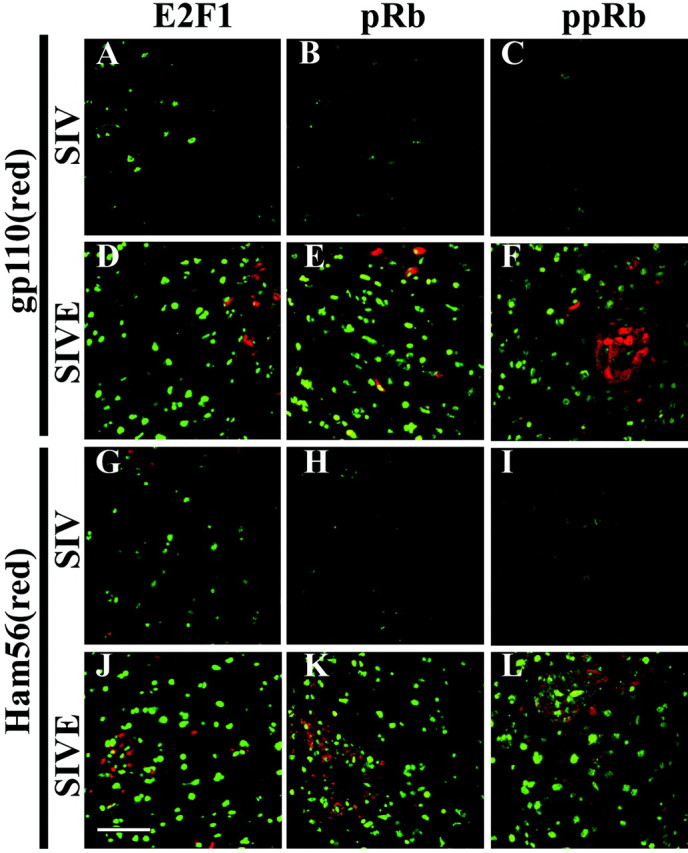
E2F1, pRb, and ppRb staining was present near SIV-infected and activated macrophages. Tissue sections from SIV and SIVE necropsies were stained for E2F1 (left column,green), pRb (middle column,green), or ppRb (right column, green) and gp110 (A–F, red) or HAM56 (G–L, red). Shown is staining from the midfrontal cortex, but similar results were seen when the caudate or putamen was used (data not shown). Little E2F1, pRb, or ppRb staining was observed in the SIV cases (A–C,G–I). Abundant staining for E2F1, pRb, and ppRb was observed near SIV-infected cells [gp110(red); D–F]. Abundant staining for E2F1, pRb, and ppRb was also observed near activated macrophages [HAM56(red);J–L]. Scale bar, 50 μm (for all images).
Table 3.
E2F1, pRb, and ppRb immunostainings correlated with macrophages, whereas only pRb and ppRb correlated with virus in the simian model of HIVE
| SIVE | A or T | Midfrontal cortex | Caudate | Putamen | |||
|---|---|---|---|---|---|---|---|
| gp110 | HAM56 | gp110 | HAM56 | gp110 | HAM56 | ||
| E2F1 | A | −0.1725 | 0.8357** | −0.2334 | 0.7887** | 0.7601** | 0.8936** |
| T | −0.1253 | 0.8101** | −0.2417 | 0.7803** | 0.7137** | 0.9169** | |
| pRb | A | 0.7917** | 0.6568** | 0.4656* | 0.7983** | 0.5382* | −0.5082* |
| T | 0.7890** | 0.6593** | 0.6525** | 0.8050** | 0.5526* | −0.4712 | |
| ppRb | A | 0.8898** | 0.6253** | 0.7329** | 0.5473** | 0.7272** | 0.7130** |
| T | 08052** | 0.6769** | 0.6374** | 0.3661 | 0.7076** | 0.6508** | |
Fluorescent immunostainings for E2F1, pRb, and ppRb were quantified in midfrontal cortex, caudate, and putamen of SIVE macaques. For each cell cycle protein, double-label immunofluorescent staining for the SIV viral envelope protein gp110 (columns 1, 3, and 5) and HAM56 for activated macrophages (columns 2, 4, and 6) were also quantified in the same fields as the cell cycle proteins. Pearson's correlation coefficients (r) were determined for each cell cycle protein compared with either SIV virus (gp110) or macrophages (HAM56) staining area (A) or total fluorescence (T). The rvalues are indicated in the table. *p ≤ 0.01; **p≤ 0.001.
In SIVE, the pRb staining area correlated moderately with both gp110 and HAM56 in midfrontal cortex and caudate, but not in putamen (Table3) (r > 0.65). The ppRb staining area correlated with both gp110 and HAM56 pixel number in all regions, but correlated more strongly with gp110 than HAM56 in midfrontal cortex and caudate (Table3). Neither pRb nor ppRb colocalizes with virus or macrophages and rarely exhibits positive staining in gp110- or HAM56-postive cells, indicating that any observed correlation was not caused by pRb-positive infiltrating macrophages but by increased staining of cells in the vicinity of the infiltrated and infected macrophages (Fig.5E,F,K,L). These results show that changes in pRb staining correlate with the presence of both virus and macrophages in SIVE, but the inactive form of pRb, ppRb, correlates more strongly with virus.
In the human autopsy tissue, the spatial relationships between E2F1 and ppRb staining were compared with the presence of activated (HLA-DR) or HIV-infected (p24) macrophages. Because HIV infection of human brain tissue was most pronounced in deep gray matter, the correlation between ppRb or E2F1 and p24 was limited to the basal ganglia. The correlation between cell cycle proteins and HLA-DR was assessed in both gray and white matter from midfrontal cortex, hippocampus, and basal ganglia from each of six cases. By double-label immunofluorescent laser confocal microscopy, E2F1, ppRb, HLA-DR, and p24 staining was minimal in HIV cases without encephalitis in all brain regions that were assessed (Fig.6A,D) (data not shown). Consistent with our previous observations, we observed increased staining for E2F1 and ppRb in basal ganglia from HIVE cases in which there was also abundant staining for activated macrophages and HIV-infected macrophages (Fig.6B,C,E,F). Although there was abundant virus and activated macrophage staining, neither correlated with the abundant E2F1 or ppRb staining in any brain region assessed (Table 4). These results suggest that although there was abundant E2F1 and ppRb in the vicinity of virus and macrophages, the amount of staining for each marker did not change proportionally with the marker for virus or macrophages.
Fig. 6.
E2F1 and ppRb staining was present near HIV-infected and activated macrophages. Tissue sections from the basal ganglia of HIV and HIVE autopsies were stained for ppRb (A–C, green) or E2F1 (D–F, green) and HLA-DR (left and middle columns,red) or p24 (right column,red) (and data not shown). Little if any ppRb, E2F1, HLA-DR, or p24 staining was observed in HIV autopsies without encephalitis (A, D). In HIVE, abundant E2F1 and ppRb staining was noted in regions with activated macrophages (B, E) or HIVE-infected macrophages (C, F). Scale bar, 50 μm (for all images).
Table 4.
E2F1, pRb, and ppRb immunostainings did not correlate with macrophages or HIV virus in the human disease
| HIVE | A or T | Frontal cortex gray matter | Frontal cortex white matter | Basal ganglia putamen | Hippocampus CA1 | ||||
|---|---|---|---|---|---|---|---|---|---|
| p24 | HLA | p24 | HLA | p24 | HLA | p24 | HLA | ||
| E2F1 | A | NA | 0.1127 | NA | −0.4285* | −0.1876 | −0.3854 | NA | −0.3359 |
| T | NA | 0.1801 | NA | −0.3277 | −0.1674 | −0.3868 | NA | −0.3775 | |
| ppRb | A | NA | 0.1955 | NA | −0.04937 | 0.3343 | −0.005737 | NA | −0.3665 |
| T | NA | 0.2081 | NA | −0.1850 | 0.6138* | 0.01184 | NA | −0.3765 | |
Fluorescent immunostaining for E2F1 and ppRb was quantified in HIVE midfrontal cortex gray matter, frontal cortex white matter, putamen, and the CA1 region of the hippocampus. For each cell cycle protein, double-label immunofluorescent staining for the HIV viral capsid protein p24 (columns 1, 3, and 5) and HLA-DR for activated macrophages (columns 2, 4, and 6) were also quantified in the same fields as the cell cycle proteins. Pearson's correlation coefficients (r) were determined for each cell cycle protein compared with either HIV virus (p24) or macrophages (HLA-DR) staining area (A) or total fluorescence (T). The r values are indicated in the table. *p ≤ 0.05. NA, Not assessed.
E2F/DNA complexes were altered in SIVE
Our results in both the simian model and human autopsy material suggest that the staining patterns and levels for E2F1 were altered under encephalitic conditions. We also observed increased phosphorylation of E2F1 regulatory proteins, such as pRb. Because phosphorylation of pRb reduces its ability to bind and regulate E2F1 activity, the changes that we observed suggest a change in E2F1 activity. As a transcription factor, one way of assessing E2F1 activity is to determine its ability to bind DNA. Protein extracts were prepared from three macaques with SIVE, two macaques infected with SIV without encephalitis, and three macaques that were neither infected nor encephalitic. A detergent soluble “cytoplasmic” fraction was collected from each sample, leaving the nuclei to be extracted with 0.42 m sodium chloride. Cytoplasmic and nuclear fractions from both midfrontal cortex and basal ganglia were used in the EMSA. When nuclear extracts from midfrontal cortex were assayed for E2F/DNA binding activity, a slow migrating complex was observed in the control and SIV cases (Fig. 7A,C 1–C 3, SIV 1, SIV 2). In SIVE cases 2 and 3, two complexes that migrate more quickly than the complex observed in the control lanes were observed in extracts from midfrontal cortex (Fig. 7A, SIVE). Similar results were observed when extracts from the basal ganglia were used (data not shown). When the cytoplasmic extracts were used, we observed a reversal in the pattern of complexes for both brain regions (Fig. 7B, midfrontal cortex) (data not shown for basal ganglia). Cytoplasmic extracts from the control or SIV cases produced a band that migrated more quickly. In the SIVE cases, the quicker migrating band was present, but a slower migrating band appears in all three cases (Fig.7B). To demonstrate that the observed bands were specific for the E2F DNA binding site, we included increasing concentrations of unlabeled E2F oligonucleotides or an unrelated sequence (Fig. 7C). The upper complexes generated by both the nuclear and cytoplasmic extracts were competed away by the E2F sequence, but not by the unrelated sequence (Fig. 7C), suggesting that the complexes present in both nuclear and cytoplasmic extracts are specific for the E2F site. The same results were obtained when extracts from basal ganglia were used (data not shown). Taken together, these data suggest that the protein composition of the E2F/DNA complex is altered in SIV encephalitis. This alteration appears to involve a switch in the E2F DNA binding proteins present in the different subcellular fractions.
Fig. 7.

E2F/DNA complexes were altered in SIVE. E2F DNA binding activity was assessed in protein extracts from midfrontal cortex and basal ganglia of control, SIV, and SIVE cases by electrophoretic mobility shift assay. A, When nuclear extracts from the midfrontal cortex were assayed, E2F complexes were observed in all samples: control (C 1–C 3), SIV (SIV 1, SIV 2), and SIVE (SIVE 1–SIVE 3). The complexes observed in the encephalitic cases exhibited distinct properties as indicated by their further migration in the gel. Similar results were seen in extracts from basal ganglia (data not shown). B, E2F complexes were also observed in all cytoplasmic samples from the midfrontal cortex. In addition to the E2F complex present in both control and SIVE lanes, the extracts from encephalitic cases produced a slower migrating complex. Similar results were also seen in the basal ganglia (data not shown). C, To show that the complexes that we observed in the nuclear and cytoplasmic extracts were specific for the E2F DNA binding assay, we added unlabeled competitor oligonucleotides that contained the E2F site (+E2F comp) and an unrelated sequence (+unrelated comp) in increasing amounts of 50 and 150 ng. Shown are the results using the SIVE samples. The bands specific for E2F are indicated by the bracket to the left and are competed by E2F oligonucleotides but not the unrelated sequence. Experiments on control and SIV samples yielded similar results (data not shown). The first lane in each panel contains only the radiolabeled, double-stranded oligonucleotides used as probe in all other lanes.
DISCUSSION
In the simian model for HIV encephalitis, we previously demonstrated alterations in expression levels and subcellular localization of the E2F1 transcription factor with concomitant alterations in pRb subcellular localization and phosphorylation (Jordan-Sciutto et al., 2000). Here we have shown similar results in human autopsy tissue. In HIVE, we observed increased staining for E2F1 in midfrontal cortex, hippocampus, and basal ganglia. This staining localized predominantly to neurons in both the cytoplasm and nucleus. Changes in E2F1 staining were similar to changes in ppRb staining. Staining for ppRb also increased in all three brain regions of HIVE and was predominantly nuclear in neurons. Phosphorylation of pRb is believed to disrupt interaction with E2F family members, freeing them to increase transcription from E2F target promoters. Our observations suggest that such events may be occurring in response to encephalitic conditions.
The increased staining for E2F1 could result from changes in a number of cellular regulatory processes, from an increase in gene expression at the transcriptional or translational level to changes in post-translational modification leading to increased protein stability. We favor the interpretation that increased staining is caused by an alteration in the availability of the E2F1 epitope recognized by the antibody used in this study. The unique feature of this antibody is that it recognizes E2F1 through its pRb interaction domain. Thus, when E2F1 is in complex with pRb, the antibody will not bind E2F1, but when pRb is phosphorylated and no longer interacting with E2F1, the antibody detects E2F1. This implies that the observed increase in E2F1 in HIVE could be caused by an increase in “free” E2F1 and not necessarily increased E2F1 gene expression. We have tried to support this hypothesis by assessing protein levels by immunoblot analysis; however, because of variability in human tissue preservation, this was not possible. Our results in the simian model support this hypothesis (Jordan-Sciutto et al., 2000). In SIVE, we observed increased E2F1 staining (Fig. 3) but did not see changes in protein levels in SIVE midfrontal cortex. E2F1 protein levels also do not change in BDNF-, NGF-, or RANTES-treated neuroglial cultures, despite dramatic increases in E2F-1 immunostaining and altered subcellular localization (Jordan-Sciutto et al., 2001). Further support for this hypothesis will be provided by using this in vitro model to assess mechanisms regulating E2F1 subcellular localization.
Whether the increase in E2F1 staining is caused by E2F1 liberation or induced E2F1 expression, the presence of phosphorylated pRb indicates that less E2F1 will be complexed with pRb. In the nucleus, free E2F1 induces transcription of genes necessary for cell cycle control (e.g., cyclin A, cyclin E, cdc2, and p19ARF) (Bates et al., 1998; Black and Azizkhan-Clifford, 1999). Our observations indicate that the proteins binding to the E2F DNA binding site are distinct in SIV encephalitic versus control brains. These data suggest that under normal conditions, a specific set of E2F/pocket protein complexes are able to bind DNA in the nucleus; under encephalitic conditions, the E2F/pocket protein complexes bound to promoters are different. Several cell types in the brain can be contributing to the observed E2F complexes in brain extracts. In encephatlitic cases, the presence of infiltrating macrophages may account for the E2F complexes observed. However, our immunohistochemistry for E2F1 suggests that neuronal, oligodendroglial, and astrocytic elements may also exhibit altered E2F complexes in response to pathology in SIVE brain. In any case, it is likely that changes in E2F/pocket protein complexes present in the nucleus represent a change in E2F target promoters being modulated. E2F-regulated gene products can be divided into distinct subsets on the basis of their function in S phase: those needed to regulate cell cycle phase progression (i.e., cyclin E, cdc2, pRb), those needed to synthesize DNA (DNA polymerase α, proliferating cell nuclear antigen, ribonucleotide reductase), and those that regulate cell death (apoptotic activating protein, p19ARF) (Nevins, 1992; Farnham et al., 1993; LaThangue, 1994; Bates et al., 1998; Moroni et al., 2001). One hypothesis is that the change in E2F complexes will activate a distinct subset of genes needed under the conditions dictated by the environment and signaling molecules present therein. By determining which E2F family members and pocket proteins are present in the nucleus under normal verses encephalitic conditions, we can begin to determine which promoters make up the distinct subsets of E2F target promoters being regulated in the two conditions.
The role of cytoplasmic E2F1 in neurons is less easily understood. One possibility is that E2F1 is shuttled into the cytoplasm for degradation by the proteosome, but this does not explain the staining in neuronal processes. Alternatively, E2F1 may serve other functions in neurons besides transcriptional regulation. Recent reports have indicated a role for E2F1 in apoptosis independent of its ability to induce transcription (Phillips et al., 1999; Hou et al., 2001). This was shown recently in cerebellar granule neurons induced to die by dopamine-evoked apoptosis. These reports are intriguing in association with our observations regarding E2F DNA binding activity in cytoplasmic extracts. We observed distinct DNA/protein complexes in normal verses encephalitic brains in the cytoplasm. Interestingly, the DNA/protein complexes in the nucleus of control brains appeared to be in the cytoplasm of the encephalitic brains and vice versa. Finally, the presence of E2F/DNA complexes in the cytoplasm is striking, particularly in light of the cytoplasmic localization of E2F1 in the encephalitic cases. It will be interesting to determine whether the cytoplasmic E2F1 indeed retains its DNA binding activity and what function it may be serving in such a subcellular location.
These findings implicate E2F1 and ppRb in the cellular response to encephalitic conditions. In lentiviral encephalitides, there are two potential sources for neurotoxic factors leading to encephalitis: activated macrophages and the virus itself. Our results indicate that staining for pRb, ppRb, and E2F1 correlated positively with staining for macrophages present in SIVE midfrontal cortex and caudate. Staining for pRb and ppRb also correlated positively with the presence of virus in the same brain regions, whereas E2F1 did not. These results provide support for the hypothesis that changes in E2F1 in surrounding neural cells are not directly dependent on virus but on molecules secreted by macrophages, such as chemokines, cytokines, and neurotrophic factors. These data are corroborated by numerous reports implicating chemokines, cytokines, and neurotrophic factors in HIVE progression (Price et al., 1988; Lipton et al., 1991; Lo et al., 1992; Brouwers et al., 1993;Gelbard et al., 1994; Pulliam et al., 1994, 1996; Crowe, 1995; Giulian et al., 1996; Heyes et al., 1998; Power et al., 1998). Because ppRb staining correlated with both macrophages and virus, we cannot distinguish which initiating factor is responsible for observed increases in pRb staining and its hyperphosphorylation. In the putamen, E2F1 staining was similar to pRb, correlating with both macrophages and virus. It is not clear whether this is caused by a paucity of macrophages that are not also infected with SIV or whether it indicates a need for virus to induce E2F1 neuronal staining in this specific brain region.
The observations in the simian model did not reflect the results in the human disease. This is because the macaques are killed before end-stage encephalitis as part of a vaccine trial. This is in direct contrast to the human disease, in which patients are at the terminal stage. In early stages seen in the simian model, the disease is more focal, and therefore its effects are localized to the initiating lesions. In the human disease, the changes have propagated to regions more distal to the site of injury. This explains why the cell cycle protein staining is more widespread than macrophage or viral staining, which is limited to deep gray matter tracts in HIVE. This would preclude our ability to correlate initiating factors of disease with increased cell cycle protein staining. Despite the discrepancy in the human autopsy tissue, results in our simian model are consistent with previous findings in our human neuroglial cultures demonstrating that both E2F1 and ppRb respond to neurotrophic factors or chemokines with increased staining intensity and altered subcellular distribution in neurons (Jordan-Sciutto et al., 2001). The role of viral-associated proteins in cell cycle protein induction remains to be investigated in this model and may also address our observed discrepancies between the simian and human disease.
Although the virus is the etiologic entity of HIVE, a number of reports indicate that the damage induced in adjacent neurons may be attributable to factors secreted by HIV-infected or activated macrophages. Additional support for a specific role for activated macrophages in neuronal damage associated in HIVE comes from studies of other neurodegenerative diseases in which inflammatory processes have been documented. Alterations in cell cycle protein staining patterns have been observed in several neurodegenerative conditions, including Alzheimer's disease (AD), amyotrophic lateral sclerosis, and Parkinson's disease (Arendt et al., 1996; Vincent et al., 1996; McShea et al., 1997, 1999; Nagy et al., 1997a,b; Ranganathan et al., 2001; K. Jordan-Sciutto, unpublished observations). Early reports in AD demonstrated an increased presence of cyclin-dependent kinases (CDKs) and their regulators (Arendt et al., 1996; Vincent et al., 1996; McShea et al., 1997, 1999; Nagy et al., 1997a,b). Activation of CDKs usually results in phosphorylation of pRb (Kouzarides, 1995;Pines, 1995; Whyte, 1996). Our results in HIVE are consistent with findings in AD demonstrating alteration in pRb and E2F1 staining patterns in areas of inflammatory activation surrounding β amyloid-containing plaque structures. It will be interesting to correlate changes in pRb with changes in CDKs and their regulators in HIVE, to determine whether the upstream signaling cascades are also conserved between neurodegenerative responses in multiple disease states.
Our findings in HIVE are consistent with several reports studying expression patterns of E2F1 and ppRb in neurodegenerative diseases involving an inflammatory component including SIVE, and other neurodegenerative diseases (Jordan-Sciutto et al., 2000, 2001;Ranganathan et al., 2001). Our results indicate that one neuronal response to inflammatory signaling is phosphorylation of pRb, resulting in an alteration in the state of E2F1. The impact of these changes on neuronal function and survival needs to be investigated further, although preliminary evidence suggests that these changes will have consequences for nuclear gene expression. These data also implicate a novel function for E2F1 and ppRb in neurons and potentially oligodendrocytes. By studying the activities of E2F1 and pRb in neurons responding to neurodegenerative stimuli present in HIVE, we hope to gain a greater understanding of disease progression and provide targets for therapy that are specific to degenerating neurons.
Footnotes
This work was supported by National Institutes of Health Grants MH46790, MH182273, NS35731, MH01717, and NS41202, and by the Pathology Post-Doctoral Fellow Research Training Grant from the University of Pittsburgh School of Medicine. We extend our gratitude to the California NeuroAIDS Tissue Network (Grant P-50 MH45294) for providing us with paraffin-embedded tissue sections and frozen tissues from HIV patients for these studies. We also thank Dr. Kelly Stefano Cole for the kind gift of the SIV gp110 polyclonal antibody and Jonette Werley and John Caltagarone for technical support.
Correspondence should be addressed to Dr. Clayton A. Wiley, A-515 University of Pittsburgh Medical Center Presbyterian, 200 Lothrop Street, Pittsburgh, PA 15213, E-mail:wiley@np.awing.upmc.edu, or Dr. Kelly L. Jordan-Sciutto, Department of Pathology, University of Pennsylvania, 4010 Locust Street, Room 312 Levy, Philadelphia, PA 19104-6002, E-mail:jordan@path.dental.upenn.edu.
REFERENCES
- 1.Achim CL, Wiley CA. Expression of major histocompatibility complex antigens in the brains of patients with progressive multifocal leukoencephalopathy. J Neuropathol Exp Neurol. 1992;51:257–263. doi: 10.1097/00005072-199205000-00003. [DOI] [PubMed] [Google Scholar]
- 2.Achim CL, Wiley CA. Inflammation in AIDS and the role of the macrophage in brain pathology. Curr Opin Neurol. 1996;9:221–225. doi: 10.1097/00019052-199606000-00013. [DOI] [PubMed] [Google Scholar]
- 3.Achim CL, Wang R, Miners DK, Wiley CA. Brain viral burden in HIV-infection. J Neuropathol Exp Neurol. 1994;53:284–294. doi: 10.1097/00005072-199405000-00010. [DOI] [PubMed] [Google Scholar]
- 4.Adams P, Kaelin WJ. Transcriptional control by E2F. Semin Cancer Biol. 1995;6:99–108. doi: 10.1006/scbi.1995.0013. [DOI] [PubMed] [Google Scholar]
- 5.Arendt T, Rodel L, Gartner U, Holzer M. Expression of the cyclin-dependent kinase inhibitor p16 in Alzheimer's disease. NeuroReport. 1996;7:3047–3049. doi: 10.1097/00001756-199611250-00050. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology, Vol 2 (Janssen K, ed), pp 12.11.11–12.11.18. Wiley; New York: 1994. [Google Scholar]
- 7.Bacellar H, Munoz A, Miller EN, Cohen BA, Besley D, Selnes OA, Becker JT, McArthur JC. Temporal trends in the incidence of HIV-1-related neurologic diseases: multicenter AIDS cohort study, 1985–1992. Neurology. 1994;44:1892–1900. doi: 10.1212/wnl.44.10.1892. [DOI] [PubMed] [Google Scholar]
- 8.Baskin GB, Murphey-Corb M, Roberts ED, Didier PJ, Martin LN. Correlates of SIV encephalitis in rhesus monkeys. J Medic Primatol. 1992;21:59–63. [PubMed] [Google Scholar]
- 9.Bates S, Phillips A, Clark P, Scott F, Peters G, Ludwig R, Vousden K. p14ARF links the tumour suppressors pRb and p53. Nature. 1998;39:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 10.Black AR, Azizkhan-Clifford J. Regulation of E2F: a family of transcription factors involved in proliferation control. Gene. 1999;237:281–302. doi: 10.1016/s0378-1119(99)00305-4. [DOI] [PubMed] [Google Scholar]
- 11.Brouwers P, Heyes MP, Moss HA, Wolters PL, Poplack DG, Markey SP, Pizzo PA. Quinolinic acid in the cerebrospinal fluid of children with symptomatic human immunodeficiency virus type 1 disease: relationships to clinical status and therapeutic response. J Infect Dis. 1993;168:1380–1386. doi: 10.1093/infdis/168.6.1380. [DOI] [PubMed] [Google Scholar]
- 12.Budka H. Neuropathology of human immunodeficiency virus infection. Brain Pathol. 1991;1:163–175. doi: 10.1111/j.1750-3639.1991.tb00656.x. [DOI] [PubMed] [Google Scholar]
- 13.Budka H, Wiley CA, Kleihues P, Artigas J, Asbury AK, Cho E-S, Cornblath DR, Dal Canto MC, DeGirolami U, Dickson D, Epstein LG, Esiri MM, Giangaspero F, Gosztonyi G, Gray F, Griffin JW, Henin D, Iwasaki Y, Janssen RS, Johnson RT. HIV-associated disease of the nervous system: review of nomenclature and proposal for neuropathology-based terminology. Brain Pathol. 1991;1:143–152. doi: 10.1111/j.1750-3639.1991.tb00653.x. [DOI] [PubMed] [Google Scholar]
- 14.Chellappan S, Hiebert S, Mudryj M, Horowitz J, Nevins J. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991;65:1053–1061. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 15.Chellappan S, Kraus V, Droger B, Munger K, Howley P, Phelps W, Nevins J. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crowe SM. Role of macrophages in the pathogenesis of human immunodeficiency virus (HIV) infection. Aust N Z J Med. 1995;25:777–783. doi: 10.1111/j.1445-5994.1995.tb02881.x. [DOI] [PubMed] [Google Scholar]
- 17.Czub S, Muller JG, Czub M, Muller-Hermelink HK. Impact of various simian immunodeficiency virus variants on induction and nature of neuropathology in macaques. Res Virol. 1996;147:165–170. doi: 10.1016/0923-2516(96)80231-7. [DOI] [PubMed] [Google Scholar]
- 18.Davies J, Everall IP, Weich S, McLaughlin J, Scaravilli J, Lantos PL. HIV-associated brain pathology in the United Kingdom: an epidemiological study. AIDS. 1997;11:1145–1150. doi: 10.1097/00002030-199709000-00010. [DOI] [PubMed] [Google Scholar]
- 19.Farnham P, Slansky J, Kollmar R. The role of E2F in the mammalian cell cycle. Biochim Biophys Acta. 1993;1155:125–131. doi: 10.1016/0304-419x(93)90001-s. [DOI] [PubMed] [Google Scholar]
- 20.Gelbard HA, Nottet HSLM, Swindells S, Jett M, Dzenko KA, Genis P, White R, Wang L, Choi Y-B, Zhang D, Lipton SA, Tourtellotte WW, Epstein LG, Gendelman HE. Platelet activating factor: a candidate HIV-1-induced neurotoxin. J Virol. 1994;68:4628–4635. doi: 10.1128/jvi.68.7.4628-4635.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giovanni A, Wirtz-Bruger F, Keramaris E, Slack R, Park D. Involvement of cell cycle elements, cyclin-dependent kinases, pRb, and E2F-DP, in B-amyloid-induced neuronal death. J Biol Chem. 1999;274:19011–19016. doi: 10.1074/jbc.274.27.19011. [DOI] [PubMed] [Google Scholar]
- 22.Giovanni A, Keramaris E, Morris E, Hou S, O'Hare M, Dyson N, Robertson G, Slack R, Park D. E2F1 mediates death of B-amyloid-treated cortical neurons in a manner independent of p53 and dependent on Bax and Caspase 3. J Biol Chem. 2000;275:11553–11560. doi: 10.1074/jbc.275.16.11553. [DOI] [PubMed] [Google Scholar]
- 23.Giulian D, Yu JH, Li X, Tom D, Li J, Wendt E, Lin SN, Schwarcz R, Noonan C. Study of receptor-mediated neurotoxins released by HIV-1-infected mononuclear phagocytes found in human brain. J Neurosci. 1996;16:3139–3153. doi: 10.1523/JNEUROSCI.16-10-03139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glass JD, Fedor H, Wesselingh SL, McArthur JC. Immunocytochemical quantitation of human-immunodeficiency-virus in the brain: correlations with dementia. Ann Neurol. 1995;38:755–762. doi: 10.1002/ana.410380510. [DOI] [PubMed] [Google Scholar]
- 25.Guy C, Zhou W, Kaufman S, Robinson M. E2F1 blocks terminal differentiation and causes proliferation in transgenic megakaryocytes. Mol Cell Biol. 1996;16:685–693. doi: 10.1128/mcb.16.2.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas S, Gordon J, Khalili J. A developmentally regulated DNA-binding protein from mouse brain stimulates myelin basic protein gene expression. Mol Cell Biol. 1993;13:3103–3112. doi: 10.1128/mcb.13.5.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall DJ. Regulation of c-myc transcription in vitro: dependence on the luanine-rich promoter element ME1a1. Oncogene. 1990;5:47–54. [PubMed] [Google Scholar]
- 28.Heyes MP, Saito K, Lackner A, Wiley CA, Achim CL, Markey SP. Sources of the neurotoxin quinolinic acid in the brain of HIV-1 infected patients and retrovirus-infected macaques. FASEB J. 1998;12:881–896. doi: 10.1096/fasebj.12.10.881. [DOI] [PubMed] [Google Scholar]
- 29.Hou ST, Cowan E, Walker T, Ohan N, Dove M, Tasqinha I, MacManus JP. The transcription factor E2F1 promotes dopamine-evoked neuronal apoptosis by a mechanism independent of transcriptional activation. J Neurochem. 2001;78:287–297. doi: 10.1046/j.1471-4159.2001.00402.x. [DOI] [PubMed] [Google Scholar]
- 30.Joag SV, Stephens EB, Galbreath D, Zhu GW, Li Z, Foresman L, Zhao LJ, Pinson DM, Narayan O. Simian immunodeficiency virus SIVmac chimeric virus whose env gene was derived from SIV-encephalitic brain is macrophage-tropic but not neurovirulent. J Virol. 1995;69:1367–1369. doi: 10.1128/jvi.69.2.1367-1369.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jordan KL, Haas AR, Logan TJ, Hall DJ. Detailed analysis of the basic domain of the E2F1 transcription factor indicates that it is unique among bHLH proteins. Oncogene. 1994;9:1177–1185. [PubMed] [Google Scholar]
- 32.Jordan-Sciutto K, Wang G, Murphey-Corb M, Wiley C. Induction of cell cycle regulators in simian immunodeficiency virus encephalitis. Am J Pathol. 2000;157:497–507. doi: 10.1016/S0002-9440(10)64561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jordan-Sciutto K, Fenner B, Wiley C, Achim C. Response of cell cycle proteins to neurotrophic factor and chemokine stimulation in human neuroglia. Exp Neurol. 2001;167:205–214. doi: 10.1006/exnr.2000.7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kouzarides T. Transcriptional control by the retinoblastoma protein. Semin Cancer Biol. 1995;6:91–98. doi: 10.1006/scbi.1995.0012. [DOI] [PubMed] [Google Scholar]
- 35.LaThangue N. DRTF1/E2F: an expanding family of heterodimeric transcription factors implicated in cell-cycle control. Trends Biochem Sci. 1994;19:108–114. doi: 10.1016/0968-0004(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 36.Lipton SA, Sucher NJ, Kaiser PK, Dreyer EB. Synergistic effects of HIV coat protein and NMDA receptor-mediated neurotoxicity. Neuron. 1991;7:111–118. doi: 10.1016/0896-6273(91)90079-f. [DOI] [PubMed] [Google Scholar]
- 37.Lo TM, Fallert CJ, Piser TM, Thayer SA. HIV-1 envelope protein evokes intracellular calcium oscillations in rat hippocampal neurons. Brain Res. 1992;594:189–196. doi: 10.1016/0006-8993(92)91125-x. [DOI] [PubMed] [Google Scholar]
- 38.Maehlen J, Dunlop O, Liestol K, Dobloug JH, Goplen AK, Torvik A. Changing incidence of HIV-induced brain lesions in Oslo, 1983–1994: effects of zidovudine treatment. AIDS. 1995;9:1165–1169. [PubMed] [Google Scholar]
- 39.Mankowski JL, Flaherty MT, Spelman JP, Hauer DA, Didier PJ, Amedee AM, Murphey-Corb M, Kirstein LM, Munoz A, Clements JE, Zink MC. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055–6060. doi: 10.1128/jvi.71.8.6055-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Masliah E, Achim CL, Ge N, DeTeresa R, Terry RD, Wiley CA. Spectrum of human immunodeficiency virus-associated neocortical damage. Ann Neurol. 1992;32:321–329. doi: 10.1002/ana.410320304. [DOI] [PubMed] [Google Scholar]
- 41.Masliah E, DeTeresa RM, Mallory ME, Hansen LA. Changes in pathological findings at autopsy in AIDS cases for the last 15 years. AIDS. 2000;14:69–74. doi: 10.1097/00002030-200001070-00008. [DOI] [PubMed] [Google Scholar]
- 42.McShea A, Harris P, Webster K, Wahl A, Smith M. Abnormal expression of the cell cycle regulators p16 and cdk4 in Alzheimer's disease. Am J Pathol. 1997;150:1933–1939. [PMC free article] [PubMed] [Google Scholar]
- 43.McShea A, Wahl A, Smith M. Re-entry into the cell cycle: a mechanism for neurodegeneration in Alzheimer's disease. Med Hypotheses. 1999;52:525–527. doi: 10.1054/mehy.1997.0680. [DOI] [PubMed] [Google Scholar]
- 44.Moroni M, Hickman E, Lazzerini Denchi E, Caprara G, Colli E, Cecconi F, Muller H, Helin K. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–558. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 45.Nagy Z, Esiri M, Cato A-M, Smith AD. Cell cycle markers in the hippocampus in Alzheimer's disease. Acta Neuropathol. 1997a;94:6–15. doi: 10.1007/s004010050665. [DOI] [PubMed] [Google Scholar]
- 46.Nagy Z, Esiri M, Smith A. Expression of cell division markers in the hippocampus in Alzheimer's disease and other neurodegenerative conditions. Acta Neuropathol. 1997b;93:294–300. doi: 10.1007/s004010050617. [DOI] [PubMed] [Google Scholar]
- 47.Nevins J. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science. 1992;258:424–429. doi: 10.1126/science.1411535. [DOI] [PubMed] [Google Scholar]
- 48.Pan H, Yin C, Dyson N, Harlow E, Yamasaki L, Van Dyke T. Key roles for E2F1 in signaling p53-dependent apoptosis and in cell division within developing tumors. Mol Cell. 1998;2:283–292. doi: 10.1016/s1097-2765(00)80273-7. [DOI] [PubMed] [Google Scholar]
- 49.Park D, Morris E, Greene L, Geller H. G1/S cell cycle blockers and inhibitors of cyclin-dependent kinases suppress camptothecin-induced neuronal apoptosis. J Neurosci. 1997;17:1256–1270. doi: 10.1523/JNEUROSCI.17-04-01256.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park D, Morris E, Bremmer R, Keramaris E, Padmanabhan J, Rosenbaum M, Shelanski M, Geller H, Greene L. Involvement of retinoblastoma family members and E2F/DP complexes in the death of neurons by DNA damage. J Neurosci. 2000;20:3104–3114. doi: 10.1523/JNEUROSCI.20-09-03104.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Phillips AC, Ernst MK, Bates S, Rice NR, Vousden KH. E2F-1 potentiates cell death by blocking antiapoptotic signaling pathway. Mol Cell. 1999;4:771–781. doi: 10.1016/s1097-2765(00)80387-1. [DOI] [PubMed] [Google Scholar]
- 52.Pines J. Cyclins, CDKs and cancer. Semin Cancer Biol. 1995;6:63–72. doi: 10.1006/scbi.1995.0009. [DOI] [PubMed] [Google Scholar]
- 53.Power C, Kong PA, Crawford TO, Wesselingh S, Glass JD, McArthur JC, Trapp BD. Cerebral white matter changes in acquired immunodeficiency syndrome dementia: alterations of the blood-brain barrier. Ann Neurol. 1993;34:339–350. doi: 10.1002/ana.410340307. [DOI] [PubMed] [Google Scholar]
- 54.Power C, McArthur JC, Nath A, Wehrly K, Mayne M, Nishio J, Langelier T, Johnson RT, Chesebro B. Neuronal death induced by brain-derived human immunodeficiency virus type 1 envelope genes differs between demented and nondemented AIDS patients. J Virol. 1998;72:9045–9053. doi: 10.1128/jvi.72.11.9045-9053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Price RW, Brew B, Sidtis J, Rosenblum M, Scheck AC, Cleary P. The brain in AIDS: central nervous system HIV-1 infection and AIDS dementia complex. Science. 1988;239:586–592. doi: 10.1126/science.3277272. [DOI] [PubMed] [Google Scholar]
- 56.Pulliam L, Clarke JA, McGuire D, McGrath MS. Investigation of HIV-infected macrophage neurotoxin production from patients with AIDS dementia. Adv Neuroimmunol. 1994;4:195–198. doi: 10.1016/s0960-5428(06)80257-3. [DOI] [PubMed] [Google Scholar]
- 57.Pulliam L, Clarke JA, McGrath MS, Moore D, McGuire D. Monokine products as predictors of AIDS dementia. AIDS. 1996;10:1495–1500. doi: 10.1097/00002030-199611000-00006. [DOI] [PubMed] [Google Scholar]
- 58.Ranganathan S, Scudiere S, Bowser R. Hyperphosphorylation of the retinoblastoma gene product and altered subcellular distribution of E2F1 in Alzheimer's disease and amyotrophic lateral sclerosis. J Alzheimer's Dis. 2001;3:377–385. doi: 10.3233/jad-2001-3403. [DOI] [PubMed] [Google Scholar]
- 59.Riley D, Nikitin A, Lee W-H. Adenovirus-mediated retinoblastoma gene therapy suppresses spontaneous pituitary melanotroph tumors in Rb+/− mice. Genes Dev. 1996;10:1870–1879. doi: 10.1038/nm1296-1316. [DOI] [PubMed] [Google Scholar]
- 60.Ross M. Cell division and the nervous system: regulating the cycle from neural differentiation to death. Trends Neurosci. 1996;19:62–68. doi: 10.1016/0166-2236(96)89622-6. [DOI] [PubMed] [Google Scholar]
- 61.Scherr C. Tumor surveillance via the ARF-p53 pathway. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 62.Sharer LR, Michaels J, Murphey-Corb M, Hu FS, Kuebler DJ, Martin LN, Baskin GB. Serial pathogenesis study of SIV brain infection. J Med Primatol. 1991;20:211–217. [PubMed] [Google Scholar]
- 63.Silverstein M, Poller D, Waisman J, Colburn W, Barth A, Gierson E, Lewinsky B, Gamagami P. Prognostic classification of breast ductal carcinoma-in-situ. Lancet. 1995;345:1154–1157. doi: 10.1016/s0140-6736(95)90982-6. [DOI] [PubMed] [Google Scholar]
- 64.Soontornniyomkij V, Wang G, Kapadia S, Achim C, Wiley C. Confocal assessment of lymphoid tissues with follicular hyperplasia from patients infected with human immunodeficiency virus type 1. Arch Pathol Lab Med. 1998;122:534–538. [PubMed] [Google Scholar]
- 65.Strelow LI, Watry DD, Fox HS, Nelson JA. Efficient infection of brain microvascular endothelial cells by an in vivo-selected neuroinvasive SIVmac variant. J Neurovirol. 1998;4:269–280. doi: 10.3109/13550289809114528. [DOI] [PubMed] [Google Scholar]
- 66.Tsai K, Hu Y, Macleod K, Crowley D, Yamasaki L, Jacks T. Mutation of E2F1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 67.Tyor WR, Glass JD, Baumrind N, McArthur JC, Griffin JW, Becker PS, Griffin DE. Cytokine expression of macrophages in HIV-1-associated vacuolar myelopathy. Neurology. 1993;43:1002–1009. doi: 10.1212/wnl.43.5.1002. [DOI] [PubMed] [Google Scholar]
- 68.Vincent I, Rosado M, Davies P. Mitotic mechanisms in Alzheimer's disease? J Cell Biol. 1996;132:413–425. doi: 10.1083/jcb.132.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Westmoreland SV, Halpern E, Lackner AA. Simian immunodeficiency virus encephalitis in rhesus macaques is associated with rapid disease progression. J Neurovirol. 1998;4:260–268. doi: 10.3109/13550289809114527. [DOI] [PubMed] [Google Scholar]
- 70.Whyte P. The retinoblastoma protein and its relatives. Semin Cancer Biol. 1996;6:83–90. doi: 10.1006/scbi.1995.0011. [DOI] [PubMed] [Google Scholar]
- 71.Wiley CA, Achim C. Human immunodeficiency virus encephalitis is the pathological correlate of dementia in acquired immunodeficiency syndrome. Ann Neurol. 1994;36:673–676. doi: 10.1002/ana.410360422. [DOI] [PubMed] [Google Scholar]
- 72.Wiley CA, Soontornniyomkij V, Radhakrishnan L, Masliah E, Mellors J, Herman SA, Dailey P, Achim CL. Distribution of brain HIV load in AIDS. Brain Pathol. 1998;8:277–284. doi: 10.1111/j.1750-3639.1998.tb00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wiley CA, Achim CL, Christopherson C, Kidane Y, Kwok S, Masliah E, Mellors J, Radhakrishnan L, Wang G, Soontornniyomkij V. HIV mediates a productive infection of the brain. AIDS. 1999;13:2055–2059. doi: 10.1097/00002030-199910220-00007. [DOI] [PubMed] [Google Scholar]
- 74.Zink MC, Amedee AM, Mankowski JL, Craig L, Didier P, Carter DL, Munoz A, Murphey-Corb M, Clements JE. Pathogenesis of SIV encephalitis. Selection and replication of neurovirulent SIV. Am J Pathol. 1997;151:793–803. [PMC free article] [PubMed] [Google Scholar]