Abstract
Measuring the states of cell signaling pathways in tumor samples promises to advance understanding of oncogenesis and identify response biomarkers. Here, we describe the use of Reverse Phase Protein Arrays (RPPAs or RPLAs) to profile signaling proteins in 56 breast cancers and matched normal tissue. In RPPAs, hundreds to thousands of lysates are arrayed in dense regular grids and each grid is probed with a different antibody (100 in the current work, of which 71 yielded strong signals with breast tissue). Although RPPA technology is quite widely used, measuring changes in phosphorylation reflective of protein activation remains challenging. Using repeat deposition and well-validated antibodies we show that diverse patterns of phosphorylation can be monitored in tumor samples and changes mapped onto signaling networks in a coherent fashion. The patterns are consistent with biomarker-based classification of breast cancers and known mechanisms of oncogenesis. We explore in detail one tumor-associated pattern that involves changes in the abundance of the Axl receptor tyrosine kinase (RTK) and phosphorylation of the cMet RTK. Both cMet and Axl have been implicated in breast cancer, or in resistance to anti-cancer drugs, but the two RTKs are not known to be linked functionally. Protein depletion and over-expression studies in a “triple-negative” breast cells line reveal crosstalk between Axl and cMet involving Axl-mediated modification of cMet, a requirement for cMet in efficient and timely signal transduction by the Axl ligand Gas6 and the potential for the two receptors to interact physically. These findings have potential therapeutic implications since they imply that bi-specific receptor inhibitors (e.g. ATP-competitive small kinase inhibitors such as GSK1363089, BMS-777607 or MP470) may be more efficacious than the monospecific therapeutic antibodies currently in development (e.g. MetMAb).
Keywords: reverse phase protein arrays, breast cancer, tumor lysate, cell signaling, MET/AXL
Oncogenic selection functions at the level of networks and pathways rather than individual genes (Vogelstein and Kinzler 2004). To date, most multiplex analyses of clinical specimens has involved genomic data because measurement of gene sequences and expression levels is reliable and relatively simple. However, expression profiling does not report directly on regulation at the level of protein abundance or posttranslational modification, both of which are required to understand the activities of signaling pathways (Kolch and Pitt 2010). Protein state can be assayed using conventional immunoblotting but this technique is relatively low throughput. The throughput of mass spectrometry (LC/MS) is much greater in terms of total number of data-points but relatively large samples are required, a problem when working with clinical specimens, and assaying many samples remains slow. In the past few years ‘Reverse-phase’ protein microarrays (RPPAs) have emerged as a way to perform high-throughput immune-based assays on small amounts of material. In an RPPA, thousands of lysates are arrayed in a dense, regular grid onto glass-supported nitrocellulose pads mounted on a microscope slide that is then probed with a different antibody (Grubb et al 2003, Hennessy et al 2010a, Paweletz et al 2001, Sevecka and MacBeath 2006, Tibes et al 2006, VanMeter et al 2007). Subsequent visualization of the bound antibody on each spot provides a quantitative measure of specific antigens in immobilized samples. A drawback of this approach is that only a small subset of antibodies are sufficiently selective to work in an RPPA format (Sevecka et al 2011b) largely because off-target binding by antibodies contributes to the overall signal. Nonetheless, several studies have shown that RPPA technology is effective in mapping intracellular signaling networks in cell lines (Jiang et al 2006, Lin et al 2003, Nishizuka et al 2003, Sevecka and MacBeath 2006, Shankavaram et al 2007). Here, we ask if RPPAs can also be used to analyze phosphorylation-mediated signal transduction in human tumor samples.
The current study is a collaboration between a company specializing in rapid processing of surgical tissues and an academic group experienced in RPPA analysis (samples of the lysates analyzed in this paper are available from www.proteinbiotechnologies.com for those who wish to follow up our experiments). Analyzing post-translational modifications in clinical samples requires that biopsies be processed rapidly to minimize degradation and dephosphorylation: tissue ischemia alters the expression of 10–15% of all genes within 15 min of resection, and ~30% of all proteins change in abundance within 30 min. (Spruessel et al 2004). To minimize changes in protein abundance and phosphorylation, tumors were flash-frozen in liquid nitrogen within 5–10 min of resection. Adjacent normal tissue was also collected and processed in parallel. Frozen tissue was minced and homogenized in cold modified RIPA buffer and total protein levels were quantified (Bio-Rad Laboratories). Tumors included the major histotypes and stages of breast cancer: most cases (n = 48) were classified as ductal carcinoma of varying grade; mucinous and intraductal cancers were represented by 3 samples each; and lobular and a metaplastic tumors by one sample (Supplementary Table S1).
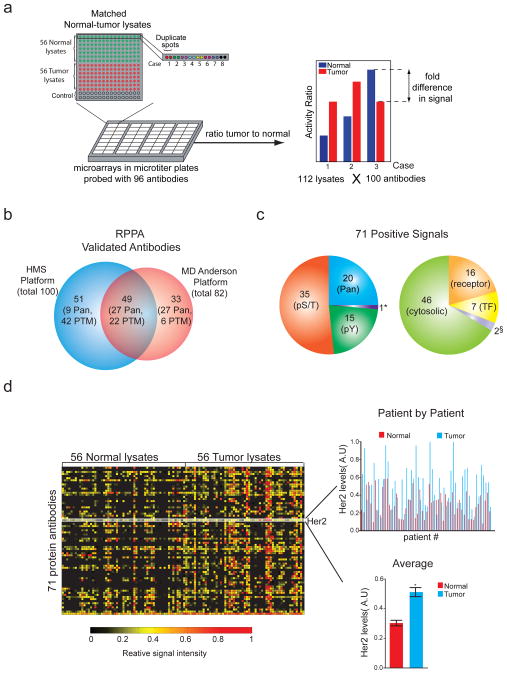
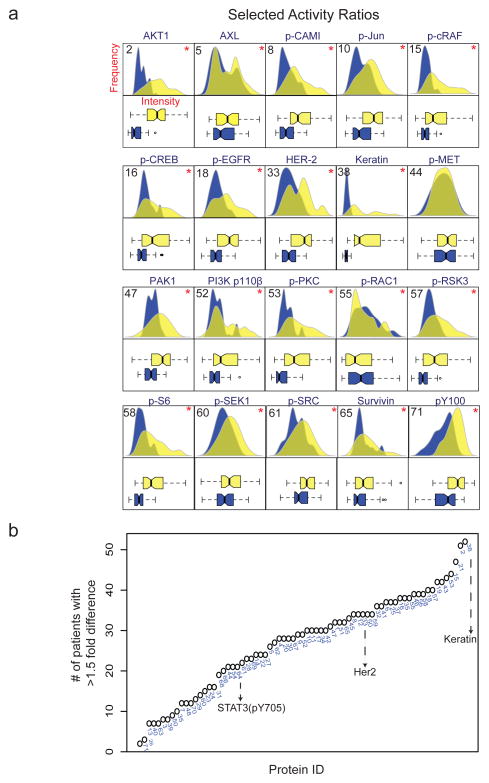
Approximately 100 arrays were printed from normal and tumor-derived extracts (~10 μg total protein per extract), with each array receiving 8 depositions per spot to increase the signal to noise ratio. Arrays were adhered to bottomless microtiter plates, allowing rapid processing of 100 arrays with 100 different primary antibodies (Figure 1a). Slides were incubated with dye-labeled secondary antibodies and scanned to quantify fluorescence levels on a spot-by spot basis. Primary antibodies were directed against proteins, or phosphorylated forms of proteins, known to play a role in oncogenic signal transduction. Several thousand antibodies have been screened by us and by others (Hennessy et al 2010b, Sevecka et al 2011b) to identify the ~5% of commercial antibodies that exhibit sufficient specificity in multiple cell lines (Figure 1b) particularly for the phosphorylated forms of receptor tyrosine kinases (RTKs) and the cytosolic and nuclear proteins they regulate (Figure 1c). In approach to validation, antibodies are screened against a variety of “biological contexts” using lysate microarrays. Each context represents a specific combination of cellular type and treatment conditions. Based on the statistical significance of the resulting measurements, promising antibody-context pairs are further evaluated by quantitative Western blotting. If the two data sets agree (R2≥0.7), the then antibody is considered “validated” for use. Using this strategy, we screened over 400 commercial antibodies and successfully validated 100 of them in one or more biological context (Luckert et al 2012, Sevecka et al 2011a). Our RPPA antibody panel overlaps, but is not identical to, panels developed by investigators at M.D. Anderson Cancer Center who are active in the Cancer Genome Atlas project (Hennessy et al 2010b, Sevecka et al 2011b) (Figure 1b). With breast cancer samples, 71 members our 100-antibody panel yielded signals above background and were used in the current work to generate a set of ~8,000 intensity measurements, each of which was performed in duplicate (Figure 1c,d). We estimate the technical error in the measurements to be ~ ±5% (See Supplementary Materials for details). Normalizing the activity of specific measurements to total protein or to a panel of housekeeping proteins has been proven to be the most effective approach and we applied the former (Gonzalez-Angulo et al 2011) (Carey et al 2010, Nanjundan et al 2010). The activities of phospho-antibodies were therefore normalized to total protein levels or pan-specific antibodies against the same protein (e.g. phospho- and total FGFR receptor) when the reagents were available. RPPA data were standardized by dividing intensity values for each set of duplicate spots by the maximum intensity value for that particular antibody across all normal and cancer samples, thereby generating a set of “activity ratios.” In general, distributions of activity ratios were narrower for normal tissue than for tumor tissue (Figure 2a). For measurements such as the total levels of the Her2/ErbB2 receptor (antibody ID 33), the distribution was unimodal for normal tissue and multimodal for tumor tissue (Figure 2a). “Fold-difference” is a robust metric for scoring the abundance or activities of signaling proteins (Goentoro and Kirschner 2009, Janes et al 2008) and we therefore calculated fold-change data by dividing activity ratios for tumor samples by ratios for matched normal tissue on an antibody by antibody basis. When data were ranked by the frequency of >1.5-fold difference, we observed significant changes between matched normal and tumor tissue for 54 out of 71 protein signals (p<0.05, Kolmogorov-Smirnov test (KS-test); Figure 2b). For example, Keratin (Protein ID:38) was overexpressed in tumor tissue in 55/56 patients (98%), whereas TCF1 (Protein ID:20) was overexpressed in 20/56 patients (36%) (see Supplementary Table S2 for ID assignments). Protein biomarkers used to classify breast cancers clinically were elevated to a similar extent in RPPA data and in breast cancers in general: HER2/neu2 levels (which are predictive of responsiveness to Trastuzumab (Burstein et al 2003) were elevated in 24/56 samples (43%) as compare to ~30% of patients based on the literature (Slamon et al 1987). Similarly, estrogen or progesterone receptor levels, which are predictive of responsiveness to anti-hormonal therapy, were elevated in 22/56 (40%) of samples compared to ~60% of patients (Isola 1993). 12/56 (21%) samples appeared to derive from triple negative tumors, as compared to ~20% of patients overall. Thus, RPPA data captures significant variation between tumor and normal tissue and among different tumors, consistent with current understanding of breast cancer as a complex disease involving multiple subtypes.
Figure 1. Studying signal transduction in clinical samples by RPPA analysis in patient-matched normal and tumor lysates from 56 cases of breast cancer.
(a) Schematic of the RPPA screen. Lysates from 56 tumor and corresponding normal samples were arrayed onto nitrocellulose pads, assembled into microtiter plates, and probed with a collection of 100 primary antibodies. A “fold-difference” in protein measurements was calculated by dividing the activity ratio of a tumor sample by the activity ratio of its matched normal sample. (b) Venn diagram of antibodies validated for RPPAs at HMS and MD Anderson (Hennessy et al 2010b, Sevecka et al 2011b). (c) Breakdown of the 71 antibodies yielding positive signals for breast cancer samples by target and localization. 70% of all antibodies (50/71) were phospho-specific, recognizing modifications that are known to be involved in protein activation. * denotes ‘caspase cleavage’; § denotes ‘other’subcellular localization (d) A heatmap of “activity ratios” of 71 protein measurements in 56 normal and corresponding tumor samples. The protein level distribution of HER2/neu across normal (red) and tumor (blue) samples is also highlighted. Lower bar graph showing an average relative intensity of HER2/neu in all cases of normal and tumor samples. Error bars represent SEM. * denotes P<0.05.
Figure 2. Proteomic measurement for 71 signaling proteins in 56 normal and breast tumor lysates.
(a) Violin and box plots of probability distributions for a subset of protein measurements. * denotes P<0.05 in Kolmogorov-Smirnov test. (b) Activity ratios for all 71 protein measurements ranked by the frequency of a 1.5-fold difference across the 56 matched patient samples. Numbers denoted Protein ID for each measurement; see supplementary materials for the ID code.
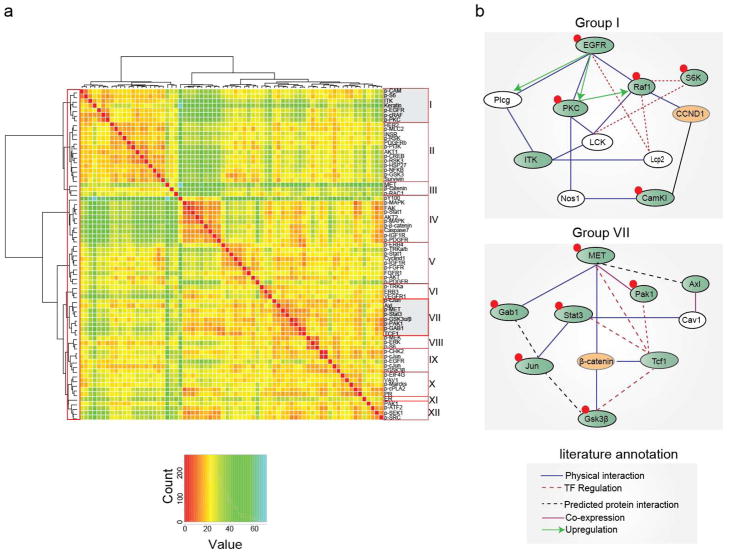
As a first step in uncovering patterns of protein co-regulation, we performed unsupervised hierarchical clustering (Herrero et al 2001) and then drew a threshold in the dendrogram to highlight 12 clusters (i.e., with an average cluster size of six samples; Figure 3a). Diagnostic biomarkers were observed to fall into distinct groups: HER2/neu co-clustered in group II with pro-survival signaling molecules such as pAkt1, pPI3K, pRSK3, pGSK3β, PDGFRβ and insulin receptor; progesterone receptor co-clustered in group X with Vav1, peIF4G, pPLA2; and ER clustered by itself (group XI). It has been shown that mapping clusters of co-expressed genes onto pathways increases information content and intepretability (Makretsov et al 2004, Pe’er and Hacohen 2011, Storey and Tibshirani 2003, Subramanian et al 2005, Vaske et al 2010). We therefore mapped RPPA clusters onto pathways derived from Gene Network Central Pro (GNCPro); an on-line resource of literature-curated interaction graphs derived from protein-protein interaction and gene co-expression data (Liu et al 2010). In general, co-clustering RPPA measurements mapped onto known RTK signaling networks. For example, the cluster containing phosphorylated EGF receptor (pY845, group I) also contained the phosphorylated forms of kinases known to function downstream of EGFR such as Raf1-pS289/296/301, S6K-pS235/236, CAM1-pS81 and PKC-pS660 (Figure 3b). Phosphorylated forms of ERK (pT202/pY204), MEK (pS217/pS221) and S6K (pS240/pS244) kinases made up group VIII, consistent with the known topology of the MAPK signaling cascade and its importance in breast cancer (Menashe et al 2010) (Supplementary Figure S1).
Figure 3. Proteomic profiling of clinical breast cancer specimens reveals distinct network topologies.
(a) Unsupervised clustermap showing correlation between 71 protein measurements. The map is graded to generate twelve distinct clusters (labeled I – XII) based on activity ratios across all samples. The clusters containing EGFR (Cluster I) and cMet (Cluster VII) are highlighted. (b) Protein-Interaction Networks of clusters containing EGFR and cMet. Protein-interaction networks were constructed for each of these clusters using the GNCPro tool. The proteins measured in this study that clustered together are highlighted in green. Red circle denotes phosphorylation measurement. First neighbors of selected proteins are shown in white. Proteins that were measured but do not fall within the cluster are highlighted in orange. Three receptor tyrosine kinases involved in breast cancer, EGFR, Her2, and cMet, form three distinct clusters. The EGFR network cluster (cluster I) includes pPLCγ, pPKC, ITK, pRAF, pS6 and pCAMI kinases. The data show that the phosphorylation of cMet kinase correlated with phosphorylation of its downstream substrates, Gab1, and Stat3, consistent with established network topology of cMet signaling(cluster VII), (Birchmeier et al 2003, Mood et al 2006, Sam et al 2007, Wojcik et al 2006). Phosphorylation of cMet also correlated with phosphorylation of c-Jun and protein levels of Tcf1. In addition to these known components of cMet pathway, measurements for the total levels of p21-activated kinase (Pak1) and Axl fell in the same cluster.
We focused follow-on analysis on the cluster containing the RTK cMet (whose ligand is Hepatocyte Growth Factor (HGF)): cMet levels are not currently used as a prognostic biomarker in breast cancer patients, but overexpression of the receptor is associated with poor clinical outcome, independent of HER2/neu status (Lengyel et al 2005), and several drugs targeting cMet are currently in clinical development (Underiner et al 2010). We observed that levels of active cMet -pY1349 (group VII) clustered away from total HER2, ER and PR and from pEGFR (pY845, and pY1173) but correlated well with the total level of the Axl receptor (R2 = 0.870, p<0.0001) and the phosphorylated form of the Stat3 transcription factor (R2 = 0.883, p<0.0001), generating a well-connected GNCPro network that also contained the phospho forms of the Gab1 adapter (pY627), kinases such as Pak1 (pS199/pS204) and GSK3β (pS21/pS9) and the transcription factor cJun (pT91/pT93). Total levels of Tcf1 also fell in this group. Axl is a member of the TAM (Tyro-Axl-Mer) family of receptor tyrosine kinases and has previously been shown to play a role in the motility and invasiveness of breast cancer cells (Gjerdrum et al 2010, Zhang et al 2008b). Large-scale network reconstruction suggests an interaction between cMet and Axl (Bowers et al 2004, Marcotte et al 1999) and the two receptors are co-regulated by miR-34a in breast cancer cell lines (Mackiewicz et al 2011).
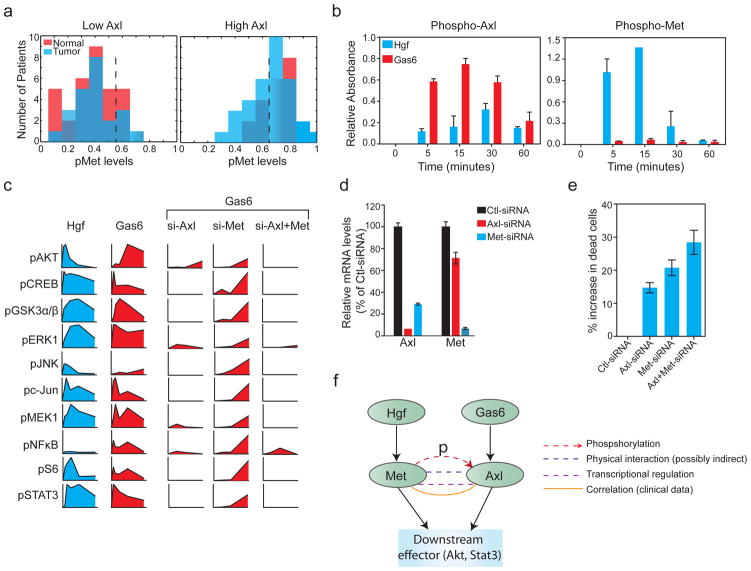
As a statistically rigorous way to investigate possible co-regulation of Axl and cMet in RPPA data, we turned to structured Bayesian inference involving eleven assays for proteins known to be involved in cMet and Axl signal transduction (ten phospho-proteins and one total protein, most of which clustered in group 7). We considered 15 network topologies, all of which involved the class random variable cancer versus normal; 11 naive networks capture the prior assumption that all measurements vary independently. The performance of these models was compared to that of four networks in which the cMet and Axl receptors were presumed to co-vary with cannonical downstream effectors such as Raf and Akt. To learn the parameters of the Bayesian networks, data on RPPA activity ratios were discretized (Supplementary Figure S2) and inference performed on a training- and test-set. The greatest generalization accuracy (82%) was acheived with a network containing the interactions pcMet →Axl, pcMet →pRaf and pRaf→pAkt; adding more interactions decreased the quality of the inference, showing that simpler networks captured all of the available data. Co-variation captured by Bayesian inference is not evident from simple inspection of individual probability distributions for p-cMet and total Axl (Figure 2a): p-cMet levels are unimodal and nearly identical in normal and tumor samples whereas total Axl levels are bimodally distributed in tumor samples (and have a higher mean value; p<0.05, KS-test; Figure 2b). Bayesian network analysis captures the fact that higher p-cMet levels correlate with higher levels of total Axl and this co-variation also correlates with activation of Raf and Akt (Figure 4a, Supplementary Figure S2).
Figure 4. cMet plays a role in Gas6-Axl-mediated activation of downstream signaling pathways.
(a) Frequency distribution of cMet phosphorylation when total Axl levels were low (below the median; left) or high (above the median; right) in both normal and tumor samples. The dashed line denotes the median threshold. (b) Bar graphs showing phosphorylation of Axl (left) and cMet (right) upon stimualtion of MDA-MB-231 cells with HGF or Gas6 for the indicated times. Phosphorylation of cMet and Axl receptors were measured using a sandwich ELISA kit (from Cell Signaling Technology). (c) Time plots showing phosphorylation of ten proteins following treatment of MDA-MB-231 cells 0–120 min. after treatment with HGF or Gas6 (as inidicated) and assayed using xMAP assays. Time courses to the right show cells treated with siRNA against Axl, cMet or both and exposed to Gas6. (d) Bar graph showing relative mRNA levels of Axl and cMet in MDA-MB-231 cells transfected with Axl-siRNA and cMet-siRNA. All values were normalized to control samples transfected with Ctl-siRNA. (e) Dual knockdown of Axl and cMet causes increases in cell death, as shown by increase in the number dead cells upon knockdown of Axl, cMet individually or together. Bars represent mean of three independent experiments and error bars represent SEM. (f) Pathway diagram summarizing proposed mechanisms of cross talk between Axl and cMet kinases based on this work and the literature. Levels Axl and p-cMet are correlated in breast tumor samples (yellow) and we have shown that treatment of cells with HGF results in Axl phosphorylation (red line); over-expressed Axl and cMet can be co-precipitated (blue line) and the two receptors are transcriptionally co-regulated (Yeh et al 2011). The dashes are intended to convey the idea that interactions might be indirect.
To determine if cMet and Axl receptors interact functionally, we turned to breast cancer cell that express both receptors. Exposing MDA-MB-231 triple-negative breast cancer cells to the Axl ligand Gas6 (Growth arrest–specific 6) activated multiple downstream signaling cascades including the PI3K-Akt, MAP kinase, NF-κB, and JAK-STAT pathways (Hafizi and Dahlback 2006). Exposing these cells to cMet HGF activated many of the same signaling molecules. To determine if Gas6- and HGF-mediated signaling are interdependent at the receptor level, MDA-MB-231 cells were exposed to either Gas6 or HGF, and Axl and cMet receptor phosphorylation assayed by ELISA. We observed that Gas6 caused transient phosphorylation of Axl, peaking at 15 min and falling to baseline levels by 60 min (Figure 4b). No increase in phospho-cMet levels were observed. In contrast, exposure of cells to HGF resulted in transient phosphorylation of both cMet and Axl receptors: p-cMet peaked at 15 min and p-Axl peaked at 30 min (Figure 4b). This suggests crosstalk between the two receptors, as has previously been demonstrated for cMet and EGFR in lung tumor cells that have acquired resistance to the EGFR inhibitors gefitinib or between IGF1 recepor and EGFR in breast cancers resistant to erlotinib (Bean et al 2007, Morgillo et al 2006).
To investigate the consequences of Axl and cMet interaction on downstream signaling, we used xMAP micro-sandwich assays (Lim and Zhang 2007) to measure the phosphorylation status of 19 signaling proteins at 4–5 time points ranging from 0 to 120 min after exposure of MDA-MB-231 cells to HGF or Gas6 (Figure 4c, Supplementary Figure S3). Both ligands activated Akt (pS473), CREB (pS133), GSK3α/β (pS21/pS9), Erk1 (pT202/pY204), c-Jun (pS63), MEK1 (pS217/pS221), S6 (pT421/pS424) and Stat3 (pS727), but NFκB phosphorylation (pS536) was observed only after Gas6 exposure, as previously described (Demarchi et al 2001). We then used RNAi to deplete cells of one receptor or the other and exposed them to Gas6 or HGF. Axl mRNA and protein levels were depleted ~10-fold (Figure 4d, Supplementary Figure S4) and we observed 5–10 fold reductions in Gas6-induced phosphorylation of downstream proteins. Knocking down cMet by RNAi was less efficient (four-fold depletion at the mRNA level and 2-fold at the protein level, Figure 4d, Supplementary Figure S4), resulting in a 2–4-fold reduction and a delay in both HGF and Gas6-mediated phosphorylation of downstream proteins (Figure 4c, Supplementary Figure S3). Knocking down cMet also resulted in a reduction in Axl mRNA levels, consistent with previous reports that the two receptors are co-regulated and consistent with delays in Gas6-mediated phosphorylation of downstream signaling proteins (Figure 4d). The converse was not true, however: Axl depletion did not alter the dynamics or magnitude of HGF-mediated signal transduction. Several studies have shown that knockdown of Met or Axl reduced motility, invasiveness and tumorogenicity in mice (Previdi et al 2012), (Vuoriluoto et al 2011), (Holland et al 2010), (Zhang et al 2008a)). In our hands, dual knockdown of Axl and cMet in MDA-MB-231 cells also increased cell death. (Figure 4e) and knockdown of either gene reduced cell migration in a wound-healing assay (Supplementary Figure S5). Thus, both Axl and cMet are involved in typical oncogenic processes in MDA-MB-231 and they may act cooperatively in pro-survival signaling. Co-immunoprecipitation of ectopically expressed Axl and cMet also suggested that the receptors may associate physically: we detected the presence of myc-cMet in HA-Axl immuno-precipitates prepared from lysates of HEK 293 cells over-expressing HA-tagged Axl and myc-tagged cMet (Supplementary Figure S6). Co-purification has been observed in some but not all previously described heterotypic RTK interactions: for example, IGF1R and EGFR, but not cMet and EGFR, co-purify in drug resistant cell lines (Nahta et al 2005). The details of the interaction between cMet and Axl remain to be elucidated and are likely to be complex: although HGF exposure causes both cMet and Axl phosphorylation, knocking down Axl does not appear to alter HGF-mediated phosphorylation of downstream proteins. In contrast, Gas6 exposure results only in Axl phosphorylation, but knocking down cMet alters the timing and magnitude of Gas6-mediated signaling (Figure 4f). Despite these complexities, cell line data support findings from RPPA analysis showing that Axl and cMet receptors exhibit crosstalk and may interact physically interact in breast cancer cells.
In this paper, we show that RPPA analysis can be used to assay the activation state of signaling proteins in lysates derived from primary tumors. Patterns of protein phosphorylation are remarkably diverse from one tumor to the next, but are consistent with the known topologies of oncogenic signaling pathways and with biomarker-based classification of breast cancer subtypes. The current study benefits from the availability of high quality lysates, but suffers from a lack of access to genomic data or the clinical histories of the individual patients. We have therefore focused on pathway inference, specifically with regard to the cMet and Axl RTKs. It is highly probable that additional information can be extracted from our RPPA data using more sophisticated analytical methods, and we encourage others to download the data.
If cMet and Axl function coordinately in cancer, as we hypothesize, then it may be advantageous to develop bi-specific antibodies that block both receptors simultaneously. Therapeutic antibodies currently in development that target cMet, such as MetMAb are not expected to block Axl but small molecule cMet kinase inhibitors (GSK1363089, BMS-777607, MP470) do appear to inhibit Axl as well (Kataoka et al 2011, Mahadevan et al 2007, Underiner et al 2010). More generally, positive results from the current study provide an impetus to clinical investigators, drug companies, and institutional review boards to include phosphorylation-rich RPPA analysis in the analysis of clinical data. We note that relatively little work will be required to create a superset of 133 well-validated antibodies spanning both the HMS and MD Anderson RPPA platforms.
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institutes of Health (R33 CA128726, R21 CA126720, and 5RC1-HG005354) and from the Stand Up to Cancer Project (AACR-SU2C-DT0409). TSG is a Human Frontier Science Program Fellow. RLK is partially supported by NSF 0856285. Supplementary Information accompanies the paper on the Oncogene website
Footnotes
Conflict of Interest Statement
G.M. is a Vice President and co-founder, and P.K.S a co-founder of Merrimack Pharmaceuticals, a biotechnology company that develops anti-cancer drugs. P.E.S is a President and CEO of Protein Biotechnologies, Inc.
References
- Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier C, Birchmeier W, Gherardi E, Woude GFV. Met, metastasis, motility and more. Nature Reviews Molecular Cell Biology. 2003;4:915–925. doi: 10.1038/nrm1261. [DOI] [PubMed] [Google Scholar]
- Bowers PM, Pellegrini M, Thompson MJ, Fierro J, Yeates TO, Eisenberg D. Prolinks: a database of protein functional linkages derived from coevolution. Genome Biol. 2004;5:R35. doi: 10.1186/gb-2004-5-5-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burstein HJ, Harris LN, Marcom PK, Lambert-Falls R, Havlin K, Overmoyer B, et al. Trastuzumab and vinorelbine as first-line therapy for HER2-overexpressing metastatic breast cancer: multicenter phase II trial with clinical outcomes, analysis of serum tumor markers as predictive factors, and cardiac surveillance algorithm. J Clin Oncol. 2003;21:2889–2895. doi: 10.1200/JCO.2003.02.018. [DOI] [PubMed] [Google Scholar]
- Carey MS, Agarwal R, Gilks B, Swenerton K, Kalloger S, Santos J, et al. Functional proteomic analysis of advanced serous ovarian cancer using reverse phase protein array: TGF-beta pathway signaling indicates response to primary chemotherapy. Clin Cancer Res. 2010;16:2852–2860. doi: 10.1158/1078-0432.CCR-09-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarchi F, Verardo R, Varnum B, Brancolini C, Schneider C. Gas6 anti-apoptotic signaling requires NF-kappa B activation. J Biol Chem. 2001;276:31738–31744. doi: 10.1074/jbc.M104457200. [DOI] [PubMed] [Google Scholar]
- Gjerdrum C, Tiron C, Høiby T, Stefansson I, Haugen H, Sandal T, et al. Axl is an essential epithelial-to-mesenchymal transition-induced regulator of breast cancer metastasis and patient survival. Proceedings of the National Academy of Sciences. 2010;107:1124. doi: 10.1073/pnas.0909333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goentoro L, Kirschner MW. Evidence that fold-change, and not absolute level, of beta-catenin dictates Wnt signaling. Mol Cell. 2009;36:872–884. doi: 10.1016/j.molcel.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Angulo AM, Hennessy BT, Meric-Bernstam F, Sahin A, Liu W, Ju Z, et al. Functional proteomics can define prognosis and predict pathologic complete response in patients with breast cancer. Clin Proteomics. 2011;8:11. doi: 10.1186/1559-0275-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb RL, Calvert VS, Wulkuhle JD, Paweletz CP, Linehan WM, Phillips JL, et al. Signal pathway profiling of prostate cancer using reverse phase protein arrays. Proteomics. 2003;3:2142–2146. doi: 10.1002/pmic.200300598. [DOI] [PubMed] [Google Scholar]
- Hafizi S, Dahlback B. Signalling and functional diversity within the Axl subfamily of receptor tyrosine kinases. Cytokine Growth Factor Rev. 2006;17:295–304. doi: 10.1016/j.cytogfr.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clin Proteomics. 2010a;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clin Proteomics. 2010b;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Valencia A, Dopazo J. A hierarchical unsupervised growing neural network for clustering gene expression patterns. Bioinformatics. 2001;17:126–136. doi: 10.1093/bioinformatics/17.2.126. [DOI] [PubMed] [Google Scholar]
- Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–1554. doi: 10.1158/0008-5472.CAN-09-2997. [DOI] [PubMed] [Google Scholar]
- Isola JJ. Immunohistochemical demonstration of androgen receptor in breast cancer and its relationship to other prognostic factors. J Pathol. 1993;170:31–35. doi: 10.1002/path.1711700106. [DOI] [PubMed] [Google Scholar]
- Janes KA, Reinhardt HC, Yaffe MB. Cytokine-induced signaling networks prioritize dynamic range over signal strength. Cell. 2008;135:343–354. doi: 10.1016/j.cell.2008.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Mircean C, Shmulevich I, Cogdell D, Jia Y, Tabus I, et al. Pathway alterations during glioma progression revealed by reverse phase protein lysate arrays. Proteomics. 2006;6:2964–2971. doi: 10.1002/pmic.200500555. [DOI] [PubMed] [Google Scholar]
- Kataoka Y, Mukohara T, Tomioka H, Funakoshi Y, Kiyota N, Fujiwara Y, et al. Foretinib (GSK1363089), a multi-kinase inhibitor of MET and VEGFRs, inhibits growth of gastric cancer cell lines by blocking inter-receptor tyrosine kinase networks. Invest New Drugs. 2011 doi: 10.1007/s10637-011-9699-0. [DOI] [PubMed] [Google Scholar]
- Kolch W, Pitt A. Functional proteomics to dissect tyrosine kinase signalling pathways in cancer. Nature Reviews Cancer. 2010;10:618–629. doi: 10.1038/nrc2900. [DOI] [PubMed] [Google Scholar]
- Lengyel E, Prechtel D, Resau JH, Gauger K, Welk A, Lindemann K, et al. C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer. 2005;113:678–682. doi: 10.1002/ijc.20598. [DOI] [PubMed] [Google Scholar]
- Lim CT, Zhang Y. Bead-based microfluidic immunoassays: the next generation. Biosens Bioelectron. 2007;22:1197–1204. doi: 10.1016/j.bios.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Lin Y, Huang R, Cao X, Wang SM, Shi Q, Huang RP. Detection of multiple cytokines by protein arrays from cell lysate and tissue lysate. Clin Chem Lab Med. 2003;41:139–145. doi: 10.1515/CCLM.2003.023. [DOI] [PubMed] [Google Scholar]
- Liu G, Fong E, Zeng X. GNCPro: Navigate Human Genes and Relationships Through Net-Walking. Advances in Computational Biology. 2010:253–259. doi: 10.1007/978-1-4419-5913-3_29. [DOI] [PubMed] [Google Scholar]
- Luckert K, Gujral TS, Chan M, Joos TO, Sorger PK, Macbeath G, et al. A dual array-based approach to assess the abundance and posttranslational modification state of signaling proteins. Sci Signal. 2012;5 doi: 10.1126/scisignal.2002372. pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH, Ambs S, Caplen NJ. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res Treat. 2011;130:663–679. doi: 10.1007/s10549-011-1690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan D, Cooke L, Riley C, Swart R, Simons B, Della Croce K, et al. A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene. 2007;26:3909–3919. doi: 10.1038/sj.onc.1210173. [DOI] [PubMed] [Google Scholar]
- Makretsov NA, Huntsman DG, Nielsen TO, Yorida E, Peacock M, Cheang MCU, et al. Hierarchical clustering analysis of tissue microarray immunostaining data identifies prognostically significant groups of breast carcinoma. Clinical cancer research. 2004;10:6143. doi: 10.1158/1078-0432.CCR-04-0429. [DOI] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- Menashe I, Maeder D, Garcia-Closas M, Figueroa JD, Bhattacharjee S, Rotunno M, et al. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer research. 2010;70:4453. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mood K, Saucier C, Bong YS, Lee HS, Park M, Daar IO. Gab1 is required for cell cycle transition, cell proliferation, and transformation induced by an oncogenic met receptor. Molecular biology of the cell. 2006;17:3717. doi: 10.1091/mbc.E06-03-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgillo F, Woo JK, Kim ES, Hong WK, Lee HY. Heterodimerization of insulin-like growth factor receptor/epidermal growth factor receptor and induction of survivin expression counteract the antitumor action of erlotinib. Cancer research. 2006;66:10100–10111. doi: 10.1158/0008-5472.CAN-06-1684. [DOI] [PubMed] [Google Scholar]
- Nahta R, Yuan LX, Zhang B, Kobayashi R, Esteva FJ. Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer research. 2005;65:11118–11128. doi: 10.1158/0008-5472.CAN-04-3841. [DOI] [PubMed] [Google Scholar]
- Nanjundan M, Byers LA, Carey MS, Siwak DR, Raso MG, Diao L, et al. Proteomic profiling identifies pathways dysregulated in non-small cell lung cancer and an inverse association of AMPK and adhesion pathways with recurrence. J Thorac Oncol. 2010;5:1894–1904. doi: 10.1097/JTO.0b013e3181f2a266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka S, Charboneau L, Young L, Major S, Reinhold WC, Waltham M, et al. Proteomic profiling of the NCI-60 cancer cell lines using new high-density reverse-phase lysate microarrays. Proc Natl Acad Sci U S A. 2003;100:14229–14234. doi: 10.1073/pnas.2331323100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paweletz CP, Charboneau L, Bichsel VE, Simone NL, Chen T, Gillespie JW, et al. Reverse phase protein microarrays which capture disease progression show activation of pro-survival pathways at the cancer invasion front. gene. 2001;20:1981–1989. doi: 10.1038/sj.onc.1204265. [DOI] [PubMed] [Google Scholar]
- Pe’er D, Hacohen N. Principles and Strategies for Developing Network Models in Cancer. Cell. 2011;144:864–873. doi: 10.1016/j.cell.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previdi S, Abbadessa G, Dalo F, France DS, Broggini M. Breast cancer-derived bone metastasis can be effectively reduced through specific c-MET inhibitor tivantinib (ARQ 197) and shRNA c-MET knockdown. Mol Cancer Ther. 2012;11:214–223. doi: 10.1158/1535-7163.MCT-11-0277. [DOI] [PubMed] [Google Scholar]
- Sam MR, Elliott BE, Mueller CR. A novel activating role of SRC and STAT3 on HGF transcription in human breast cancer cells. Mol Cancer. 2007;6:69. doi: 10.1186/1476-4598-6-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevecka M, MacBeath G. State-based discovery: a multidimensional screen for small-molecule modulators of EGF signaling. Nat Methods. 2006;3:825–831. doi: 10.1038/NMETH931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevecka M, Wolf-Yadlin A, MacBeath G. Lysate microarrays enable high-throughput, quantitative investigations of cellular signaling. Mol Cell Proteomics. 2011a;10:M110 005363. doi: 10.1074/mcp.M110.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevecka M, Wolf-Yadlin A, Macbeath G. Lysate Microarrays Enable High-throughput, Quantitative Investigations of Cellular Signaling. Mol Cell Proteomics. 2011b;10:M110 005363. doi: 10.1074/mcp.M110.005363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6:820–832. doi: 10.1158/1535-7163.MCT-06-0650. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Spruessel A, Steimann G, Jung M, Lee SA, Carr T, Fentz AK, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. Biotechniques. 2004;36:1030–1037. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibes R, Qiu YH, Lu Y, Hennessy B, Andreeff M, Mills GB, et al. Reverse phase protein array: validation of a novel proteomic technology and utility for analysis of primary leukemia specimens and hematopoietic stem cells. Molecular cancer therapeutics. 2006;5:2512. doi: 10.1158/1535-7163.MCT-06-0334. [DOI] [PubMed] [Google Scholar]
- Underiner TL, Herbertz T, Miknyoczki SJ. Discovery of small molecule c-Met inhibitors: Evolution and profiles of clinical candidates. Anticancer Agents Med Chem. 2010;10:7–27. doi: 10.2174/1871520611009010007. [DOI] [PubMed] [Google Scholar]
- VanMeter A, Signore M, Pierobon M, Espina V, Liotta LA, Petricoin I, et al. Reverse-phase protein microarrays: application to biomarker discovery and translational medicine. Expert review of molecular diagnostics. 2007;7:625–633. doi: 10.1586/14737159.7.5.625. [DOI] [PubMed] [Google Scholar]
- Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nature medicine. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Vuoriluoto K, Haugen H, Kiviluoto S, Mpindi JP, Nevo J, Gjerdrum C, et al. Vimentin regulates EMT induction by Slug and oncogenic H-Ras and migration by governing Axl expression in breast cancer. Oncogene. 2011;30:1436–1448. doi: 10.1038/onc.2010.509. [DOI] [PubMed] [Google Scholar]
- Wojcik E, Sharifpoor S, Miller N, Wright T, Watering R, Tremblay E, et al. A novel activating function of c-Src and Stat3 on HGF transcription in mammary carcinoma cells. Oncogene. 2006;25:2773–2784. doi: 10.1038/sj.onc.1209306. [DOI] [PubMed] [Google Scholar]
- Yeh CY, Shin SM, Yeh HH, Wu TJ, Shin JW, Chang TY, et al. Transcriptional activation of the Axl and PDGFR-alpha by c-Met through a ras- and Src-independent mechanism in human bladder cancer. BMC Cancer. 2011;11:139. doi: 10.1186/1471-2407-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YX, Knyazev PG, Cheburkin YV, Sharma K, Knyazev YP, Orfi L, et al. AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer Res. 2008a;68:1905–1915. doi: 10.1158/0008-5472.CAN-07-2661. [DOI] [PubMed] [Google Scholar]
- Zhang YX, Knyazev PG, Cheburkin YV, Sharma K, Knyazev YP, rfi L, et al. AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer research. 2008b;68:1905. doi: 10.1158/0008-5472.CAN-07-2661. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.