Abstract
Macroautophagy has been implicated in a variety of pathological processes. Hypoxic/ischemic cellular injury is one such process in which autophagy has emerged as an important regulator. In general, autophagy is induced after an hypoxic/ischemic insult; however, whether the induction of autophagy promotes cell death or recovery is controversial and appears to be context dependent. We have developed C. elegans as a genetically tractable model for the study of hypoxic cell injury. Both necrosis and apoptosis are mechanisms of cell death following hypoxia in C. elegans. However, the role of autophagy in hypoxic injury in C. elegans has not been examined. Here, we found that RNAi knockdown of the C. elegans homologs of beclin 1/Atg6 (bec-1) and LC3/Atg8 (lgg-1, lgg-2), and mutation of Atg1 (unc-51) decreased animal survival after a severe hypoxic insult. Acute inhibition of autophagy by the type III phosphatidylinositol 3-kinase inhibitors, 3-methyladenine and Wortmannin, also sensitized animals to hypoxic death. Hypoxia-induced neuronal and myocyte injury as well as necrotic cellular morphology were increased by RNAi knockdown of BEC-1. Hypoxia increased the expression of a marker of autophagosomes in a bec-1-dependent manner. Finally, we found that the hypoxia hypersensitive phenotype of bec-1(RNAi) animals could be blocked by loss-of-function mutations in either the apoptosis or necrosis pathway. These results argue that inhibition of autophagy sensitizes C. elegans and its cells to hypoxic injury and that this sensitization is blocked or circumvented when either of the two major cell death mechanisms is inhibited.
Keywords: Autophagy, Cell death, Hypoxia, Apoptosis, Necrosis
INTRODUCTION
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved mechanism to recycle proteins and organelles.1, 2 Autophagy has been most extensively studied in yeast where it functions in adaptation to and survival during nutrient deprivation.3 In metazoans, autophagy has been implicated in a variety of physiological and pathological processes. In C. elegans, RNAi-knockdown of homologs of yeast autophagy genes shortens lifespan and interferes with formation of stress resistant dauer larvae.4 In Drosophila, autophagy is induced by starvation and redistributes nutrients during larval development.5, 6 In mammals, autophagy has been functionally linked to survival during growth factor deprivation in mouse bone marrow cell lines7 and during the early mouse neonatal period where autophagy is upregulated in tissues with increased metabolic demands.8 More recently, autophagy has been implicated in a variety of pathological conditions including neurodegenerative diseases, myocardial and cerebral hypoxia/ischemia, tumor formation, and bacterial pathogenicity.2, 9 In each of these disease states, the precise role of the autophagy pathway is unclear. In particular, whether autophagy is contributing to and/or an adaptive reaction against the cellular pathology is an unresolved and fundamental question.
Hypoxic/ischemic injury is one such pathological condition, in which autophagy may play a role. Multiple studies have reported an increase in markers of autophagy after hypoxic/ischemic injury of mammalian cells.10–15 In particular, increases of autophagic activity have been well documented after hypoxia/ischemia of cardiac myocytes and cerebral neurons.16, 17 In some models, autophagy appears to promote cell death following hypoxia/ischemia whereas in others it may function to promote survival. What determines whether autophagy promotes death or survival is not yet clear. One hypothesis with some experimental support is that normally autophagy inhibits apoptotic cell death. Thus, in tissues or conditions where apoptosis is the dominant form of cell death following hypoxic injury, autophagy would be expected to promote survival.
C. elegans has made crucial contributions towards our understanding of the fundamental mechanisms of cell death. The core pathway for apoptosis was defined in C. elegans using elegant classical genetic techniques.18 Potent modulators of necrotic cell death have also been identified in C. elegans by mutagenesis screens.19–24 Motivated by these successes, we and others have developed C. elegans as genetically tractable model for hypoxic adaptation and injury and have identified genes that either promote or suppress hypoxic cell death.25–35 C. elegans is relatively hypoxia resistant compared to most mammalian cells. Six hours of near anoxic incubation of C. elegans is required to produce permanent behavioral deficits and a measurable level of whole animal death.32 About half of adult animals are killed by 12 hours of hypoxia and all by 24 hours. Both neurons and myocytes die prior to and contribute to whole animal death. Morphological criteria indicate that necrosis is one mechanism of cell death. Mutations in the C. elegans programmed cell death pathway produce whole animal hypoxia resistance implicating apoptosis as also mediating a substantial fraction of the hypoxic cell death.36, 37 The role of autophagy in hypoxic injury in invertebrates has yet to be defined. Here, we use genetic and pharmacological reagents to examine the role of autophagy in hypoxic injury in C. elegans.
MATERIALS AND METHODS
Strains and cultivation
Except for RNAi experiments, all strains were grown at 20° on nematode growth medium NGM agar plates seeded with OP50 bacteria.38 The wild-type strain used was N2 var Bristol and was the background for all strains and for all RNAi experiments unless otherwise indicated. PD4251 containing [nls::pmyo-3::GFP]39 and SK4005 containing zdIs4[pmec-4::GFP;lin-15(+)] were kindly provided by Monica Driscoll and Scott Clark.40 QU1 containing izEx1[Plgg-1::GFP::lgg-1 + rol-6(su1006)] was kindly provided by Alicia Melendez and Beth Levine.4 MT1522 ced-3(n717), MT3002 ced-3(n1286), MT2547 ced-4(n1162), MT4770 ced-9(n1950gf), ZB1028 crt-1(bz29), CB369 unc-51(e369), and CB1189 unc-51(e1189) were obtained from the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR).
RNAi experiments
RNAi bacterial strains were obtained from the Ahringer Library41 and were grown overnight in Luria Broth with 100 µg/ml ampicillin at 37° and then diluted 1:100 in 2xYT medium with the same antibiotics and grown at 37° with shaking until reaching an OD of 0.4. 150 µl of the RNAi bacteria plus 1mM IPTG was then added to each NGM plate containing 50µg/ml of ampicillin. After 2 days of bacterial growth, animals were added to the RNAi plates. Animals were grown for two generations from egg to adult on RNAi plates plus 1mM IPTG; the second generation was age synchronized by transferring adults to a new RNAi plate for four hours then removing the parents to produce a synchronous brood of approximately 100 animals/plate. The empty vector L4440 was used as the negative control for all RNAi experiments and L4440 was grown and induced identically to other RNAi bacteria.
Hypoxic exposure
Synchronized populations of well-fed animals were transferred from their agar plates to a 1.5 ml eppendorf tube with 1 ml M9 buffer; the buffer was exchanged three times to remove bacteria and the final wash was removed down to 100 µl; the tubes were placed into the hypoxia chamber as described previously32 and incubated for the specified period at 26° then removed from the chamber, transferred to NGM agar plates seeded with OP50 bacteria to recover for 24 four hours in normoxia at 20° before scoring. Animals without pharyngeal pumping and without spontaneous or touch-evoked movement were scored as dead. One day post L4 animals were used for scoring whole animal death assays; one day post egg animals were used for scoring cell pathology and GFP::LGG-1 density.
Autophagy inhibition with 3MA and wortmannin
A frozen 10 µl aliquot of 100 mM 3-methyladenine (3MA – Sigma #M9281) or 1mM wortmannin (Sigma #W1628) in DMSO was thawed and diluted into 90 µl of M9 buffer in minifuge tubes with about 100 adult worms to achieve the specified 3MA concentrations. The control incubations were in 10% DMSO in M9. The tubes were immediately placed into the hypoxia chamber for the hypoxic incubation for the specified time, and the worms recovered and scored as usual.
Neuronal and muscular degeneration assay
L1 and L2 larvae underwent twelve hours of hypoxic incubations as described above. All pathologies were scored on surviving animals after 5 hours of recovery by an observer blinded to condition. Nomarski optics was used for scoring necrotic cells. Markedly swollen cells with no apparent nuclei were scored as necrotic. For muscle pathology, muscle nuclei were visualized and counted using nuclear-localized GFP driven by a muscle-specific promoter - pmyo-3::GFP as described previously.32 Axonal beading pathology was scored in mechanosensory neurons visualized with pmec-4::GFP as described previously.32
Quantitative Real-time PCR analysis
A synchronous population of wild-type animals was treated for two generations with bec-1, lgg-1, or L4440 empty vector RNAi. RNA was then isolated from adult animals (one day post L4) by a Trizol freeze-cracking method. cDNA was synthesized with a RETROscript random decamer kit (Ambion, Austin, TX) with 2 µg of total RNA as template. Quantitative real-time PCR was performed with SYBR green PCR master mix (Applied Biosystems, Foster City, CA) in an Applied Biosystems 7500-fast RT PCR instrument with a Rox passive-reference dye. Primers were constructed to amplify a 100 bp fragment of the bec-1 transcript, a 97 bp fragment of the lgg-1 transcript, or 114 bp fragment of the act-1 β-actin transcript, which was used as the endogenous control (housekeeping gene). Standard PCR amplification with the primer sets produced single bands migrating at the correct size. Fold-expression changes were calculated with the formula 2−ΔΔCT, where ΔΔCT is the RNAi ΔCT [RNAi cycle threshold(CT) value – ACT-1 CT value] subtracted by the control ΔCT [L4440 cycle threshold(CT) value – ACT-1 CT value]. In all cases, raw SYBR fluorescence values were normalized to the passive-reference dye ROX. Primer sequences were as follows:
bec-1 – Forward: CCCATCTGATGCTCCAGTTT
Reverse: CAACTGCAAGAATCGACGAA
lgg-1 – Forward: TCCAACTTCGTCCAGAAGATGCTC
Reverse: TGCTGATGGTCCTGGTAGAGTTGT
act-1 – Forward: GCTGGACGTGATCTTACTGATTACC
Reverse: GTAGCAGAGCTTCTCCTTGATGTC
RESULTS
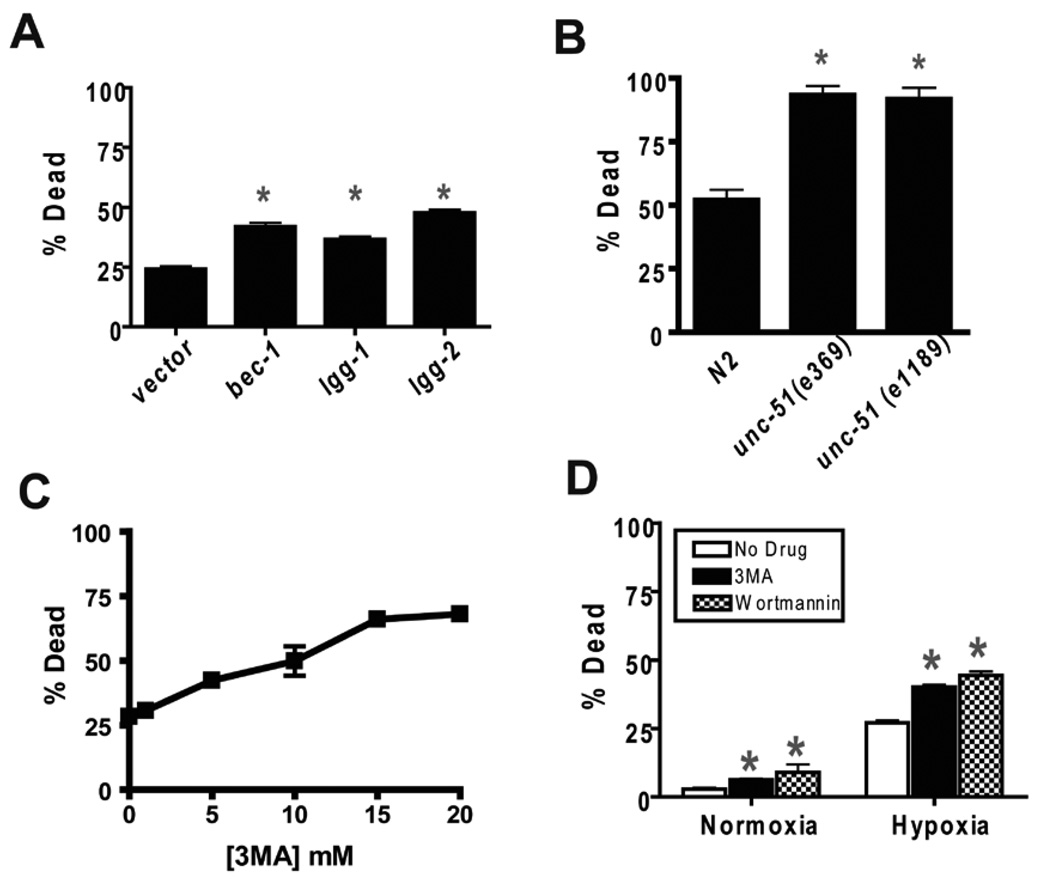
Two distinct steps in the autophagy pathway were disrupted in order to test the role of autophagy in hypoxic injury in C. elegans. bec-1 encodes the C. elegans homolog of beclin1/Atg6.4, 42, 43 unc-51 encodes an ortholog of Atg14, 44. Beclin1 and Atg1 promote initiation of autophagy, and their C. elegans homologs have been shown to be required for normal autophagy4, 44. lgg-1 and lgg-2 encode C. elegans homologs of LC3/Atg8. LC3 is critical for autophagosome formation and maturation. To test the role of these genes in hypoxic sensitivity, we used RNAi constructs against bec-1, lgg-1, and lgg-2 and tested unc-51 loss-of-function mutants. bec-1(RNAi) significantly increased hypoxia-induced death of wild type C. elegans compared to the empty vector control (Fig. 1A). Similarly, RNAi knockdown of lgg-1 and lgg-2 increased hypoxic death (Fig. 1A). Two unc-51(lf) mutants also had a hypoxia hypersensitivity phenotype (Fig. 1B). Thus, the autophagy genes, bec-1, lgg-1, lgg-2, and unc-51, promote organismal survival after severe hypoxia.
Figure 1.
Inhibition of autophagy increases hypoxic sensitivity. (A) Hypoxic sensitivity of animals treated with RNAi against bec-1, lgg-1, and lgg-2 autophagy genes compared to empty vector control. Second generation RNAi-treated adult animals were scored as alive or dead after a 24 hour recovery from a twelve hour hypoxic incubation. Error bars are mean ± sem of 5 trials. * p < 0.01 vs vector control, two-tailed t-test. (B) Hypoxic sensitivity of loss-of-function mutants of unc-51. % dead following recovery from a twelve hour hypoxic incubation. The N2 control animals were scored contemporaneously with the unc-51 mutants. * p < 0.01 vs N2, two-tailed t-test. (C) Concentration/response curve for the effect of 3-methyladenine (3MA) on hypoxic sensitivity. Adult animals were transferred into M9 buffer containing the indicated concentrations of 3-MA then immediately placed into the hypoxia chamber for twelve hours, recovered on agar plates without 3MA for 24 hours then scored as alive or dead. Error bars are mean ± sem of 5 trials. (D) Effect of 3MA (10 mM) and wortmannin (100 µM) on death after normoxic or hypoxic incubation. Wortmannin experiments are otherwise identical to that described for 3MA. Bars are mean ± sem of 9 trials for 3MA and 3 trials for wortmannin. * p < 0.01 vs No Drug, two-tailed t-test.
Because bec-1 is required for normal development,42 the hypoxic hypersensitivity of bec-1 inactivation might not be due to an acute requirement for BEC-1 but rather to some temporally remote developmental effect. To inactivate autophagy acutely, we used the type III phosphatidylinsositol-3 kinase (PI-3 kinase) inhibitor 3-methyladenine (3MA). 3MA is a widely-used inhibitor of autophagy.45–47 When given to adults just prior to hypoxic incubation, 3MA dose dependently increased the lethality of hypoxia with an EC50 of approximately 10 mM (Fig. 1C); this concentration is similar to that required to inhibit autophagy in mammalian cells.47 Multiple trials with 10 mM 3MA confirmed that it significantly increased hypoxic death (Fig. 1D). Wortmannin, another PI-3 kinase inhibitor that inhibits autophagy,46, 48 also increased hypoxic lethality in adults (Fig. 1D). These data demonstrate that autophagy is required just prior to, during, or after hypoxia for normal recovery from a hypoxic insult. Notably, both 3MA and Wortmannin produced a relatively small but nevertheless significant increase in death following normoxic incubation as well. The normoxic incubation is performed in liquid at 26°C but the tubes are placed in a normoxic incubator. Thus, both 3MA and Wortmannin sensitize to this temperature stress as well. Thus, the increased death observed after hypoxic incubation in 3MA and Wortmannin likely represents sensitization to both temperature and hypoxia.
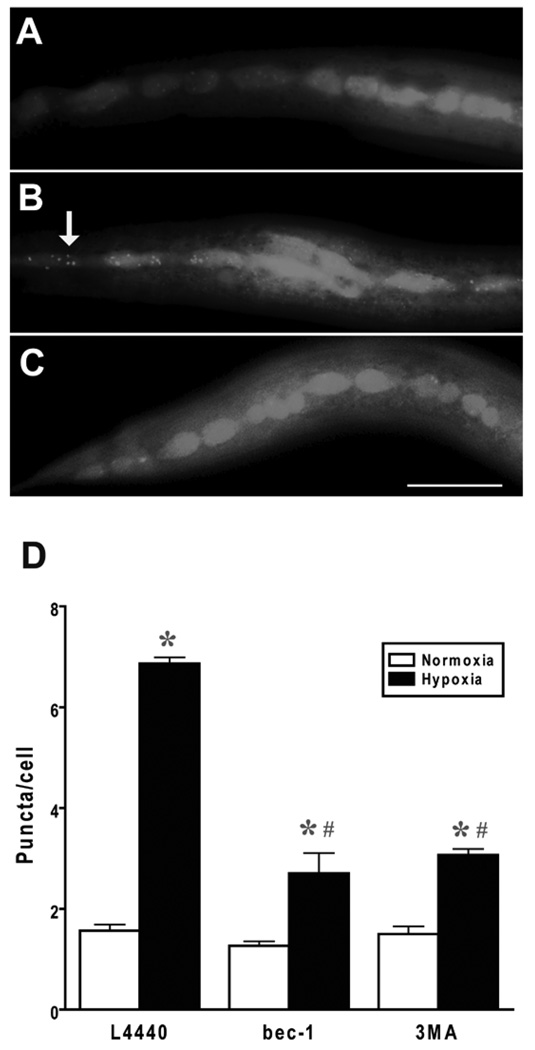
Autophagy is required for a normal recovery from hypoxic injury in C. elegans but does autophagic activity respond in some way to hypoxic injury? To address this question, we used GFP-labeled LGG-1, which is concentrated on autophagosomes and has been shown in several studies to be a reliable indicator of autophagosome number in C. elegans.4, 44, 49–51 GFP::LGG-1 puncta were markedly increased by a sublethal hypoxic insult (Fig. 2). This induction of puncta was diminished but not abolished by both bec-1(RNAi) and 3MA consistent with the puncta being a marker of autophagosomes. To quantitate the level of knockdown of bec-1 or lgg-1 expression by the respective RNAi, we performed quantitative RT-PCR on adult worms that had been cultured for two generations on bec-1(RNAi) or lgg-1(RNAi). We were not able to get reliable data with primers against lgg-2. Compared to growth on empty vector, the abundance of bec-1 transcript was 57% and 49% in bec-1(RNAi)-treated animals in two independent experiments. For lgg-1(RNAi), the transcript levels were 55% and 74% relative to empty vector. Knockdown is clearly incomplete; thus, a reduced, but not absent, level of autophagy is responsible for the hypoxic hypersensitivity phenotype.
Figure 2.
Effect of hypoxia on autophagosome density. Autophagosomes were labeled by LGG-1::GFP as described previously 4 and the number of GFP-labeled puncta were scored after recovery from a twelve hour normoxic or hypoxic incubation by an observer blinded to condition. (A) Normoxic vector-treated animal with diffuse LGG-1::GFP expression in hypodermal seam cells. (B) Hypoxic vector-treated animal with multiple LGG-1::GFP puncta visible (arrow). (C) Hypoxic bec-1(RNAi) animal with a reduction in LGG-1::GFP puncta. (D) Quantification of puncta/seam cell. # animals: Vector normoxia – 21, Vector hypoxia – 20, bec-1(RNAi) normoxia – 7, bec-1(RNAi) hypoxia – 9. * p < 0.01 vs normoxia; # p < 0.01 vs vector. Scale bar = 20 µm.
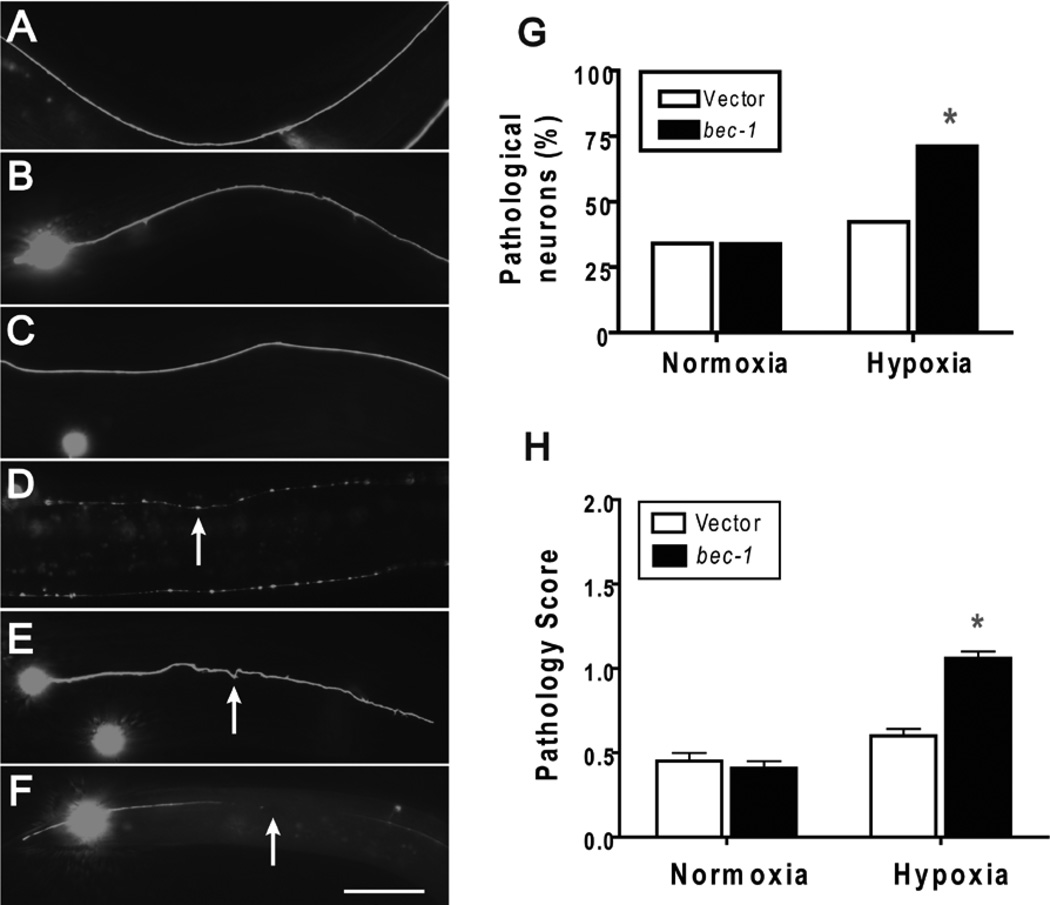
Neurons and myocytes are two cell types that are particularly prone to hypoxic injury in other organisms. In addition, the determinants of hypoxic injury of these cell types are of considerable clinical interest because they are injured in common human diseases such as stroke and ischemic heart disease. In the case of neuronal injury in C. elegans, previously we have observed axonal beading in animals that survive a sublethal hypoxic incubation.32, 37 We tested whether reduction of bec-1 activity influences this axonal pathology by examining mechanosensory axons labeled with GFP. bec-1(RNAi) increased hypoxia-induced axonal beading of the type observed previously (Fig. 3A–D), but we also observed severe axonal tortuousity and large gaps in the axons (Fig. 3E,F). Pathology of any of three types was significantly more prevalent (Fig. 3G) and more severe (Fig. 3H) in animals treated with bec-1(RNAi). Beading, tortuousity, and gaps are typical of traumatic and ischemic axonal pathology in mammalian models,52–55 but only beading or complete loss of the neuron had been previously observed in C. elegans. All three axonal pathological features are thought to be ultimately a consequence of loss of membrane integrity and dysfunctional axonal transport.56, 57
Figure 3.
Effect of bec-1 knockdown on hypoxic axonal pathology. Wild type larval animals with vector or bec-1 RNAi treatment underwent a twelve hour hypoxic or normoxic incubation then scored after recovery for axonal pathology by an observer blinded to condition. pMEC-4::GFP was used to visualize mechanosensory axons. Axonal beading, tortuosity, and gaps were scored. (A) Normoxic vector-treated animal showing normal axonal morphology. (B) Hypoxic vector-treated animal showing little pathological changes. (C) bec-1(RNAi) normoxic animal with normal axonal morphology. (D) bec-1(RNAi) hypoxic animal with axonal beading. Arrow indicates a bead. (E) bec-1(RNAi) hypoxic animal with tortuous axon pathology. Arrow indicates tortuous axon. (F) bec-1(RNAi) hypoxic animal with large axon gap. Arrow indicates gap. (G) The prevalence of pathological neurons following hypoxia is increased by bec-1(RNAi). % of neurons with any pathology was scored. * - p < 0.01, Fisher’s exact test. (H) The severity of axonal pathology following hypoxia is increased in bec-1(RNAi) animals. The severity of the axonal pathology for each axon was scored on a 0 – 2 scale. Severe beading, tortuosity, and/or large gaps were scored as a 2, milder but present pathology as a 1, and no pathology a 0. Data are mean ± sem of at least 100 animals/condition scored by an observer blinded to condition. * - p < 0.01 vs vector control, two-tailed t-test. Scale bar = 20 µm.
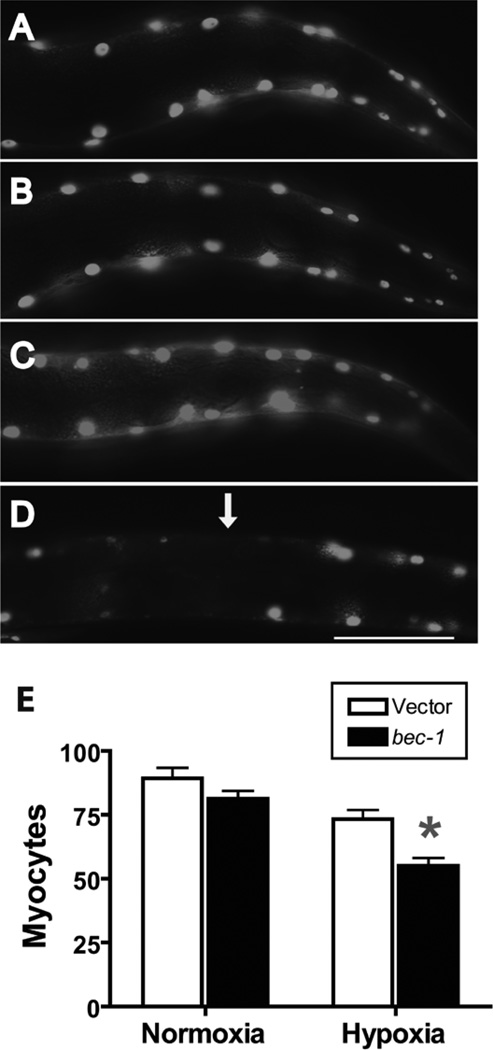
C. elegans has approximately 90 mononucleated myocytes that comprise the striated body wall muscle. Using a nuclear-localized GFP driven by a muscle-specific promoter, we have previously observed nuclear fragmentation and disappearance following a sublethal hypoxic insult32. bec-1(RNAi) significantly worsened the hypoxia-induced loss of myocyte nuclei (Fig. 4). Thus, BEC-1 functions to protect both neurons and myocytes from hypoxic injury and cell death.
Figure 4.
Effect of bec-1 knockdown on hypoxic muscle pathology. Wild type larval animals with vector or bec-1 RNAi treatment underwent a twelve hour hypoxic or normoxic incubation then scored after recovery for myocyte number. Nuclear-localized GFP expressed in body wall muscles was used to visualize and count myocytes. (A) Normoxic vector-treated animal showing normal myocyte nuclear morphology and number. (B) Hypoxic vector-treated animal showing normal myocyte nuclear morphology and number. (C) Normoxic bec-1(RNAi)-treated animal showing normal myocyte nuclear morphology and number. (D) Hypoxic bec-1(RNAi)-treated animal showing pale or absent nuclei (arrow). (E) Comparison of myocyte number in bec-1(RNAi) vs vector-treated animals after normoxic or hypoxic incubation. Data are mean ± sem of 35 animals/condition. * - p < 0.01 vs vector control. Scale bar = 20 µm.
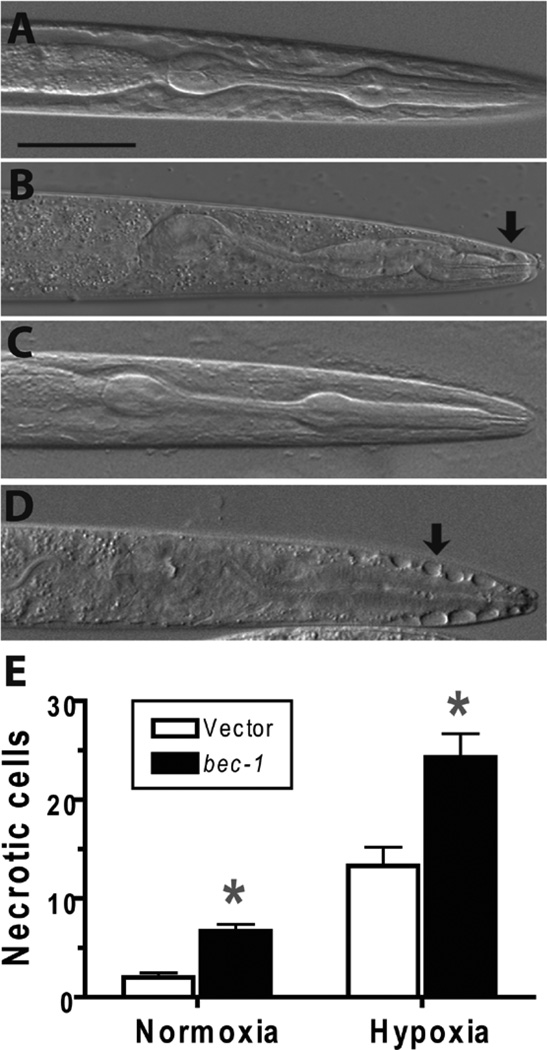
Mammalian cells die from both necrotic and apoptotic mechanisms after hypoxic/ischemic injury. Early after a hypoxic insult, most cells die a necrotic death whereas later apoptotic death tends to be more prevalent.58 In C. elegans, hypoxia results in necrosis of a subset of C. elegans cells particularly those within and around the pharynx.32 Necrotic cell death of neurons in C. elegans has been extensively studied using gain-of-function mutants in ion channel subunits.21 Interestingly, inhibition of autophagy suppresses ion channel-mediated necrosis of neurons.49 However the role of autophagy in hypoxia-induced necrotic cell death has not been examined in C. elegans and is relatively unexplored in mammalian models as well. After hypoxic incubation, bec-1(RNAi) animals had an increased number of necrotic cells compared to vector controls (Fig. 5). Given the distortion of the anatomy by the necrotic process, the type of cells dying by necrosis was not determined. Nevertheless, we can conclude that BEC-1 activity inhibits necrotic cell death after a hypoxic insult.
Figure 5.
Effect of bec-1 knockdown on necrotic pathology. Wild type larval animals with vector or bec-1 RNAi treatment underwent a twelve hour hypoxic or normoxic incubation then scored after recovery for myocyte number. Necrotic cells were visualized and counted using Nomarski optics. Cells with markedly swollen morphology and lack of nuclei were scored as necrotic. (A) Normoxic vector-treated animal showing normal cellular morphology in the pharyngeal region. (B) Hypoxic vector-treated animal showing a few necrotic-appearing cells (arrow). (C) Normoxic bec-1(RNAi) animal showing normal cellular morphology. (D) Hypoxic bec-1(RNAi)-treated animal showing multiple necrotic cells adjacent to the anterior pharynx (arrow). (E) Comparison of necrotic cell number in bec-1(RNAi) vs vector-treated animals after normoxic or hypoxic incubation. Data are mean ± sem of 35 animals/condition. * - p < 0.01 vs vector control. Scale bar = 20 µm.
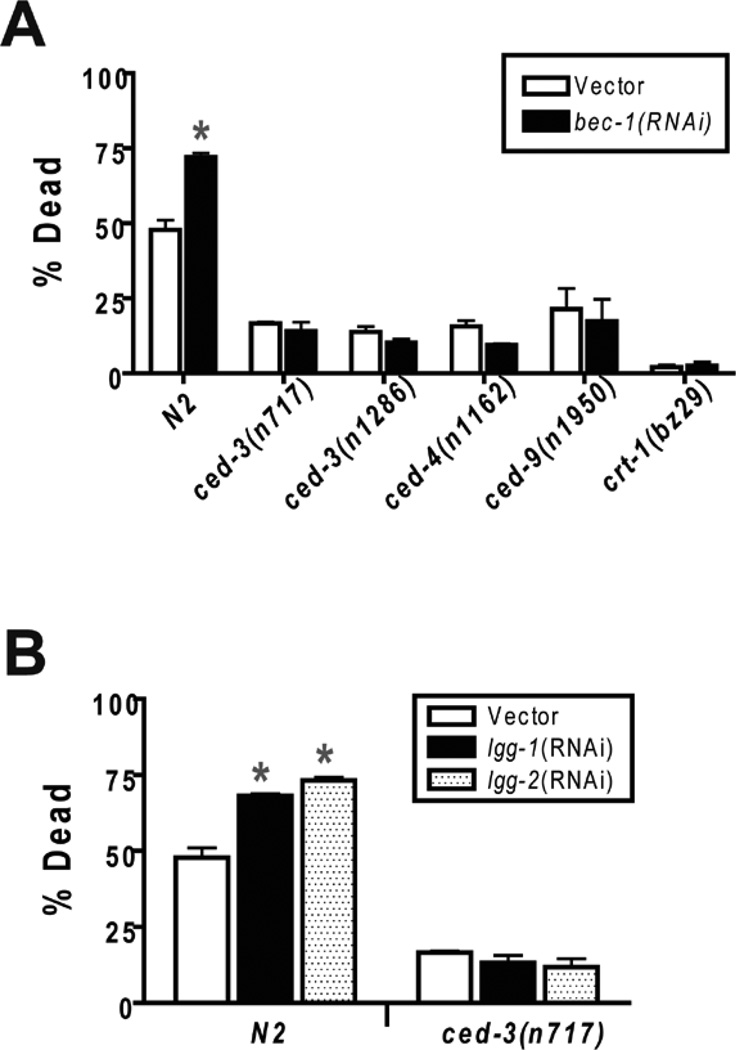
Apoptosis and autophagy interact in a complex manner.59 Cellular insults can induce both apoptosis and autophagy. Apoptosis and autophagy appear in many cases to be mutually inhibitory. Inhibition of autophagy unmasks apoptosis, and likewise apoptosis inhibition may enhance autophagic cell death. However, in other models, the pro-apoptotic protein Bnip3 appears to promote autophagy. Consistent with mutual inhibition, reduction of BEC-1 activity in C. elegans was found to increase the number of apoptotic cell corpses in both the germline and in developing embryos.42 The increase in apoptosis in bec-1 mutant animals could be blocked by a loss-of-function mutation of ced-3, the canonical C. elegans caspase gene. We have previously shown that loss-of-function mutations in the pathway leading to CED-3-mediated apoptosis reduces C. elegans hypoxic death.37 Thus, a reasonable hypothesis for the mechanism of the hypoxia hypersensitive phenotype of bec-1(RNAi) animals is that knockdown of BEC-1 activity increases apoptotic cell death and thereby sensitize animals to hypoxic injury. We tested this hypothesis by measuring hypoxic death in bec-1(RNAi) animals in wild type versus apoptosis-defective mutant backgrounds. As reported previously, a ced-9(gf) mutant, a ced-4(lf) mutant, and two ced-3(lf) mutants were hypoxia resistant relative to the wild type strain N2 (Fig. 6A). Importantly, the hypoxia resistance of all of the tested apoptosis mutants was not diminished by bec-1(RNAi) treatment. Thus, the hypoxic hypersensitivity of bec-1(RNAi) is not apparent when apoptosis is blocked. Similarly, neither lgg-1 nor lgg-2 RNAi increased the hypoxic sensitivity of a ced-3(lf) mutant (Fig. 6B).
Figure 6.
Epistatic relationship between autophagy pathway RNAi’s and apoptosis pathway and calreticulin mutants. Second generation RNAi- or vector control-treated adult wild type or mutant animals were scored as alive or dead after recovery from a twelve hour hypoxic incubation. (A) Apoptosis pathway and calreticulin mutants are hypoxia resistant and are epistatic to bec-1(RNAi). Error bars are mean ± sem of at least 4 trials for each condition and genotype. * different from vector control, p < 0.01, two-tailed t-test. (B) Epistatic relationship of lgg-1 and lgg-2 RNAi with a ced-3(lf) mutant. Error bars are mean ± sem of at least 4 trials for each condition and genotype. * different from vector control, p < 0.01, two-tailed t-test.
To ask whether this epistatic relationship is unique to apoptosis mutants or might also be shared by mutations that suppress necrotic cell death, we tested the effect of bec-1(RNAi) in a loss-of-function mutant of crt-1(bz29). crt-1 encodes a calreticulin, and crt-1 mutants have been shown to suppress ion-channel-mediated necrosis24. crt-1(bz29lf) was markedly hypoxia resistant, and bec-1(RNAi) was not effective at suppressing its resistance (Fig. 6A).
DISCUSSION
We have shown that inhibition of autophagy is deleterious to the survival of C. elegans following a severe hypoxic insult. A reduction in the activity of autophagy genes not only increases animal death but also worsens the cellular pathology seen after hypoxia. Following a damaging hypoxic incubation, we observed an increase in punctated LGG-1 expression in presumptive autophagosomes, an observation consistent with the induction of autophagic activity in response to hypoxic injury. Finally, we found that the hypoxic hypersensitivity produced by knockdown of autophagy in wild type animals is not observed in apoptosis pathway mutants or in a mutant that suppresses ion-channel-mediated necrosis. For discussion, we want to place ours and other autophagy findings in C. elegans in the context of what is known about autophagy and hypoxia/ischemia in other organisms.
A number of recent studies have examined the relationship between autophagy and hypoxic/ischemic injury in mammalian cells. Hypoxia and/or ischemia has been shown consistently to result in an increase in the number of autophagosomes in various models. For example, Yan and coinvestigators showed that ischemia increased markers of autophagy in pig myocardium.11 Similarly, autophagic activity was increased after cerebral hypoxia/ischemia in mice.60 However, whether induction of autophagy is pro-adaptive or maladaptive was not addressed in these studies. Subsequent studies indicate that autophagy can either promote or prevent cell death following hypoxia/ischemia. Simulated ischemia and reperfusion of rat primary cardiac myocytes was shown to induce autophagosome-like vesicles, and cardiac myocytes death following simulated ischemia/reperfusion was ameliorated by both 3MA and beclin1 siRNA treatments.14 Consistent with the promotion of hypoxic/ischemic cell death by autophagy, this study also showed that beclin1 overexpression increased myocyte death. Mice challenged with carotid ligation followed by a mild hypoxic exposure were found to have increased autophagosome numbers in hippocampal neurons, and a strain with selective neuronal knockout of the Atg7 gene had reduced hippocampal neuronal death following hypoxia/ischemia.15 Finally, in a pure hypoxia-induced cell death protocol, multiple mammalian cell lines were made hypoxia resistant by 3MA, beclin1 siRNA, or Atg5 siRNA.61
The evidence for a protective role of autophagy in mammalian cells against hypoxic/ischemic cell death is less extensive. In a simulated hypoxia/ischemia experimental paradigm, a mouse cardiac permanent cell line was protected from apoptotic cell death by both beclin-1 and Atg5 overexpression.13, 62 These same investigators also showed that transfection of an Atg5 dominant negative construct increased apoptotic cell death.12, 13 Similarly, 3MA treatment resulted in an increased death of primary neonatal rat cardiac myocytes after hypoxia and reoxygenation.63 While extensive evidence supports the conclusion that basal levels of autophagy are important for the health of neurons,64–66 whether autophagy can protect mammalian neurons following hypoxic/ischemic insults is unclear. Thus, the available evidence in mammalian cells indicates that autophagy can serve either a destructive or protective role following a hypoxic/ischemic insult. The factors that regulate whether activation of autophagy enhances or inhibits cell death following hypoxia are unknown.
As in mammalian cell types, autophagy in C. elegans can either be pro-adaptive or maladaptive. A regulatory role for autophagy has been particularly well established for C. elegans lifespan where autophagy appears to promote long life. The long lifespan phenotype of reduction-of-function mutations in the daf-2 gene, which encodes an insulin/IGF receptor homolog that limits lifespan, is strongly suppressed by reduction-of-function mutations and RNAi knockdown of C. elegans autophagy genes.4 Additionally, caloric restriction-induced lifespan extension requires an intact autophagy pathway.51, 67, 68
For survival from severe caloric restriction, that is starvation, the role of autophagy appears to be more complex. Prolonged and severe starvation is lethal to developing C. elegans,69, 70 and the lethality of starvation is increased by inhibition of bec-1. 70 However, C. elegans mutants where muscarinic signaling in the pharynx is hyperactivated have increased death during starvation, and this hypersensitivity to starvation can be ameliorated by bec-1 RNAi.70 Hyperactivation of muscarinic signaling was shown to increase markers of autophagy above background levels, consistent with the hypothesis that excessive autophagy can promote death from starvation. Thus, as for hypoxia-induced death in mammals, autophagy can either prevent or promote death from starvation depending on regulatory factors, perhaps one of which is second messenger pathways such as that activated by muscarinic receptors in C. elegans.
Finally, we would like to consider our finding that the hypoxic hypersensitivity produced by a reduction in autophagy is dependent on the activity the apoptosis and necrosis pathways. A direct interaction between autophagy and apoptosis was initially demonstrated by the finding that beclin-1 binds to Bcl-2 in mammalian cells.71 These investigators subsequently showed that Bcl-2 acts to inhibit beclin-1-mediated autophagy.72 In C. elegans, BEC-1 was shown to interact with the Bcl-2 homolog CED-9, and inactivation of bec-1 produced ectopic apoptotic cell death.42 We found that the hypoxic hypersensitivity phenotype of bec-1(RNAi) animals is suppressed by loss-of-function mutants in the apoptosis pathway. Thus, our results are consistent with bec-1 inactivation enhancing apoptotic death following hypoxia, perhaps through a direct disinhibition of the canonical apoptosis pathway. However, a hypoxia-resistant necrosis-defective mutation also suppressed the hypoxic hypersensitivity of bec-1(RNAi). Thus, the suppression by the apoptosis mutants can just as easily be explained by an indirect mechanism where a block of terminal cell death effectors, whether necrotic or apoptotic, negates the sensitization produced by a partial inhibition of autophagy. These complex genetic interactions suggest that the activity of apoptotic and necrotic mediators in cells injured by hypoxia will influence the effect of the autophagy pathway in cell death and survival.
Acknowledgements
Supported by NINDS (R01 NS045905), an American Heart Association Established Investigator Award, and a McKnight Endowment Fund for Neuroscience, Neuroscience of Brain Disorders Award.
Abbreviations
- 3MA
3-methyladenine
- GFP
Green fluorescent protein
- RNAi
Double-stranded RNA-mediated interference
REFERENCES
- 1.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Lum JJ, DeBerardinis RJ, Thompson CB. Autophagy in metazoans: Cell survival in the land of plenty. Nat Rev Mol Cell Biol. 2005;6:439–448. doi: 10.1038/nrm1660. [DOI] [PubMed] [Google Scholar]
- 3.Nair U, Klionsky DJ. Molecular mechanisms and regulation of specific and nonspecific autophagy pathways in yeast. J Biol Chem. 2005;280:41785–41788. doi: 10.1074/jbc.R500016200. [DOI] [PubMed] [Google Scholar]
- 4.Melendez A, Talloczy Z, Seaman M, Eskelinen E-L, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in c. Elegans. Science. 2003;301:1387–1391. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 5.Rusten TE, Lindmo K, Juhasz G, Sass M, Seglen PO, Brech A, Stenmark H. Programmed autophagy in the drosophila fat body is induced by ecdysone through regulation of the pi3k pathway. Dev Cell. 2004;7:179–192. doi: 10.1016/j.devcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 6.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the drosophila fat body. Dev Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rami A, Langhagen A, Steiger S. Focal cerebral ischemia induces upregulation of beclin 1 and autophagy-like cell death. Neurobiol Dis. 2008;29:132–141. doi: 10.1016/j.nbd.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 11.Yan L, Vatner DE, Kim SJ, Ge H, Masurekar M, Massover WH, Yang G, Matsui Y, Sadoshima J, Vatner SF. Autophagy in chronically ischemic myocardium. Proc Natl Acad Sci USA. 2005;102:13807–13812. doi: 10.1073/pnas.0506843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamacher-Brady A, Brady NR, Gottlieb RA. Enhancing macroautophagy protects against ischemia/reperfusion injury in cardiac myocytes. J Biol Chem. 2006;281:29776–29787. doi: 10.1074/jbc.M603783200. [DOI] [PubMed] [Google Scholar]
- 13.Hamacher-Brady A, Brady NR, Logue SE, Sayen MR, Jinno M, Kirshenbaum LA, Gottlieb RA, Gustafsson AB. Response to myocardial ischemia/reperfusion injury involves bnip3 and autophagy. Cell Death Differ. 2007;14:146–157. doi: 10.1038/sj.cdd.4401936. [DOI] [PubMed] [Google Scholar]
- 14.Valentim L, Laurence KM, Townsend PA, Carroll CJ, Soond S, Scarabelli TM, Knight RA, Latchman DS, Stephanou A. Urocortin inhibits beclin1-mediated autophagic cell death in cardiac myocytes exposed to ischaemia/reperfusion injury. Journal of Molecular and Cellular Cardiology. 2006;40:846–852. doi: 10.1016/j.yjmcc.2006.03.428. [DOI] [PubMed] [Google Scholar]
- 15.Koike M, Shibata M, Tadakoshi M, Gotoh K, Komatsu M, Waguri S, Kawahara N, Kuida K, Nagata S, Kominami E, Tanaka K, Uchiyama Y. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am J Pathol. 2008;172:454–469. doi: 10.2353/ajpath.2008.070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu CT. Eaten alive: Autophagy and neuronal cell death after hypoxia-ischemia. Am J Pathol. 2008;172:284–287. doi: 10.2353/ajpath.2008.071064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gustafsson AB, Gottlieb RA. Eat your heart out: Role of autophagy in myocardial ischemia/reperfusion. Autophagy. 2008;4 doi: 10.4161/auto.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzstein MM, Stanfield GM, Horvitz HR. Genetics of programmed cell death in c. Elegans: Past, present and future. Trends Genet. 1998;14:410–416. doi: 10.1016/s0168-9525(98)01573-x. [DOI] [PubMed] [Google Scholar]
- 19.Artal-Sanz M, Samara C, Syntichaki P, Tavernarakis N. Lysosomal biogenesis and function is critical for necrotic cell death in caenorhabditis elegans. J Cell Biol. 2006;173:231–239. doi: 10.1083/jcb.200511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Syntichaki P, Samara C, Tavernarakis N. The vacuolar h+-atpase mediates intracellular acidification required for neurodegeneration in c. Elegans. Curr Biol. 2005;15:1249. doi: 10.1016/j.cub.2005.05.057. [DOI] [PubMed] [Google Scholar]
- 21.Syntichaki P, Tavernarakis N. The biochemistry of neuronal necrosis: Rogue biology? Nat Rev Neurosci. 2003;4:672–684. doi: 10.1038/nrn1174. [DOI] [PubMed] [Google Scholar]
- 22.Driscoll M, Gerstbrein B. Dying for a cause: Invertebrate genetics takes on human neurodegeneration. Nat Rev Genet. 2003;4:181–194. doi: 10.1038/nrg1018. [DOI] [PubMed] [Google Scholar]
- 23.Syntichaki P, Xu K, Driscoll M, Tavernarakis N. Specific aspartyl and calpain proteases are required for neurodegeneration in c. Elegans. Nature. 2002;419:939–944. doi: 10.1038/nature01108. [DOI] [PubMed] [Google Scholar]
- 24.Xu K, Tavernarakis N, Driscoll M. Necrotic cell death in c. Elegans requires the function of calreticulin and regulators of ca(2+) release from the endoplasmic reticulum. Neuron. 2001;31:957–971. doi: 10.1016/s0896-6273(01)00432-9. [DOI] [PubMed] [Google Scholar]
- 25.Sugimoto T, Mori C, Takanami T, Sasagawa Y, Saito R, Ichiishi E, Higashitani A. Caenorhabditis elegans par2.1/mtssb-1 is essential for mitochondrial DNA replication and its defect causes comprehensive transcriptional alterations including a hypoxia response. Exp Cell Res. 2008;314:103–114. doi: 10.1016/j.yexcr.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Shen C, Shao Z, Powell-Coffman JA. The caenorhabditis elegans rhy-1 gene inhibits hif-1 hypoxia-inducible factor activity in a negative feedback loop that does not include vhl-1. Genetics. 2006 doi: 10.1534/genetics.106.063594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers C, Persson A, Cheung B, de Bono M. Behavioral motifs and neural pathways coordinating o2 responses and aggregation in c. Elegans. Curr Biol. 2006;16:649. doi: 10.1016/j.cub.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 28.Mendenhall AR, LaRue B, Padilla PA. Glyceraldehyde-3-phosphate dehydrogenase mediates anoxia response and survival in caenorhabditis elegans. Genetics. 2006;174:1173–1187. doi: 10.1534/genetics.106.061390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen C, Nettleton D, Jiang M, Kim SK, Powell-Coffman JA. Roles of the hif-1 hypoxia-inducible factor during hypoxia response in caenorhabditis elegans. J Biol Chem. 2005;280:20580–20588. doi: 10.1074/jbc.M501894200. [DOI] [PubMed] [Google Scholar]
- 30.Yuan A, Santi CM, Wei A, Wang ZW, Pollak K, Nonet M, Kaczmarek L, Crowder CM, Salkoff L. The sodium-activated potassium channel is encoded by a member of the slo gene family. Neuron. 2003;37:765–773. doi: 10.1016/s0896-6273(03)00096-5. [DOI] [PubMed] [Google Scholar]
- 31.Shen C, Powell-Coffman JA. Genetic analysis of hypoxia signaling and response in c elegans. Ann N Y Acad Sci. 2003;995:191–199. doi: 10.1111/j.1749-6632.2003.tb03222.x. [DOI] [PubMed] [Google Scholar]
- 32.Scott BA, Avidan MS, Crowder CM. Regulation of hypoxic death in c. Elegans by the insulin/igf receptor homolog daf-2. Science. 2002;296:2388–2391. doi: 10.1126/science.1072302. [DOI] [PubMed] [Google Scholar]
- 33.Padilla PA, Nystul TG, Zager RA, Johnson AC, Roth MB. Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in caenorhabditis elegans. Mol Biol Cell. 2002;13:1473–1483. doi: 10.1091/mbc.01-12-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang H, Guo R, Powell-Coffman JA. The caenorhabditis elegans hif-1 gene encodes a bhlh-pas protein that is required for adaptation to hypoxia. Proc Natl Acad Sci USA. 2001;98:7916–7921. doi: 10.1073/pnas.141234698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. Elegans egl-9 and mammalian homologs define a family of dioxygenases that regulate hif by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 36.Jia B, Crowder CM. Volatile anesthetic preconditioning present in the invertebrate caenorhabditis elegans. Anesthesiology. 2008;108:426–433. doi: 10.1097/ALN.0b013e318164d013. [DOI] [PubMed] [Google Scholar]
- 37.Dasgupta N, Patel AM, Scott BA, Crowder CM. Hypoxic preconditioning requires the apoptosis protein ced-4 in c. Elegans. Curr Biol. 2007;17:1954–1959. doi: 10.1016/j.cub.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brenner S. The genetics of caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded rna in caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 40.Gitai Z, Yu TW, Lundquist EA, Tessier-Lavigne M, Bargmann CI. The netrin receptor unc-40/dcc stimulates axon attraction and outgrowth through enabled and, in parallel, rac and unc-115/ablim. Neuron. 2003;37:53–65. doi: 10.1016/s0896-6273(02)01149-2. [DOI] [PubMed] [Google Scholar]
- 41.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the caenorhabditis elegans genome using rnai. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 42.Takacs-Vellai K, Vellai T, Puoti A, Passannante M, Wicky C, Streit A, Kovacs AL, Muller F. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in c. Elegans. Curr Biol. 2005;15:1513–1517. doi: 10.1016/j.cub.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 43.Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. Presynaptic terminals independently regulate synaptic clustering and autophagy of gabaa receptors in caenorhabditis elegans. J Neurosci. 2006;26:1711–1720. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aladzsity I, Toth ML, Sigmond T, Szabo E, Bicsak B, Barna J, Regos A, Orosz L, Kovacs AL, Vellai T. Autophagy genes unc-51 and bec-1 are required for normal cell size in caenorhabditis elegans. Genetics. 2007;177:655–660. doi: 10.1534/genetics.107.075762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stroikin Y, Dalen H, Loof S, Terman A. Inhibition of autophagy with 3-methyladenine results in impaired turnover of lysosomes and accumulation of lipofuscin-like material. Eur J Cell Biol. 2004;83:583–590. doi: 10.1078/0171-9335-00433. [DOI] [PubMed] [Google Scholar]
- 46.Blommaart EFC, Krause U, Schellens JPM, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and ly294002 inhibit autophagy in isolated rat hepatocytes. European Journal of Biochemistry. 1997;243:240–246. doi: 10.1111/j.1432-1033.1997.0240a.x. [DOI] [PubMed] [Google Scholar]
- 47.Seglen PO, Gordon PB. 3-methyladenine: Specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. PNAS. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petiot A, Ogier-Denis E, Blommaart EFC, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in ht-29 cells. J Biol Chem. 2000;275:992–998. doi: 10.1074/jbc.275.2.992. [DOI] [PubMed] [Google Scholar]
- 49.Toth ML, Simon P, Kovacs AL, Vellai T. Influence of autophagy genes on ion-channel-dependent neuronal degeneration in caenorhabditis elegans. J Cell Sci. 2007;120:1134–1141. doi: 10.1242/jcs.03401. [DOI] [PubMed] [Google Scholar]
- 50.Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of beta-amyloid peptide in c. Elegans. Autophagy. 2007;3 doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 51.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in c. Elegans. PLoS Genetics. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beirowski B, Berek L, Adalbert R, Wagner D, Grumme DS, Addicks K, Ribchester RR, Coleman MP. Quantitative and qualitative analysis of wallerian degeneration using restricted axonal labelling in yfp-h mice. J Neurosci Methods. 2004;134:23–35. doi: 10.1016/j.jneumeth.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 53.Bareyre FM, Kerschensteiner M, Misgeld T, Sanes JR. Transgenic labeling of the corticospinal tract for monitoring axonal responses to spinal cord injury. Nat Med. 2005;11:1355–1360. doi: 10.1038/nm1331. [DOI] [PubMed] [Google Scholar]
- 54.McCarran WJ, Goldberg MP. White matter axon vulnerability to ampa/kainate receptor-mediated ischemic injury is developmentally regulated. J Neurosci. 2007;27:4220–4229. doi: 10.1523/JNEUROSCI.5542-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kerschensteiner M, Schwab ME, Lichtman JW, Misgeld T. In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat Med. 2005;11:572–577. doi: 10.1038/nm1229. [DOI] [PubMed] [Google Scholar]
- 56.Medana IM, Esiri MM. Axonal damage: A key predictor of outcome in human cns diseases. Brain. 2003;126:515–530. doi: 10.1093/brain/awg061. [DOI] [PubMed] [Google Scholar]
- 57.Stys PK. General mechanisms of axonal damage and its prevention. J Neurol Sci. 2005;233:3–13. doi: 10.1016/j.jns.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 58.Dirnagl U, Simon RP, Hallenbeck JM. Ischemic tolerance and endogenous neuroprotection. Trends Neurosci. 2003;26:248–254. doi: 10.1016/S0166-2236(03)00071-7. [DOI] [PubMed] [Google Scholar]
- 59.Galluzzi L, Miguel J, Kepp VO, Tasdemir E, Maiuri MC, Kroemer G. To die or not to die: That is the autophagic question. Curr Mol Med. 2008;8:78–91. doi: 10.2174/156652408783769616. [DOI] [PubMed] [Google Scholar]
- 60.Zhu C, Wang X, Xu F, Bahr BA, Shibata M, Uchiyama Y, Hagberg H, Blomgren K. The influence of age on apoptotic and other mechanisms of cell death after cerebral hypoxia-ischemia. Cell Death Differ. 2005;12:162–176. doi: 10.1038/sj.cdd.4401545. [DOI] [PubMed] [Google Scholar]
- 61.Azad MB, Chen Y, Henson ES, Cizeau J, McMillan-Ward E, Israels SJ, Gibson SB. Hypoxia induces autophagic cell death in apoptosis-competent cells through a mechanism involving bnip3. Autophagy. 2007;4 doi: 10.4161/auto.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hamacher-Brady A, Brady NR, Gottlieb RA, Gustafsson AB. Autophagy as a protective response to bnip3-mediated apoptotic signaling in the heart. Autophagy. 2006;2:307–309. doi: 10.4161/auto.2947. [DOI] [PubMed] [Google Scholar]
- 63.Dosenko VE, Nagibin VS, Tumanovska LV, Moibenko AA. Protective effect of autophagy in anoxia-reoxygenation of isolated cardiomyocyte? Autophagy. 2006;2:305–306. doi: 10.4161/auto.2946. [DOI] [PubMed] [Google Scholar]
- 64.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 65.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 66.Komatsu M, Wang QJ, Holstein GR, Friedrich VL, Jr, Iwata J, Kominami E, Chait BT, Tanaka K, Yue Z. Essential role for autophagy protein atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci USA. 2007;104:14489–14494. doi: 10.1073/pnas.0701311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in c. Elegans. Autophagy. 2007;3:597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 68.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in caenorhabditis elegans. Autophagy. 2008;4:330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 69.You YJ, Kim J, Cobb M, Avery L. Starvation activates map kinase through the muscarinic acetylcholine pathway in caenorhabditis elegans pharynx. Cell Metab. 2006;3:237–245. doi: 10.1016/j.cmet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–2171. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal sindbis virus encephalitis by beclin, a novel bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]