Abstract
Narezushi, a derivation of sushi, is a traditional Japanese food made by fermenting salted fish meat and cooked rice together. In this study, the microbial diversity of saba-narezushi (narezushi of mackerel, Scomber japonicus) was analyzed by the 16S ribosomal RNA gene clone library method. Chemical composition was also analyzed to compare with different kinds of narezushi. The chemical composition of the narezushi was similar to those obtained from samma-narezushi. Ninety-four clones were randomly selected and DNA sequences of cloned fragments (approx. 890 bp) were analyzed. The DNA sequences obtained were phylogenetically analyzed. The expected operational taxonomy units (OTUs) by Chao1 estimates and Shannon-Wiener index (H′) at 97% identity threshold were 48 and 1.822, respectively. The sequence similarity of the cloned fragment was equal to or higher than 98% of the sequence of cultivated bacterial species in the public database. Most of the clones (85%) belonged to lactic acid bacteria (LAB). Lactobacillus curvatus was the most abundant species followed by Lactococcus piscium and Leuconostoc gasicomitatum, suggesting that these bacteria play important roles in the fermentation of saba-narezushi.
Keywords: 16S rDNA clone library, Fermented food, Lactic acid bacteria (LAB), Mackerel (Scomber japonicus), Narezushi
Introduction
Fish proteins have an excellent essential amino acid composition, which is widely recommended for a well balanced and healthy diet (Zhang et al. 2010). Fermented fish foods are popular in many regions of Southeast and Northeast Asia (Ishige 1993). In this preparation, fish is processed as a preserved food mainly by lactic acid fermentation. This process allows a longer preservation period for perishable proteinaceous food by lowering the pH, and also allows transporting the product to large distances (Cooke et al. 1993). This type of food is also found in Japan and is called narezushi. Narezushi is made by fermenting salted fish meat with cooked rice for a certain period. The prototypical narezushi has long fermentation periods. In Japan, this type of narezushi is fermented for several months (Isobe et al. 2002; Itou et al. 2006). The fermentation period of narezushi was reduced during the history of the development of this food product. Currently, narezushi is fermented from 5 d to 2 months in various regions of Japan. In Mie Prefecture, Japan, several narezushi of various kinds of fish, such as narezushi of samma (saury, Cololabis saira) (Matsui et al. 2008), ayu (sweetfish, Plecoglossus altivelis altivelis), konoshiro (spotted sardine, Konosirus punctatus), and saba (mackerel, Scomber japonicus), are produced as ritual or celebratory foods. Since narezushi is empirically known to be an intestinal regulator, it is recognized as a functional food as well as culturally important food in the region where it is consumed.
Analysis of microbial ecology by the culture-dependent method is time-consuming and laborious. Because vast majority of microorganisms are uncultivable, this has been a considerable handicap to the microbial ecology (Head et al. 1998). Recent advances in molecular biology enable us to use genetic information for the analysis of microbial communities. The PCR-derived clone library method using 16S ribosomal RNA gene (16S rDNA) is often used as the first step in the analysis of unknown microbial communities in various environments, such as the digestive tract of humans (Hold et al. 2002), aquatic and terrestrial biospheres (Bernhard et al. 2005; Chan et al. 2006), and fermented foods (Abriouel et al. 2008; Kim and Chun 2005). The advantage of the PCR-derived clone library method over the culture dependent method is ability to uncultivated microorganisms. The exhaustive analysis of microbial diversity can be achieved by the PCR-derived clone library method. This is advantageous to other culture-independent methods such as denaturing gradient gel electrophoresis (DGGE), terminal-restriction fragment length polymorphism (T-RFLP) or fluorescent in situ hybridization (FISH). To date, studies on the diversity of microorganisms involved in the fermentation of narezushi have been done using the culture-dependent technique (Fujii et al. 1992; Isobe et al. 2002). Recently, we have published a microbial diversity analysis of samma-narezushi (narezushi of saury) by using the 16S rDNA clone library technique. We also demonstrated the usefulness of this technique in analyzing microbial diversity in narezushi (Matsui et al. 2008). Our results showed that Lactobacillus sakei is the main organism involved in the fermentation of samma-narezushi. Furthermore, we have detected uncultured LAB in ayu narezushi by using the 16S rDNA clone library technique (Matsui et al. 2010). Thus the advantage of the 16S rDNA clone library technique is obvious.
The purpose of this study was to analyze bacterial diversity in saba-narezushi (narezushi of mackerel) produced in Mie Prefecture. Bacteria involved in the fermentation of narezushi were estimated by the culture-independent 16S rDNA clone library technique.
Materials and methods
Narezushi sample
Saba-narezushi was obtained from a local manufacturer in Kiho town, Mie Prefecture, Japan, where it was produced as follows. After removing the head and internal organs, the mackerel was salted for 1 month and then pickled in a tub together with cooked rice for about 3 weeks in winter.
Analysis of chemical composition
Fish meat and rice were separately homogenized with a food processor. The proximate composition of the sample was measured by the general method of food analysis. Moisture content was determined by dryness at 105 °C. Protein content was calculated from the nitrogen content by the micro-Kjeldahl method, using a conversion factor of 6.24. Lipid content was determined using the Soxhlet method. Ash content was determined by incineration at 450 °C. Carbohydrate content was calculated by subtracting the moisture, ash, protein, and lipid content from the total weight. These determinations were carried out using 6 replicates. Lactic acid contents were determined by HPLC analysis with postcolumn detection. Triplicate samples were used for this determination.
DNA extraction, cloning, and sequencing
The sample was homogenized with a sterilized food processor, immediately suspended in acetone at a final concentration of 70% to avoid DNA degradation (Fukatsu 1999), and stored at −80 °C until analysis.
The stored samples were washed twice with sterilized physiological saline by centrifugation before DNA extraction. DNA was extracted by the bead-beating method, using the FastPrep instrument (Bio 101, Vista, CA, USA) as described by Godon et al. (1997). The crude DNA was purified with Genomic-tip 100/G (QIAGEN, Hilden, Germany) and dissolved in TE buffer. DNA concentration was adjusted to 10 ng μL−1.
The 16S ribosomal RNA gene fragments were amplified by PCR with a set of universal primers (530f and 1392r) (Matsui et al. 2008). PCR was performed using TaKaRa Ex Taq (TaKaRa, Otsu, Japan) with the PCR Thermal Cycler Dice TP600 (TaKaRa, Otsu, Japan). A total of 20 μL of the PCR reaction mixture contained 1 μL of template DNA, 1× Ex Taq reaction buffer, 200 μM each deoxynucleoside triphosphate (dNTP mixture), 0.5 unit of Ex Taq DNA polymerase, 0.5 μM of each primer, and 0.5 g L−1 bovine serum albumin. The following PCR cycle was used: an initial denaturation at 95 °C for 3 min, followed by 15 cycles of 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min with a final extension step at 72 °C for 10 min. After separation by using 1.0% agarose gel electrophoresis (in TAE buffer), the PCR products were confirmed by visualization with ethidium bromide staining and then cloned into TOP10 competent Escherichia coli by using the TA cloning kit (Invitrogen, Carlsbad, CA, USA) as per the manufacturer’s protocol. Positive clones were randomly selected from the clone library. The cloned DNA fragments were commercially sequenced from both directions with M13 reverse primer (5′-CAGGAAACAGCTATGAC-3′) and M13 forward (−20) primer (5′-GTAAAACGACGGCCAG-3′) by Shimadzu Corporation (Kyoto, Japan).
Homology search, phylogenetic analysis, and diversity analysis of nucleotide sequence
DNA sequences were examined for homology with the BLAST program (Altschul et al. 1997). Chimeric artifacts of PCR were checked with the CHECK_CHIMERA online program of the Ribosomal Database Project (RDP-II) and omitted from analysis (Cole et al. 2003). Operational taxonomy unit (OTU) assignment and calculation of Chao1 and Shannon-Wiener index (H′) were carried out with the FastGroupII online program (Yu et al. 2006). The DNA sequences of the cloned fragments were aligned with CLUSTAL X ver. 2.0 (Larkin et al. 2007), and phylogenetic trees were constructed using the neighbor-joining method (Saitou and Nei 1987). The stability of branches was checked by bootstrapping (1,000 resampling). Njplot ver. 2.3 was used to draw phylogenetic tree.
Nucleotide sequence accession numbers
All nucleic acid sequences obtained in this study were deposited in the DDBJ, EMBL, and GenBank databases under accession numbers AB326516–AB326609.
Results and discussion
In Japan, a wide variety of fermented foods such as pickles, rice wine (sake), soy sauce (shoyu), fermented soybean paste (miso), and natto are quite popular. These fermented products have attracted worldwide attention as foods that might promote longevity (Murooka and Yamashita 2008). Narezushi is a traditional fermented food containing live lactic acid bacteria (LAB). The LAB plays important roles in fermentation and possibly in human health. Therefore, it is important to clarify LAB in the narezushi to study their functionality.
Chemical composition
The chemical composition of saba-narezushi was analyzed to compare nutrient composition and degree of fermentation with different kinds of narezushi which have been previously reported (Matsui et al. 2008, 2010). In both fish meat and rice, the moisture content was more than 70%. The fish meat contained 20% proteins and 7.6% lipids, whereas carbohydrate was not detected (Table 1). On the other hand, rice contained approximately 18% carbohydrate. The rice portion has a slightly higher lactic acid composition than fish meat. We have reported the chemical composition of narezushi in the meat of saury fish (samma-narezushi) (Matsui et al. 2008). The nutrient composition of samma-narezushi was as follows: 19.7% protein, 3.9% lipid, and 3.5% carbohydrate in fish meat, and 2.5% protein, 0.1% lipid, and 18.1% carbohydrate in rice. The amount of lactic acid, a component that provides taste, in samma-narezushi was 0.6% in fish meat and 1.0% in rice. The values obtained in the present study were quite similar to those found in samma-narezushi (Table 1). The results suggest that significant growth of LAB and lactic acid fermentation occurred in saba-narezushi. The lactic acid would contribute sour taste of saba-narezushi. Fujii et al. (1992) analyzed the chemical composition of saba-narezushi produced in Wakayama Prefecture in Japan. The concentration of lactic acid in the fish meat and rice portions of the saba-narezushi fermented for 20 d in January was 0.37% and 0.49%, respectively. Although the fermentation period of saba-narezushi reported by Fujii et al. (1992) and that used in this study was similar, the lactic acid concentration was 2 times higher in the present report. This may be due to the differences in the composition of LAB. Itou et al. (2006) reported the chemical composition of mackerel narezushi, which uses the same kind of fish but is fermented for a much longer period (4 months) during summer, in Fukui Prefecture in Japan. The concentration of lactic acid in mackerel narezushi was 5% in fish meat and 3.3% in rice. The difference may be due to the variation in preparation: mackerel narezushi (another type of saba-narezushi) was pickled under high temperatures in summer (from May to September) for a longer period.
Table 1.
Chemical composition (%) of saba-narezushi (narezushi of mackerel). Values are mean ± standard deviation (n = 5)
| Moisture | Protein | Lipid | Ash | Carbohydrate | Lactic acid | |
|---|---|---|---|---|---|---|
| Fish meat | 72.6 ± 0.08 | 19.7 ± 0.89 | 7.6 ± 0.07 | 1.3 ± 0.06 | nd1 | 0.77 ± 0.22 |
| Rice | 78.2 ± 0.27 | 1.3 ± 0.40 | 0.12 ± 0.07 | 1.8 ± 0.05 | 18.7 | 0.99 ± 0.01 |
1Not detected
Since the carbohydrate content was less than the detectable level, the sum of moisture, protein, lipid, and ash was greater than 100% in fish meat
Diversity analysis by 16S ribosomal RNA clone library
A total of 94 clones were randomly selected from the 16S rDNA clone library of saba-narezushi, and DNA sequences of cloned fragments (approx. 890 bp) were analyzed. Homology search, phylogenetic analysis, and calculation of Chao-1 and Shannon-Wiener index (H′) were carried out. To the best of our knowledge, this is the first report on microbial diversity analysis of saba-narezushi by the PCR-derived clone library method. No chimeric sequence was found using the CHECK_CHIMERA program. Cloned sequences were classified into 16 OTUs at the 97% identity threshold and summarized in Table 2. All sequences in the library were eubacterial 16S rDNA sequences, and no archaeal or eukaryotic sequence was recovered. The sequence similarity of the cloned fragment was equal to or higher than 98% of the sequence of cultivated bacterial species in the public database. The expected OTU by Chao1 estimates and Shannon-Wiener index (H′) of saba-narezushi were 48 and 1.822, respectively. The Chao1 estimate and Shannon-Wiener index (H′) observed in saba-narezushi was higher than those in samma-narezushi (Matsui et al. 2008).
Table 2.
Abundance and similarity of OTU to known species of bacteria in saba-narezushi (narezushi of mackerel)
| OTU (number of clone) | Nearest known species (Accession Number) | Similarity (%) | Composition (%) |
|---|---|---|---|
| Lactobacillales | |||
| OTU-01 (47), OTU-02 (1) | Lactobacillus curvatus (AJ621550) | 99–100 | 51.1 |
| OTU-03 (13) | Lactococcus piscium (DQ343754) | 99 | 13.8 |
| OTU-04 (8) | Leuconostoc gasicomitatum (AF231131) | 100 | 8.5 |
| OTU-05 (3) | Lactococcus lactis subsp. lactis (X64887) | 99 | 3.2 |
| OTU-06 (3) | Vagococcus carniphilus (AY179329) | 98 | 3.2 |
| OTU-07 (2) | Carnobacterium divergens (AY543037) | 99 | 2.1 |
| OTU-08 (1), OTU-09 (1) | Lactobacillus fuchuensis (AB370875) | 99 | 2.1 |
| OTU-10 (1) | Lactobacillus sakei (EU626014) | 99 | 1.1 |
| Subtotal | 85.1 | ||
| Bacillales | |||
| OTU-11 (1), OTU-12 (1) | Brochothrix thermosphacta (AY543029) | 99–100 | 2.2 |
| Subtotal | 2.2 | ||
| γ-Proteobacteria | |||
| OTU-13 (7) | Hafnia alvei (AB244471) | 99 | 7.5 |
| OTU-14 (3) | Buttiauxella gaviniae (AJ233403) | 100 | 3.2 |
| OTU-15 (1) | Pseudomonas psychrophila (FJ950690) | 100 | 1.1 |
| OTU-16 (1) | Shewanella baltica (CP000891) | 99 | 1.1 |
| Subtotal | 12.9 | ||
| Total | 100 |
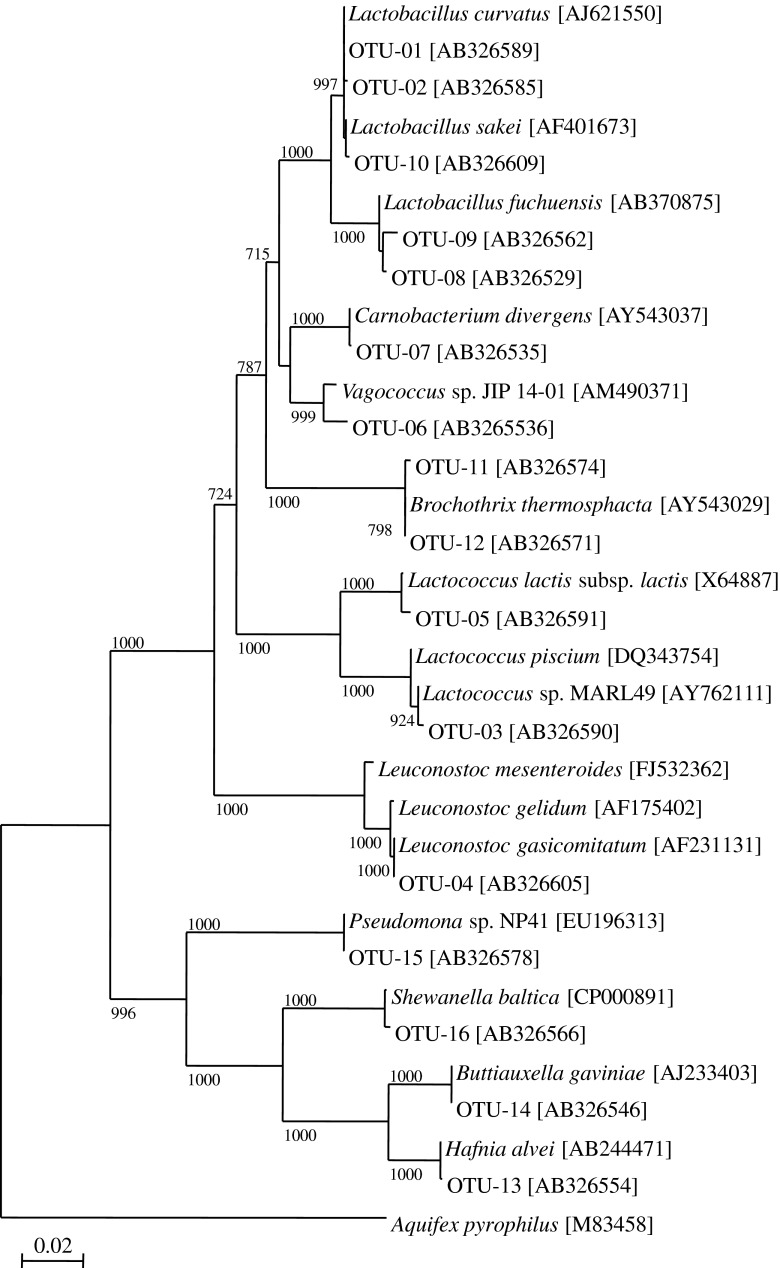
Based on phylogenetic placement and homology results, 85% of clones were assigned as LAB (Fig. 1; Table 2). These clones were classified into 10 OTUs (Table 2). OTU-01 and −02, which constitute 51.1% of the total number of clones, were classified as Lactobacillus curvatus. The second most abundant clone was OTU-03 (13.8% of the clones) and was classified as Lactococcus piscium. OTU-04 (8.5% of the clones) was classified as Leuconostoc gasicomitatum. The results suggest that these LABs play important roles in the fermentation of saba-narezushi. Although functional analysis of these LABs was not carried out in the present study, previous reports suggested that L. curvatus plays important roles in fermentation of foods (Vogel et al. 1993; Verluyten et al. 2004, Rotsatchakul et al. 2009). L. curvatus is best adapted to meat fermentations and dominate the flora during the process in the fermented sausage (Vogel et al. 1993). L. curvatus LTH1174 isolated from fermented sausage produces listericidal bacteriocin curvacinA suggesting that possible roles in suppressing Listeria growth in the sausage (Verluyten et al. 2004). Volatile compounds of nham, Thai fermented sausage, are produced by L. curvatus (Rotsatchakul et al. 2009). In a previous study, Lactobacillus plantarum, Lactobacillus alimentaria, and Lactobacillus coryniformis were isolated from saba-narezushi (Fujii et al. 1992). Another study showed that Lactobacillus buchneri was the predominant bacterial species in funazushi, which is fermented for 6 months (Isobe et al. 2002). Our results together with those of the previous studies suggest that bacteria belonging to the genus Lactobacillus play important roles in the fermentation of various narezushi. OTUs affiliated with Lactococcus lactis subsp. lactis, Vagococcus carniphilus, Carnobacterium divergens, and Lactobacillus fuchuensis were detected in low numbers among the clones (Table 2). In our previous study, 90% of the clones were LABs and most were classified as L. sakei in the clone library of samma-narezushi (Matsui et al. 2008). In contrast to this, Lactobacillus sakei was minor component in the present study. The reason for the small proportion of L. sakei in saba-narezushi is unclear.
Fig. 1.
Phylogenetic tree of the 16S rDNA fragment recovered from saba-narezushi (narezushi of mackerel). The scale bar represents 0.1 substitution per nucleotide position. Bootstrap values for 1,000 trees are shown at the branch points. Only values of 70% or above are shown. Numbers in parentheses are GenBank Accession numbers
OTU-11 and −12 were classified as Brochothrix thermosphacta (Bacillales). These OTUs composed 2.2% of the clones (Table 2). Four OTUs were assigned as γ-proteobacteria constituting 12.9% of the clones (Table 2). OTU-13 (7.5% of the clones) was classified as Hafnia alvei. OTU-14, −15, and −16 were classified as Buttiauxella, Pseudomonas psychrophila, and Shewanella baltica, respectively. These OTUs were minor components in the library. Fujii et al. (1992) detected Gram-negative rods in saba-narezushi. In the present study, Gram-negative rods such as H. alvei, Buttiauxella gaviniae, P. psychrophila, and S. baltica were also detected, suggesting that these organisms are minor but common constituents in saba-narezushi.
In our previous result, uncultured LAB was detected in ayu-narezushi (Matsui et al. 2010). Therefore, we have used culture-independent method rather than culture-dependent method. The present study shows that the PCR-derived clone library technique can be successfully applied to diversity analysis of bacteria in saba-narezushi. Because the technique is semi-quantitative analysis, the population density of saba-narezushi should be determined by quantitative technique such as real-time PCR. Lactobacillus brevis FPTLB3 isolated from freshwater fish produced bacteriocin that inhibited Escherichia coli, Enterococcus faecalis, L. sakei and Staphylococcus aureus (Banerjee et al. 2011). The isolate can potentially be used for biopreservation of the food. Isolation and phenotypic characterization of bacteria in saba-narezushi should be done to clarify their function in the fermentation process.
Acknowledgements
The present study was financially supported by Grants-in-Aid for Scientific Research, Japan Society for the Promotion of Science (15500536).
References
- Abriouel H, Martín-Platero A, Maqueda M, Valdivia E, Martínez-Bueno M. Biodiversity of the microbial community in a Spanish farmhouse cheese as revealed by culture-dependent and culture-independent methods. Int J Food Microbiol. 2008;127:200–208. doi: 10.1016/j.ijfoodmicro.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee SP, Dora KC, Chowdhury S. Detection, partial purification and characterization of bacteriocin produced by Lactobacillusibrevis FPTLB3 isolated from freshwater fish. J Food Sci Technol. 2011 doi: 10.1007/s13197-011-0240-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhard AE, Colbert D, McManus J, Field KG. Microbial community dynamics based on 16S rRNA gene profiles in a Pacific Northwest estuary and its tributaries. FEMS Microbiol Ecol. 2005;52:115–128. doi: 10.1016/j.femsec.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Chan OC, Yang X, Fu Y, Feng Z, Sha L, Casper P, Zou X. 16S rRNA gene analyses of bacterial community structures in the soils of evergreen broad-leaved forests in south-west China. FEMS Microbiol Ecol. 2006;58:247–259. doi: 10.1111/j.1574-6941.2006.00156.x. [DOI] [PubMed] [Google Scholar]
- Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–423. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke RD, Twiddy DR, Reilly APJ. Lactic fermentation of fish as a low-cost means of food preservation. In: Lee CH, Steinkraus KH, Reilly APJ, editors. Fish fermentation technology. Tokyo: United Nations University Press; 1993. pp. 291–300. [Google Scholar]
- Fujii T, Sasaki T, Okuzumi M. Chemical composition and microbial flora of saba-narezushi (fermented mackerel with rice) Nippon Suisan Gakkaishi Jpn. 1992;58:891–894. doi: 10.2331/suisan.58.891. [DOI] [Google Scholar]
- Fukatsu T. Acetone preservation: a practical technique for molecular analysis. Mol Ecol. 1999;8:1935–1945. doi: 10.1046/j.1365-294x.1999.00795.x. [DOI] [PubMed] [Google Scholar]
- Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head IM, Saunders JR, Pickup RW. Microbial evolution, diversity, and ecology: A decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- Hold GL, Pryde SE, Russell VJ, Furrie E, Flint HJ. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol. 2002;39:33–39. doi: 10.1111/j.1574-6941.2002.tb00904.x. [DOI] [PubMed] [Google Scholar]
- Ishige N. Cultural aspects of fermented fish products in Asia. In: Lee CH, Steinkraus KH, Reilly APJ, editors. Fish fermentation technology. Tokyo: United Nations University Press; 1993. pp. 13–32. [Google Scholar]
- Isobe Y, Mizuhashi T, Narita M. Microbial flora of funazushi (pickled crusian carp) Nippon Kasei Gakkaishi Jpn. 2002;53:61–64. [Google Scholar]
- Itou K, Kobayashi S, Ooizumi T, Akahane Y. Changes of proximate composition and extractive components in narezushi, a fermented mackerel product, during processing. Fish Sci. 2006;72:1269–1276. doi: 10.1111/j.1444-2906.2006.01285.x. [DOI] [Google Scholar]
- Kim M, Chun J. Bacterial community structure in kimchii, a Korean fermented vegetable food, as revealed by 16S rRNA gene analysis. Int J Food Microbiol. 2005;103:91–96. doi: 10.1016/j.ijfoodmicro.2004.11.030. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Matsui H, Tsuchiya R, Isobe Y, Maeda H, Narita M. Diversity of the bacterial community found in samma-narezushi (Saury Narezushi) revealed by the 16S rRNA gene clone library. Biocontrol Sci. 2008;13:97–102. doi: 10.4265/bio.13.97. [DOI] [PubMed] [Google Scholar]
- Matsui H, Saka E, Isobe Y, Narita M. Comparison of the bacterial community structures of ayu-narezushi produced by two different manufacturers. Biocontrol Sci. 2010;15:63–68. doi: 10.4265/bio.15.63. [DOI] [PubMed] [Google Scholar]
- Murooka Y, Yamashita M. Traditional healthful fermented products of Japan. J Ind Microbiol Biotechnol. 2008;35:791–798. doi: 10.1007/s10295-008-0362-5. [DOI] [PubMed] [Google Scholar]
- Rotsatchakul P, Visesanguan W, Smitinont T, Chaiseri S. Changes in volatile compounds during fermentation of nham (Thai fermented sausage) Int Food Res J. 2009;16:391–414. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Verluyten J, Leroy F, DeVuyst L. Effects of different spices used in production of fermented sausages on growth of and curvacin A production by Lactobacillus curvatus LTH1174. Appl Environ Microbiol. 2004;70:4807–4813. doi: 10.1128/AEM.70.8.4807-4813.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel RF, Lohmann M, Nguyen M, Weller AN, Hammes WP. Molecular characterization of Lactobacillus curvatus and Lact. sake isolated from sauerkraut and their application in sausage fermentations. J Appl Bacteriol. 1993;74:295–300. doi: 10.1111/j.1365-2672.1993.tb03029.x. [DOI] [PubMed] [Google Scholar]
- Yu Y, Breitbart M, McNairnie P, Rohwer F. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinform. 2006;7:57. doi: 10.1186/1471-2105-7-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Liu Z, Hu Z, Fang Z, Chen J, Wu D, Ye H. Effect of sucrose on the generation of free amino acids and biogenic amines in Chinese traditional dry-cured fish during processing and storage. J Food Sci Technol. 2010;48:69–75. doi: 10.1007/s13197-010-0130-1. [DOI] [PMC free article] [PubMed] [Google Scholar]