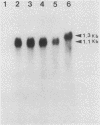
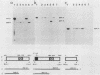
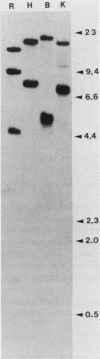
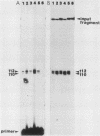
Abstract
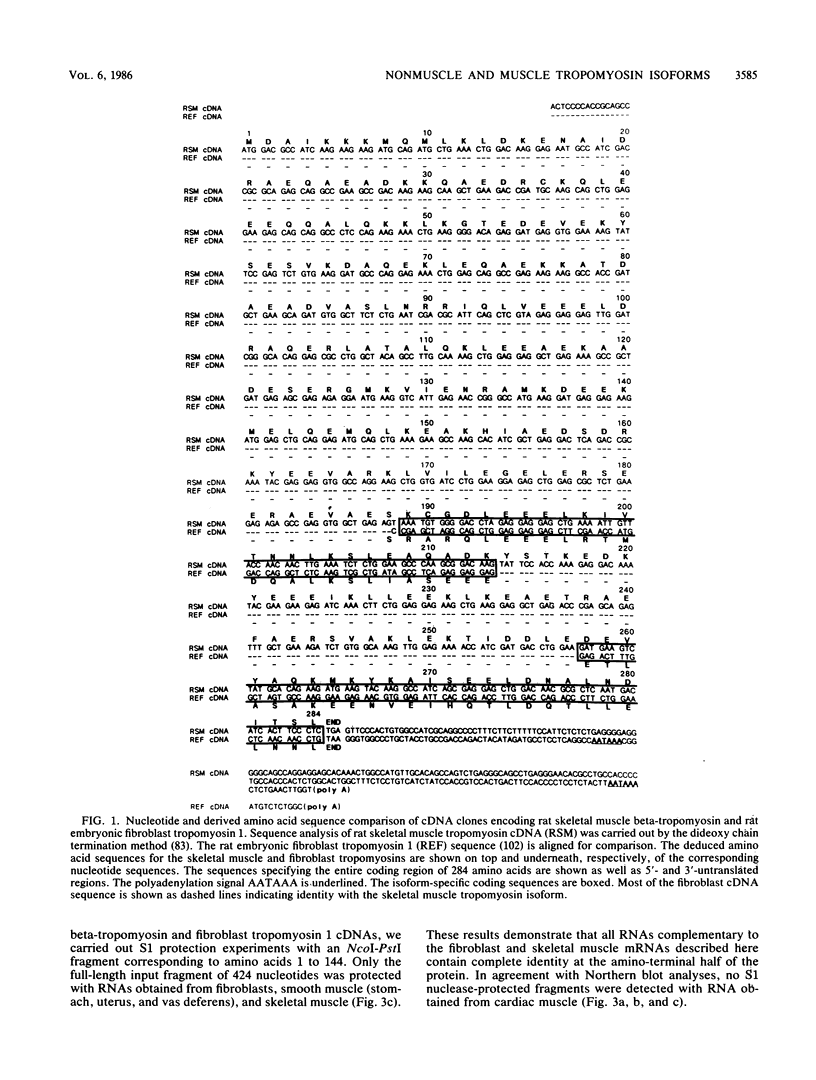
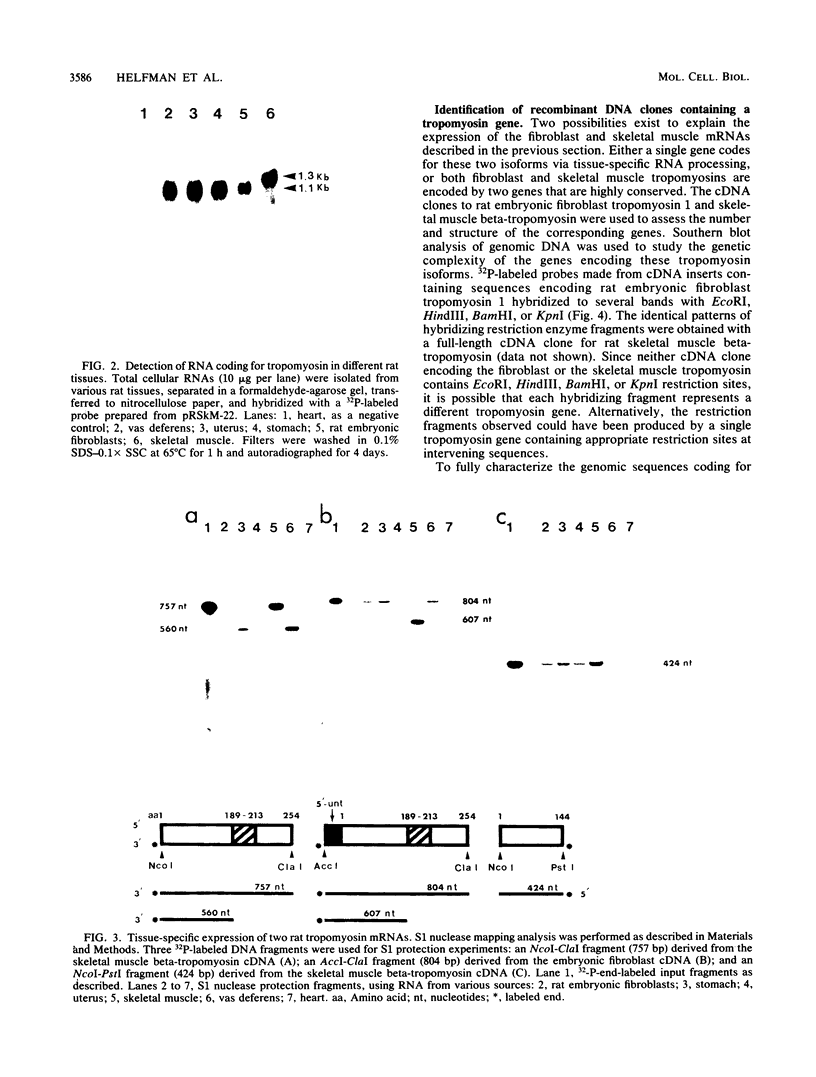
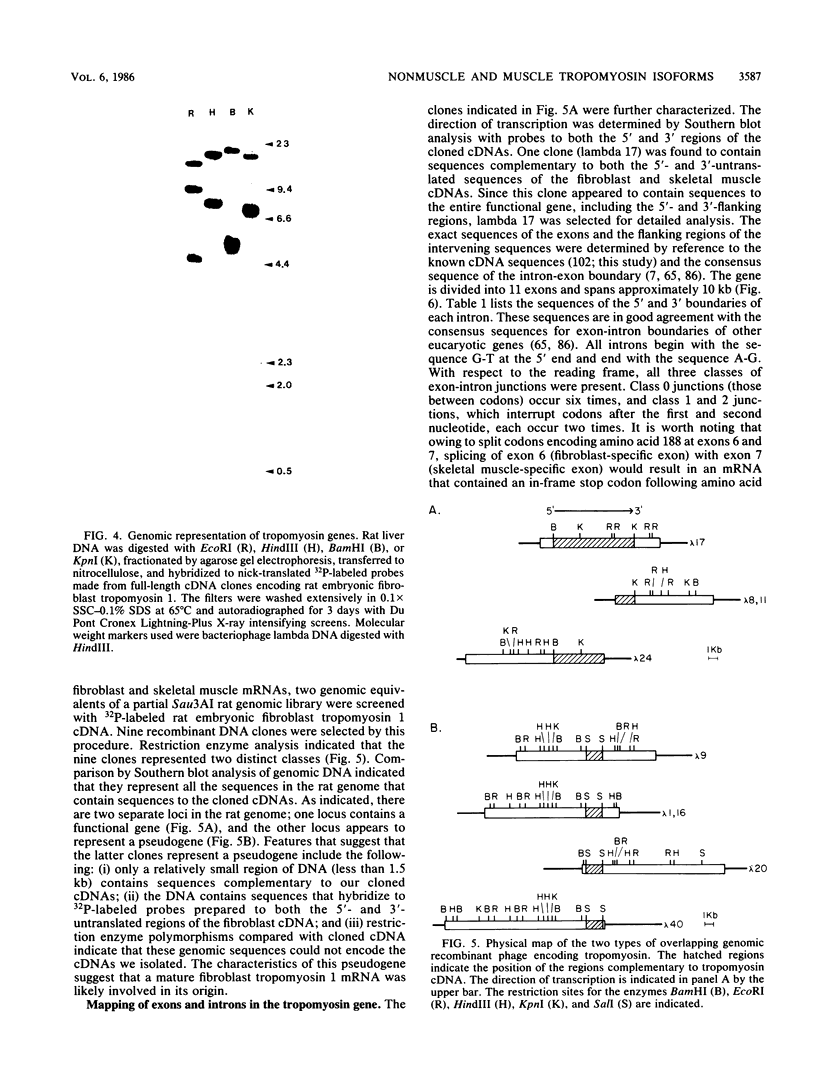
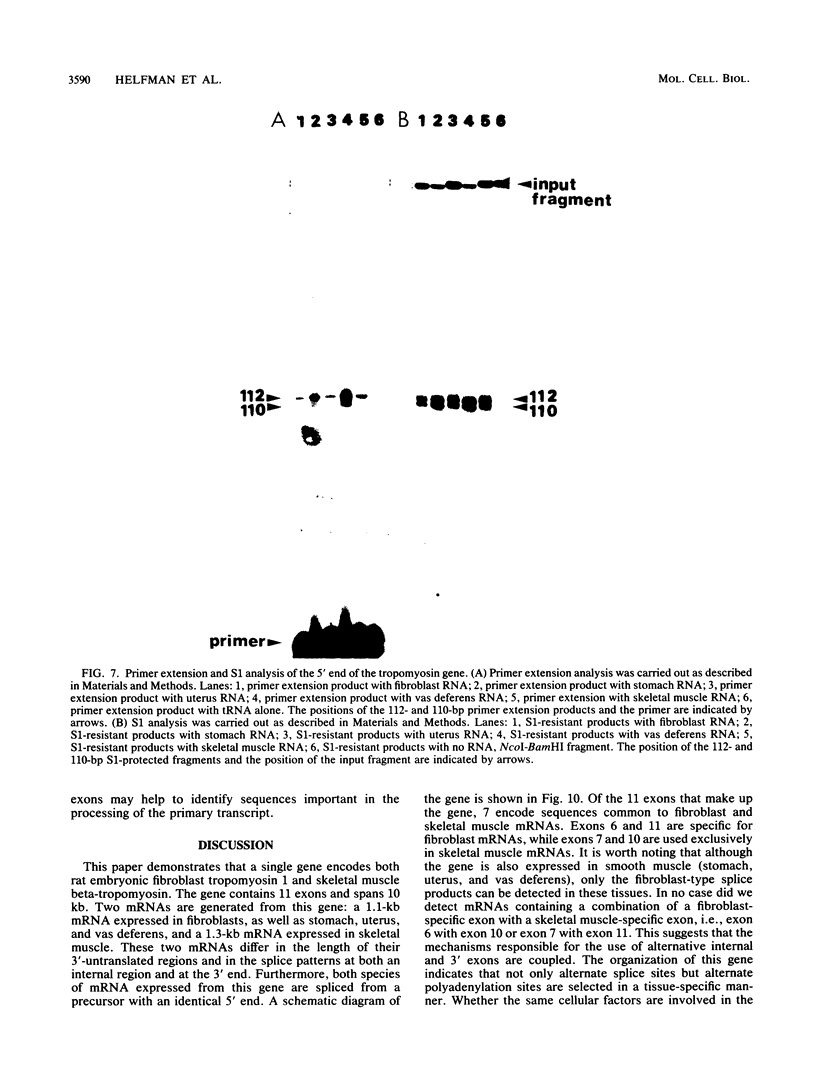
The molecular basis for the expression of rat embryonic fibroblast tropomyosin 1 and skeletal muscle beta-tropomyosin was determined. cDNA clones encoding these tropomyosin isoforms exhibit complete identity except for two carboxy-proximal regions (amino acids 189 to 213 and 258 to 284) and different 3'-untranslated sequences. The isoform-specific regions delineate the troponin T-binding domains of skeletal muscle tropomyosin. Analysis of genomic clones indicates that there are two separate loci in the rat genome that contain sequences complementary to these mRNAs. One locus is a pseudogene. The other locus contains a single gene made up of 11 exons and spans approximately 10 kilobases. Sequences common to all mRNAs were found in exons 1 through 5 (amino acids 1 to 188) and exons 8 and 9 (amino acids 214 to 257). Exons 6 and 11 are specific for fibroblast mRNA (amino acids 189 to 213 and 258 to 284, respectively), while exons 7 and 10 are specific for skeletal muscle mRNA (amino acids 189 to 213 and 258 to 284, respectively). In addition, exons 10 and 11 each contain the entire 3'-untranslated sequences of the respective mRNAs including the polyadenylation site. Although the gene is also expressed in smooth muscle (stomach, uterus, and vas deferens), only the fibroblast-type splice products can be detected in these tissues. S1 and primer extension analyses indicate that all mRNAs expressed from this gene are transcribed from a single promoter. The promoter was found to contain G-C-rich sequences, a TATA-like sequence TTTTA, no identifiable CCAAT box, and two putative Sp1-binding sites.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adelstein R. S., Pato M. D., Sellers J. R., de Lanerolle P., Conti M. A. Regulation of actin-myosin interaction by reversible phosphorylation of myosin and myosin kinase. Cold Spring Harb Symp Quant Biol. 1982;46(Pt 2):921–928. doi: 10.1101/sqb.1982.046.01.086. [DOI] [PubMed] [Google Scholar]
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi G. S., Boardman M., Storti R. V. Alternative splicing of a Drosophila tropomyosin gene generates muscle tropomyosin isoforms with different carboxy-terminal ends. Mol Cell Biol. 1984 Dec;4(12):2828–2836. doi: 10.1128/mcb.4.12.2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M. L., Busslinger M., Strub K. Transcription termination and 3' processing: the end is in site! Cell. 1985 Jun;41(2):349–359. doi: 10.1016/s0092-8674(85)80007-6. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Breitbart R. E., Nguyen H. T., Medford R. M., Destree A. T., Mahdavi V., Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated troponin T isoforms from a single gene. Cell. 1985 May;41(1):67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- Bronson D. D., Schachat F. H. Heterogeneity of contractile proteins. Differences in tropomyosin in fast, mixed, and slow skeletal muscles of the rabbit. J Biol Chem. 1982 Apr 10;257(7):3937–3944. [PubMed] [Google Scholar]
- Canaani D., Kahana C., Lavi S., Groner Y. Identification and mapping of N6-methyladenosine containing sequences in simian virus 40 RNA. Nucleic Acids Res. 1979 Jun 25;6(8):2879–2899. doi: 10.1093/nar/6.8.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin J. M., Przybyla A. E., MacDonald R. J., Rutter W. J. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979 Nov 27;18(24):5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Chong P. C., Hodges R. S. Photochemical cross-linking between rabbit skeletal troponin and alpha-tropomyosin. Attachment of the photoaffinity probe N-(4-azidobenzoyl-[2-3H]glycyl)-S-(2-thiopyridyl)-cysteine to cysteine 190 of alpha-tropomyosin. J Biol Chem. 1982 Aug 10;257(15):9152–9160. [PubMed] [Google Scholar]
- Cohen C., Caspar D. L., Parry D. A., Lucas R. M. Tropomyosin crystal dynamics. Cold Spring Harb Symp Quant Biol. 1972;36:205–216. doi: 10.1101/sqb.1972.036.01.028. [DOI] [PubMed] [Google Scholar]
- Cohen I., Cohen C. A tropomyosin-like protein from human platelets. J Mol Biol. 1972 Jul 21;68(2):383–387. doi: 10.1016/0022-2836(72)90220-3. [DOI] [PubMed] [Google Scholar]
- Cooper T. A., Ordahl C. P. A single troponin T gene regulated by different programs in cardiac and skeletal muscle development. Science. 1984 Nov 23;226(4677):979–982. doi: 10.1126/science.6095446. [DOI] [PubMed] [Google Scholar]
- Cummins P., Perry S. V. The subunits and biological activity of polymorphic forms of tropomyosin. Biochem J. 1973 Aug;133(4):765–777. doi: 10.1042/bj1330765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté G. P., Smillie L. B. Preparation and some properties of equine platelet tropomyosin. J Biol Chem. 1981 Nov 10;256(21):11004–11010. [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Dimock K., Stoltzfus C. M. Sequence specificity of internal methylation in B77 avian sarcoma virus RNA subunits. Biochemistry. 1977 Feb 8;16(3):471–478. doi: 10.1021/bi00622a021. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. Isolation of transcription factors that discriminate between different promoters recognized by RNA polymerase II. Cell. 1983 Mar;32(3):669–680. doi: 10.1016/0092-8674(83)90053-3. [DOI] [PubMed] [Google Scholar]
- Dynan W. S., Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983 Nov;35(1):79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Falkenthal S., Parker V. P., Davidson N. Developmental variations in the splicing pattern of transcripts from the Drosophila gene encoding myosin alkali light chain result in different carboxyl-terminal amino acid sequences. Proc Natl Acad Sci U S A. 1985 Jan;82(2):449–453. doi: 10.1073/pnas.82.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattoum A., Hartwig J. H., Stossel T. P. Isolation and some structural and functional properties of macrophage tropomyosin. Biochemistry. 1983 Mar 1;22(5):1187–1193. doi: 10.1021/bi00274a031. [DOI] [PubMed] [Google Scholar]
- Feramisco J. R., Smart J. E., Burridge K., Helfman D. M., Thomas G. P. Co-existence of vinculin and a vinculin-like protein of higher molecular weight in smooth muscle. J Biol Chem. 1982 Sep 25;257(18):11024–11031. [PubMed] [Google Scholar]
- Fine R. E., Blitz A. L. A chemical comparison of tropomyosins from muscle and non-muscle tissues. J Mol Biol. 1975 Jul 5;95(3):447–454. doi: 10.1016/0022-2836(75)90202-8. [DOI] [PubMed] [Google Scholar]
- Fine R. E., Blitz A. L., Hitchcock S. E., Kaminer B. Tropomyosin in brain and growing neurones. Nat New Biol. 1973 Oct 10;245(145):182–186. doi: 10.1038/newbio245182a0. [DOI] [PubMed] [Google Scholar]
- Ghosh P. K., Reddy V. B., Swinscoe J., Lebowitz P., Weissman S. M. Heterogeneity and 5'-terminal structures of the late RNAs of simian virus 40. J Mol Biol. 1978 Dec 25;126(4):813–846. doi: 10.1016/0022-2836(78)90022-0. [DOI] [PubMed] [Google Scholar]
- Giometti C. S., Anderson N. L. A variant of human nonmuscle tropomyosin found in fibroblasts by using two-dimensional electrophoresis. J Biol Chem. 1981 Nov 25;256(22):11840–11846. [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Helfman D. M., Feramisco J. R., Fiddes J. C., Thomas G. P., Hughes S. H. Identification of clones that encode chicken tropomyosin by direct immunological screening of a cDNA expression library. Proc Natl Acad Sci U S A. 1983 Jan;80(1):31–35. doi: 10.1073/pnas.80.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfman D. M., Feramisco J. R., Ricci W. M., Hughes S. H. Isolation and sequence of a cDNA clone that contains the entire coding region for chicken smooth-muscle alpha-tropomyosin. J Biol Chem. 1984 Nov 25;259(22):14136–14143. [PubMed] [Google Scholar]
- Hendricks M., Weintraub H. Multiple tropomyosin polypeptides in chicken embryo fibroblasts: differential repression of transcription by Rous sarcoma virus transformation. Mol Cell Biol. 1984 Sep;4(9):1823–1833. doi: 10.1128/mcb.4.9.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi S., Ooi T. Crystals of tropomyosin and native tropomyosin. J Mol Biol. 1968 Jun 28;34(3):699–701. doi: 10.1016/0022-2836(68)90190-3. [DOI] [PubMed] [Google Scholar]
- Hitchcock S. E., Huxley H. E., Szent-Györgyi A. G. Calcium sensitive binding of troponin to actin-tropomyosin: a two-site model for troponin action. J Mol Biol. 1973 Nov 15;80(4):825–836. doi: 10.1016/0022-2836(73)90212-x. [DOI] [PubMed] [Google Scholar]
- Ishimoda-Takagi T. Immunological purification of sea urchin egg tropomyosin. J Biochem. 1978 Jun;83(6):1757–1762. doi: 10.1093/oxfordjournals.jbchem.a132090. [DOI] [PubMed] [Google Scholar]
- Karlik C. C., Fyrberg E. A. An insertion within a variably spliced Drosophila tropomyosin gene blocks accumulation of only one encoded isoform. Cell. 1985 May;41(1):57–66. doi: 10.1016/0092-8674(85)90061-3. [DOI] [PubMed] [Google Scholar]
- Kato T., Tonomura Y. Physarum tropomyosin-troponin complex. Isolation and properties. J Biochem. 1975 Sep;78(3):583–588. doi: 10.1093/oxfordjournals.jbchem.a130943. [DOI] [PubMed] [Google Scholar]
- Lau S. Y., Sanders C., Smillie L. B. Amino acid sequence of chicken gizzard gamma-tropomyosin. J Biol Chem. 1985 Jun 25;260(12):7257–7263. [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Lewis W. G., Cote G. P., Mak A. S., Smillie L. B. Amino acid sequence of equine platelet tropomyosin. Correlation with interaction properties. FEBS Lett. 1983 Jun 13;156(2):269–273. doi: 10.1016/0014-5793(83)80511-0. [DOI] [PubMed] [Google Scholar]
- Lewis W. G., Smillie L. B. The amino acid sequence of rabbit cardiac tropomyosin. J Biol Chem. 1980 Jul 25;255(14):6854–6859. [PubMed] [Google Scholar]
- Lin J. J., Helfman D. M., Hughes S. H., Chou C. S. Tropomyosin isoforms in chicken embryo fibroblasts: purification, characterization, and changes in Rous sarcoma virus-transformed cells. J Cell Biol. 1985 Mar;100(3):692–703. doi: 10.1083/jcb.100.3.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. J., Matsumura F., Yamashiro-Matsumura S. Tropomyosin-enriched and alpha-actinin-enriched microfilaments isolated from chicken embryo fibroblasts by monoclonal antibodies. J Cell Biol. 1984 Jan;98(1):116–127. doi: 10.1083/jcb.98.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod A. R. Distinct alpha-tropomyosin mRNA sequences in chicken skeletal muscle. Eur J Biochem. 1982 Aug;126(2):293–297. doi: 10.1111/j.1432-1033.1982.tb06778.x. [DOI] [PubMed] [Google Scholar]
- MacLeod A. R., Houlker C., Reinach F. C., Smillie L. B., Talbot K., Modi G., Walsh F. S. A muscle-type tropomyosin in human fibroblasts: evidence for expression by an alternative RNA splicing mechanism. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7835–7839. doi: 10.1073/pnas.82.23.7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak A. S., Smillie L. B., Stewart G. R. A comparison of the amino acid sequences of rabbit skeletal muscle alpha- and beta-tropomyosins. J Biol Chem. 1980 Apr 25;255(8):3647–3655. [PubMed] [Google Scholar]
- Mak A. S., Smillie L. B. Structural interpretation of the two-site binding of troponin on the muscle thin filament. J Mol Biol. 1981 Jul 5;149(3):541–550. doi: 10.1016/0022-2836(81)90486-1. [DOI] [PubMed] [Google Scholar]
- Maki R., Roeder W., Traunecker A., Sidman C., Wabl M., Raschke W., Tonegawa S. The role of DNA rearrangement and alternative RNA processing in the expression of immunoglobulin delta genes. Cell. 1981 May;24(2):353–365. doi: 10.1016/0092-8674(81)90325-1. [DOI] [PubMed] [Google Scholar]
- Margossian S. S., Cohen C. Letter: Troponin subunit interactions. J Mol Biol. 1973 Dec 15;81(3):409–413. doi: 10.1016/0022-2836(73)90150-2. [DOI] [PubMed] [Google Scholar]
- Matsumura F., Lin J. J., Yamashiro-Matsumura S., Thomas G. P., Topp W. C. Differential expression of tropomyosin forms in the microfilaments isolated from normal and transformed rat cultured cells. J Biol Chem. 1983 Nov 25;258(22):13954–13964. [PubMed] [Google Scholar]
- Matsumura F., Yamashiro-Matsumura S., Lin J. J. Isolation and characterization of tropomyosin-containing microfilaments from cultured cells. J Biol Chem. 1983 May 25;258(10):6636–6644. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D., Stewart M. The troponin binding region of tropomyosin. Evidence for a site near residues 197 to 127. J Mol Biol. 1976 Oct 5;106(4):1017–1022. doi: 10.1016/0022-2836(76)90349-1. [DOI] [PubMed] [Google Scholar]
- Medford R. M., Nguyen H. T., Destree A. T., Summers E., Nadal-Ginard B. A novel mechanism of alternative RNA splicing for the developmentally regulated generation of troponin T isoforms from a single gene. Cell. 1984 Sep;38(2):409–421. doi: 10.1016/0092-8674(84)90496-3. [DOI] [PubMed] [Google Scholar]
- Montarras D., Fiszman M. Y., Gros F. Characterization of the tropomyosin present in various chick embryo muscle types and in muscle cells differentiated in vitro. J Biol Chem. 1981 Apr 25;256(8):4081–4086. [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Pettersson I., Hinterberger M., Karmas A., Steitz J. A. The U1 small nuclear RNA-protein complex selectively binds a 5' splice site in vitro. Cell. 1983 Jun;33(2):509–518. doi: 10.1016/0092-8674(83)90432-4. [DOI] [PubMed] [Google Scholar]
- Nabeshima Y., Fujii-Kuriyama Y., Muramatsu M., Ogata K. Alternative transcription and two modes of splicing results in two myosin light chains from one gene. Nature. 1984 Mar 22;308(5957):333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Nonomura Y., Drabikowski W., Ebashi S. The localization of troponin in tropomyosin paracrystals. J Biochem. 1968 Sep;64(3):419–422. doi: 10.1093/oxfordjournals.jbchem.a128912. [DOI] [PubMed] [Google Scholar]
- Ohtsuki I. Molecular arrangement of troponin-T in the thin filament. J Biochem. 1979 Aug;86(2):491–497. doi: 10.1093/oxfordjournals.jbchem.a132549. [DOI] [PubMed] [Google Scholar]
- Otsuki I. Localization of troponin in thin filament and tropomyosin paracrystal. J Biochem. 1974 Apr;75(4):753–765. doi: 10.1093/oxfordjournals.jbchem.a130448. [DOI] [PubMed] [Google Scholar]
- Pearlstone J. R., Smillie L. B. Binding of troponin-T fragments to several types of tropomyosin. Sensitivity to Ca2+ in the presence of troponin-C. J Biol Chem. 1982 Sep 25;257(18):10587–10592. [PubMed] [Google Scholar]
- Pearlstone J. R., Smillie L. B. Identification of a second binding region on rabbit skeletal troponin-T for alpha-tropomyosin. FEBS Lett. 1981 Jun 1;128(1):119–122. doi: 10.1016/0014-5793(81)81095-2. [DOI] [PubMed] [Google Scholar]
- Periasamy M., Strehler E. E., Garfinkel L. I., Gubits R. M., Ruiz-Opazo N., Nadal-Ginard B. Fast skeletal muscle myosin light chains 1 and 3 are produced from a single gene by a combined process of differential RNA transcription and splicing. J Biol Chem. 1984 Nov 10;259(21):13595–13604. [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Robert B., Daubas P., Akimenko M. A., Cohen A., Garner I., Guenet J. L., Buckingham M. A single locus in the mouse encodes both myosin light chains 1 and 3, a second locus corresponds to a related pseudogene. Cell. 1984 Nov;39(1):129–140. doi: 10.1016/0092-8674(84)90198-3. [DOI] [PubMed] [Google Scholar]
- Rogers J., Wall R. A mechanism for RNA splicing. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1877–1879. doi: 10.1073/pnas.77.4.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld M. G., Mermod J. J., Amara S. G., Swanson L. W., Sawchenko P. E., Rivier J., Vale W. W., Evans R. M. Production of a novel neuropeptide encoded by the calcitonin gene via tissue-specific RNA processing. Nature. 1983 Jul 14;304(5922):129–135. doi: 10.1038/304129a0. [DOI] [PubMed] [Google Scholar]
- Rozek C. E., Davidson N. Drosophila has one myosin heavy-chain gene with three developmentally regulated transcripts. Cell. 1983 Jan;32(1):23–34. doi: 10.1016/0092-8674(83)90493-2. [DOI] [PubMed] [Google Scholar]
- Ruiz-Opazo N., Weinberger J., Nadal-Ginard B. Comparison of alpha-tropomyosin sequences from smooth and striated muscle. Nature. 1985 May 2;315(6014):67–70. doi: 10.1038/315067a0. [DOI] [PubMed] [Google Scholar]
- Salviati G., Betto R., Danieli Betto D. Polymorphism of myofibrillar proteins of rabbit skeletal-muscle fibres. An electrophoretic study of single fibres. Biochem J. 1982 Nov 1;207(2):261–272. doi: 10.1042/bj2070261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C., Smillie L. B. Amino acid sequence of chicken gizzard beta-tropomyosin. Comparison of the chicken gizzard, rabbit skeletal, and equine platelet tropomyosins. J Biol Chem. 1985 Jun 25;260(12):7264–7275. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schibler U., Kelley D. E., Perry R. P. Comparison of methylated sequences in messenger RNA and heterogeneous nuclear RNA from mouse L cells. J Mol Biol. 1977 Oct 5;115(4):695–714. doi: 10.1016/0022-2836(77)90110-3. [DOI] [PubMed] [Google Scholar]
- Schloss J. A., Goldman R. D. Microfilaments and tropomyosin of cultured mammalian cells: isolation and characterization. J Cell Biol. 1980 Dec;87(3 Pt 1):633–642. doi: 10.1083/jcb.87.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. A. Speculations on RNA splicing. Cell. 1981 Mar;23(3):643–646. doi: 10.1016/0092-8674(81)90425-6. [DOI] [PubMed] [Google Scholar]
- Sodek J., Hodges R. S., Smillie L. B. Amino acid sequence of rabbit skeletal muscle alpha-tropomyosin. The COOH-terminal half (residues 142 to 284). J Biol Chem. 1978 Feb 25;253(4):1129–1136. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stewart M., McLachlan A. D. Structure of magnesium paracrystals of alpha-tropomyosin. J Mol Biol. 1976 May 15;103(2):251–269. doi: 10.1016/0022-2836(76)90312-0. [DOI] [PubMed] [Google Scholar]
- Stone D., Smillie L. B. The amino acid sequence of rabbit skeletal alpha-tropomyosin. The NH2-terminal half and complete sequence. J Biol Chem. 1978 Feb 25;253(4):1137–1148. [PubMed] [Google Scholar]
- Talbot K., MacLeod A. R. Novel form of non-muscle tropomyosin in human fibroblasts. J Mol Biol. 1983 Feb 15;164(1):159–174. doi: 10.1016/0022-2836(83)90091-8. [DOI] [PubMed] [Google Scholar]
- Tamkun J. W., Schwarzbauer J. E., Hynes R. O. A single rat fibronectin gene generates three different mRNAs by alternative splicing of a complex exon. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5140–5144. doi: 10.1073/pnas.81.16.5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E. W. Mechanism of actomyosin ATPase and the problem of muscle contraction. CRC Crit Rev Biochem. 1979;6(2):103–164. doi: 10.3109/10409237909102562. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa S. Somatic generation of antibody diversity. Nature. 1983 Apr 14;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wei C. M., Moss B. Nucleotide sequences at the N6-methyladenosine sites of HeLa cell messenger ribonucleic acid. Biochemistry. 1977 Apr 19;16(8):1672–1676. doi: 10.1021/bi00627a023. [DOI] [PubMed] [Google Scholar]
- Yaffe D., Nudel U., Mayer Y., Neuman S. Highly conserved sequences in the 3' untranslated region of mRNAs coding for homologous proteins in distantly related species. Nucleic Acids Res. 1985 May 24;13(10):3723–3737. doi: 10.1093/nar/13.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M., Greaser M. L., Cassens R. G. Interactions of troponin subunits with different forms of tropomyosin. J Ultrastruct Res. 1974 Jul;48(1):33–58. doi: 10.1016/s0022-5320(74)80043-2. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- Yamawaki-Kataoka Y., Helfman D. M. Rat embryonic fibroblast tropomyosin 1. cDNA and complete primary amino acid sequence. J Biol Chem. 1985 Nov 25;260(27):14440–14445. [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- der Terrossian E., Fuller S. D., Stewart M., Weeds A. G. Porcine platelet tropomyosin. Purification, characterization and paracrystal formation. J Mol Biol. 1981 Nov 25;153(1):147–167. doi: 10.1016/0022-2836(81)90531-3. [DOI] [PubMed] [Google Scholar]