Abstract
Background
Staphylococcus epidermidis is a common pathogen in medical device-associated infections and have an ability to form adherent slime. We aimed to study the effects of icaA and icaD genes on the slime formation of Staphylococcus epidermidis associated with catheter-associated infections.
Methods
S. epidermidis isolates from the central venous catheter blood of patients with catheter-associated infections, and from the nasal vestibules of healthy volunteers, intensive care unit hospital staff, and patients, were collected. Slime phenotype was determined by Congo red agar test. The icaA/D was detected by polymerase chain reaction. Slime was examined using scanning electron microscopy.
Results
A total of 82 S. epidermidis isolates were collected. We found a statistically significant difference with regards to slime production between the clinical isolates from the catheter blood specimens and those from the nasal vestibules (p<0.05). All S. epidermidis slime positive strains isolated were icaA positive. There was a greater correlation between the presence of both icaA and icaD and the slime production than the single expression of icaA or icaD and the presence of slime in all groups. The co-expression of mecA and icaD was associated with enhanced resistance to antibiotics.
Conclusion
S. epidermidis bacteria are significant nosocomial pathogens, and icaA/D can clarify the adhesion mechanism in the pathogenesis of infections associated with medical devices. This study result could be useful for the development of rapid diagnosis for slime producing and methicillin resistant S. epidermidis strains.
Keywords: Catheter-associated blood stream infections, S. epidermidis, Slime, icaA/D, Scanning electron microscopy
Background
Coagulase-negative staphylococci (CNS) are the most frequent cause of nosocomial blood stream infections [1], in particular, Staphylococcus epidermidis, which has emerged as a major pathogen [2]. S. epidermidis, a normal inhabitant of human skin and mucous membranes, is the predominant cause of foreign-body-associated infections [3,4]. In addition, S. epidermidis is isolated with increasing frequency as the causative pathogen of nosocomial sepsis, and accounts for approximately 30% of all nosocomial blood stream infections [4]. These infections are often indolent and unresponsive to antimicrobials [5], and frequently result in the removal of the adulterated device. In clinical samples, rates of methicillin resistance in intensive care unit settings have been reported to be 55–77%, or even 86% [6-8].
In an early study, the “slime” production by S. epidermidis was found to be significantly associated with clinical infections [9]. Coagulase-negative staphylococci, particularly S epidermidis, are the important cause of infections associated with foreign materials. Bacterial adhesion has been considered as the leading cause of severe nosocomial infections related to implanted medical devices [10]. In the recent years, a study has been conducted to investigate the ica gene as a marker of the S. epidermidis adhesive aptitude [11].
This study analyzed 82 S. epidermidis, collected over a 15-month period from patients, hospital staff in the intensive care unit, and volunteers. The aim of this study was to determine the presence of the icaA and icaD in S. epidermidis from catheter-related blood stream infections and to determine the correlation between the presence of icaA and icaD, biofilm production, and resistance to antibiotics.
Methods
Bacterial strains
This study analyzed 82 S. epidermidis isolates collected from May 2009 to July 2010. Twenty-two S. epidermidis studied were isolated from patients blood taken from intravascular catheters; another 60 strains of S. epidermidis isolated from the nasal vestibules of healthy non-medical volunteers, patients (same patients from whom blood specimens were obtained), and intensive care unites (ICU) hospital staff were also investigated.
All S. epidermidis isolates were characterized using vitek 2 compact (bioMe´rieux, France). Ethical approval for the study was obtained from the Central Research Ethics Committee in the Provincial Hospital and included the patients’ and volunteers’ prior written consent.
Two S. epidermidis reference strains were used, the well known slime-producing strain ATCC 35984 (RP62A) and the non-slime-producing strain ATCC 12228. The quality control strain was Staphylococcus aureus ATCC 25923 [12].
Strain storage
The S. epidermidis were stored in trypticase soy broth (TSB), with 15% glycerol added at -20C for 1 to 15 months long. The isolates were recovered when the study was initiated. The S. epidermidis recovery rate reached 100%.
Study design
This study was a prospective design. Eighty-two strains of S. epidermidis were collected over a 15 month period. Eighty-two S. epidermidis isolates were collected usng sterile swabs from patients’ central venous catheter blood, and from the nasal vestibules of non-medical volunteers, ICU hospital staff, and patients. A total of 70 volunteers and 69 staff (doctor and nurse) who haven’t had any diseases were included. Among them, the volunteers were 1st year medical students from Anhui Medical University. Only cases with bacteraemia from catheter-associated infections, confirmed by positive blood culture and clinical evidence of catheter-associated infections, were included in the study. Cases without neutropenia and cases with bacteraemia from mixed pathogens were excluded in order to avoid the contamination of S. aureus and confusion about the true pathogen. The study was conducted in the ICU of the Affiliated Provincial Hospital, Anhui Medical University, Hefei, China. The unit has the capacity to provide care for 54 patients.
Detection of slime production
Qualitative detection of the phenotypic production of biofilm formation by all strains was studied by culturing the strains on Congo red agar (CRA) plates (CRA; Sigma Chemical Company, St Louis, MO, USA) [13]. For an accurate assessment of all the possible chromatic variations exhibited by the cultured colonies, a six-color reference scale was used and interpreted, and smooth colonies were classified as slime negative strains.
Scanning electron micrograph
An additional set of experiments was performed using Scanning electron micrograph to evaluate the structure of slime formed under study. Examples of these slimes are shown in Figure 1.
Figure 1.
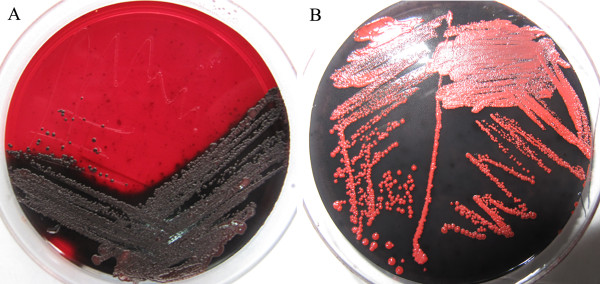
Glass-surface slime production by cultures on CRA. A) Black colonies of slime-producing S. epidermidis ATCC 35984. B) Red colonies of the non-slime-producing S. epidermidis ATCC 12228. CRA: Congo Red Agar.
Critical dried material was mounted onto an aluminum stub and coated with palladium in a scanning electron microscopy (JSM-6700 F, JEOL) [14].
Polymerase chain reaction (PCR) for icaA and icaD genes and mecA sequences
After overnight culture on brain-heart infusion agar plates, one colony was suspended in 20 ml of sterile distilled water, and the suspension was then heated at 100°C for 20 minutes. From this suspension, a 5 μl aliquot was directly used as a template for PCR amplification.
The sequences of icaA and icaD were taken from the GenBank sequence database of the National Center for Biotechnology Information [12]. Primers specific for icaA and icaD were selected based on gene sequences by the Primer 3 program. The primers were synthesized by TAKARA biotechnology (Dalian) Co, Ltd (China).
The presence of icaA, icaD, and mecA DNA were detected by polymerase chain reaction using forward and reverse primers for icaA, icaD, and mecA, as described previously [15,16]. For the detection of icaA (188-bp), the forward primer had the following sequence: 5′-TCT CTT GCA GGA GCA ATC AA; and 5′-TCA GGC ACT AAC ATC CAG CA was used as a reverse primer. The primer sequences for icaD (198-bp) were: forward, 5′-ATG GTC AAG CCC AGA CAG AG; and reverse, 5′-CGT GTT TTC AAC ATT TAA TGC AA. The primers’ detections of the mecA (224-bp) were AAA ATC GAT GGT AAA GGT TG GC forward, and AGT TCT GCA GTA CCG GAT TT GC reverse.
PCR was performed in a DNA thermal cycler and using the method described by Petrelli D et al. [17]. The Gene Ruler 100 bp DNA ladder was used as a DNA size marker, and visualized under ultraviolet transillumination.
Analysis of data
Statistical analysis was performed by means of SPSS 17.0 (SPSS Inc., Chicago, USA) software. Comparisons between groups were performed using the chi-square test. The Pearson correlation coefficient (r) and its significance (P) were calculated between groups; probability values less than 0.05 were considered statistically significant.
Results
Slime by S. epidermidis isolates
Slime-forming ability of S. epidermidis can be inferred by phenotypic effects when grown on Congo red agar. Slime producing strains form rough, black colonies such as those seen in Figure 1A, while the colonies of strains that do not produce slime are red in color, such as those seen in Figure 1B.
Detection of slime-production
Sixty five patients who provided the blood isolate were surveyed for nasal carriage and 33.85% (22/65) of them were positive. Phenotypic production of slime by all investigated strains was assessed by culture on CRA plates [13] (Figure 2). Among the clinical isolates from patient catheter blood specimens, 15 of 22 (68.18%) S. epidermidis were slime producers and the remaining 7 strains were non-slime producers. The number of slime-producing S. epidermidis from the nasal vestibules of healthy non-medical volunteers, patients, and ICU hospital staff were 8 of 21 (38.10%), 5 of 19 (26.32%), and 8 of 20 (40.00%), respectively.
Figure 2.
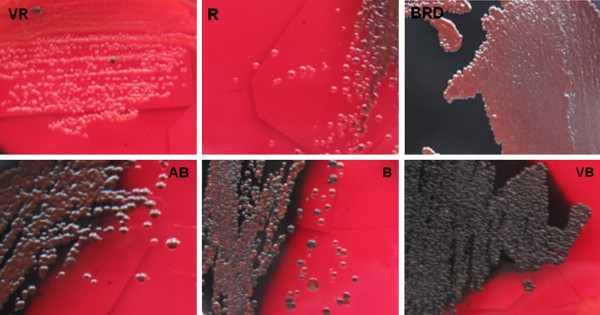
Colorimetric scale adopted for colony evaluation by Congo red agar. vr, very red; r, red; brd, Bordeaux; ab, almost black; b, black; vb, very black.
Scanning electron micrograph of biofilm
Slime is a biomass of exopolysaccharide that adheres to culture medium surfaces. It appears as clear mucus. (See Figure 3A and B) (Scale bar _2 μm).
Figure 3.
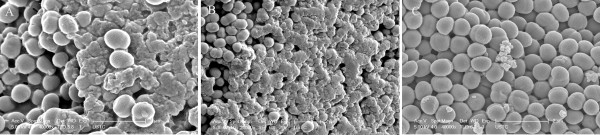
A scanning electron microscopy image shows a laboratory-grown Staphylococcus epidermidis biofilm. Image A shows normal slime-producing strains; B shows slime-producing strain ATCC 35984; C shows non-slime-producing strain ATCC 12228. Scale bar 2 μm. Image taken by Fu, Instruments’ Center for Physical Science, University of Science & Technology China.
PCR detection of icaA, icaD, and mecA
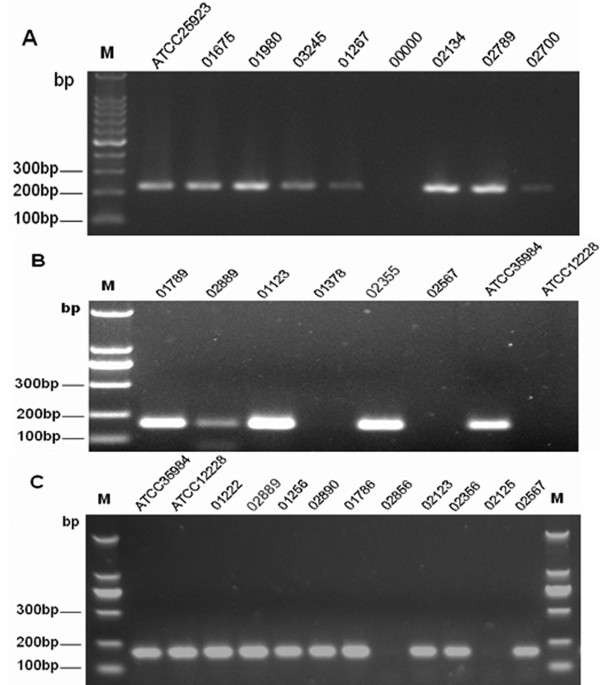
The PCR technique was performed on the 82 S. epidermidis strains isolated. Typical results are shown in the Figure 4. It was also shown that all strains which were positive for icaA were also positive for icaD, giving 188-bp and 198-bp band for the icaA and icaD, and a 224-bp band for the mecA. In our study, all isolated slime-positive S. epidermdis strains were icaA positive, and it was also demonstrated that icaD had higher positive rate than icaA in all S. epidermdis isolates. Also, in this study, a relationship between mecA and icaD was observed among all four groups (p<0.05). For the clinical isolates from catheter blood, 15 of 22 S. epidermidis were found to be positive for both icaA and icaD, and 15 strains were found to be biofilm producers using the CRA method (Table 1). All 15 slime producing S. epidermidis isolates were positive for both icaA and icaD (Table 2).
Figure 4.
PCR detection of icaA, icaD, and mecA. Lane 1, 100 bp DNA molecular size marker. Image A shows mecA locus (A, 224-bp band), lanes 2–10, PCR amplification of mecA, lanes 2, ATCC25923. Lane 7, negative control; lanes 3 to 10, PCR amplicons obtained with DNA of S. epidermidis; B shows icaA locus (B, 198-bp band),lanes 2–9, PCR amplification of icaA. Lane 8, ATCC 35984 (RP62A). Lane 9, ATCC 12228. Lanes 2–7, amplification obtained with DNA of clinical S. epidermidis isolate. C shows icaD locus (C, 198-bp band), lanes 2–13, PCR amplification of icaD. Lane 2, ATCC 35984 (RP62A). Lane 3, ATCC 12228. Lanes 4–13, amplification obtained with DNA of clinical S. epidermidis isolate.
Table 1.
Association between slime production on congo red agar and presence of icaA and icaD genes in the 82 S. epidermidis strains
| Groups | n |
Slime production |
mecA vs icaD |
||||||
|---|---|---|---|---|---|---|---|---|---|
|
icaA |
icaD |
icaA +icaD |
|||||||
| r | P value | r | P value | r | P value | r | P value | ||
| 1 |
22 |
0.894 |
0.000◇ |
0.459 |
0.036△ |
1.000 |
0.000◇ |
0.686 |
0.001☆ |
| 2 |
21 |
0.679 |
0.001◇ |
0.439 |
0.047△ |
0.748 |
0.000◇ |
0.553 |
0.009☆ |
| 3 |
19 |
0.701 |
0.001◇ |
0.406 |
0.085 |
0.782 |
0.000◇ |
0.456 |
0.049☆ |
| 4 | 20 | 0.533 | 0.015△ | 0.408 | 0.074 | 0.698 | 0.001◇ | 0.535 | 0.015☆ |
Group 1, blood isolates obtained from patients’ IV line; Group 2, nasal isolates of healthy volunteers; Group 3, nasal isolates of the same patients in Group 1; Group 4, nasal isolates of ICU staff. △The correlation between the presence of the operon and the presence of slime production in all groups (p<0.05). ◇The correlation between the presence of the operon and the presence of slime production in all groups (p<0.001). ☆The correlation between the presence of MecA and icaD (p<0.05).
Table 2.
Association between slime production on congo red agar and presence of icaA and icaD genes in the 82 S. epidermidis strains
| Groups | strains (n) | Slime Production | icaA | icaD | icaA +icaD | mecA |
|---|---|---|---|---|---|---|
| 1 |
22 |
15 (68.18%) |
16 (72.72%) |
20 (90.91%) |
15 (68.18%) |
20 |
| 2 |
21 |
8 (38.10%)△ |
12 (57.14%) |
15 (71.43%) |
11 (52.38%) |
14 |
| 3 |
19 |
5 (26.32%)◇ |
8 (42.11%) |
15 (78.95%) |
7 (36.84%) |
14 |
| 4 | 20 | 8 (40.00%)◇ | 12 (60.00%) | 17 (85.00%) | 12 (60.00%) | 14 |
Group 1, blood isolates obtained from patients’ IV line; Group 2, nasal isolates of healthy volunteers; Group 3, nasal isolates of the same patients in Group 1; Group 4, nasal isolates of ICU staff. △Values are given as slime production of S. epidermidis strains vs first group (p<0.05), ◇Values are given as slime production of S. epidermidis strains vs first group (p<0.001). There was no statistical difference among other three groups (P>0.05).
Discussion
Staphylococcus epidermidis are found ubiquitously, residing on human skin in healthy adults [18]. It has now been recognized as a key pathogen involved in adhereing to indwelling medical devices surfaces and form biofilms. The major virulence factor is that they can form biofilm on polymeric surfaces and adherence to catheters and other artificial materials during the early phase of biofilm development [13]. Slime production seems, therefore, to be a very important mechanism of adhesion onto biomaterials. In particular, the current study demonstrated that S. epidermidis should be recognized as a major cause of catheter infections. This study investigating the biofilm formation showed that slime accumulation is mediated by the chromosomal ica gene, which comprises four intercellular adhesion genes (icaA, icaB, icaC and icaD) and one regulator gene (icaR) [19].
Detection of slime-production
The results of our study are similar to previous literature, in which the percentage of slime-producing strains of S. epidermidis ranged from 31% to 89% [20,21]. In addition, we found a statistical difference with regards to slime production between the clinical isolates from the patient catheter blood specimens and isolates from the nasal vestibules of three groups (p<0.05) (Table 2). This indicates that slime is produced in higher proportion in the blood group than in the other groups. The results also indicate that the internal in vivo environment can offer a satisfactory biological environment for producing slime.
Our study also found that slime production was reduced in some S. epidermidis [43.90% (36/82)]. In this investigation, some strains were found to gradually change from black or bordeaux to red or very red in 3 to 5 generations. We could not find similar results in any literature. Two hypotheses can be proposed: (1) there are many differences in nutrients, pH, oxygen radicals, and antibiotics between in vivo and in vitro environmental conditions which may confer a positive selective pressure on slime production; and (2) certain genes such as icaA or icaD may be lost or mutated. In addition, we conjecture that the virulence factor of S. epidermidis will be weak. We believe that a future study of the presence and expression of genes (such as the icaA or icaD after passage may help clarify the relevance of slime, virulence factor of S epidermidis, and ica.
Scanning electron micrograph
In this observation, we intensely studied S. epidermidis slime by Scanning electron micrograph. In some studies, the monomeric carbohydrate moieties and several amino sugars are the most distinctive features of a microbial biofilm produced by S. epidermidis[22,23]. This is particularly true that slime layer was a suitable intermedia for S. epidermidis growth and may cause a subclinical chronic to severe fatal infection [24].
icaA and icaD and slime production
Coagulase-negative staphylococci, particularly S. epidermidis, are the major causes of infections associated with catheters and other artificial materials. Usually considered to be of low virulence, S. epidermidis is now recognized as a potential pathogen because of its ability to produce biofilm and adhere to the walls of artificial materials. In other evidences, the biofilm accumulation is mediated by certain genes, such as icaA, icaB, icaC, icaD and icaR[25,26]. The recent findings point to an important role of the icaA and icaD due to their ability to produce slime strongly in a high percentage of clinical isolates collected from patients with catheters associated infection [10]. Cafiso V reported that the co-expression of icaA with icaD can increase slime production remarkably [19].
In our investigation, all isolated S. epidermdis slime-positive strains were positive for the icaA, which is in agreement with other studies [15]. In addition, our investigation demonstrated that icaD has a higher positive detection rate. Slime production depends on the presence of both icaD and icaA. The reason for the absence of biofilm production in some icaA and icaD positive isolates in the present study may be the lack of icaC [27].
In this study we found a stronger correlation between the presence of the icaA and icaD and the presence of slime production than the single expression of icaA or icaD and the presence of slime production in all groups (Table 2), and consequently, these results support the idea that the co-expression of icaA and icaD is associated with enhanced slime production. Our experimental evidence supports the result of Cafiso V [19], and another study demonstrated that co-expression of icaA and icaD was associated with enhanced slime production [21].
MecA and icaD genes
In this study, a relationship was observed between mecA and icaD among the four groups, and in another investigation of ours, there was a significant association between a positive PCR result for icaD and a positive CRA test result for S. epidermidis, which indicates the important role of icaD as markers in S. epidermidis infections. In fact, in recent studies [28,29], S. epidermidis is the most common cause of clinical infections because of strains that are able to produce a polysaccharide slime depending on the presence of ica, such as icaD. This finding suggests the possibility that co-expression of mecA and icaD is associated with enhanced clinical isolates that produce an extracellular matrix called slime and may make them more resistant to antibiotics. The results of our study are similar to Cafiso V’s study, which suggested that the ica and mecA could be considered as important virulence markers for clinical staphylococcal isolates [19]. Therefore, the demonstration of mecA and icaD in clinical isolates is useful for the rapid determination of the severity of S. epidermidis infection.
Limitations of this study included small numbers of cases, single study site. A future study about the mechanism of the ica with prospective design and more numbers of cases will enhance our understanding of the pathogenesis of CVC-related BSI and provide useful information for the development of more effective therapeutic measures to eradicate biofilm in hospitals.
Conclusion
In summary, these data suggested that ica is typically associated with biofilm production of S. epidermidis in ICU. Both icaA and icaD can support the adhesion mechanisms of S. epidermidis involved in the infections associated with medical devices. Co-expression of mecA and icaD is associated with enhanced clinical isolates that produce slime and may be more resistant to antibiotics. This study may help the development of rapid diagnosis approach for slime producing and methicillin resistant strains in hospitals.
Abbreviations
CNS: Coagulase-negative staphylococci; CRA: Congo red agar; ICU: Intensive care unites; PCR: Polymerase chain reaction; TSB: Trypticase soy broth.
Competing interests
We declare that we have no dual or conflicting interests.
Authors’ contributions
SZ participated in the design of the study and performed the statistical analysis. XC participated in the PCR(icaA and icaD genes and MecA sequences) and Scanning electron micrograph. MF cultured the bacteria on CRA plates and assessed the Phenotypic production of slime of all investigated strains. YD carried out the collection and cultivation of bacteria. BL participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Shusheng Zhou, Email: zhouss108@163.com.
Xiaoguang Chao, Email: chaoxg1001@gmail.com.
Mingming Fei, Email: feimm202@gmail.com.
Yuanyuan Dai, Email: daiyy118@163.com.
Bao Liu, Email: az306w@gmail.com.
References
- Rogers KL, Fey PD, Rupp ME. Coagulase-negative staphylococcal infections. Infect Dis Clin North Am. 2009;23(1):73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Gomes FI, Teixeira P, Azeredo J, Oliveira R. Effect of farnesol on planktonic and biofilm cells of Staphylococcus epidermidis. Curr Microbiol. 2009;59(2):118–122. doi: 10.1007/s00284-009-9408-9. [DOI] [PubMed] [Google Scholar]
- Knobloch JK, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D. Biofilm formation by staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol. 2001;183(8):2624–2633. doi: 10.1128/JB.183.8.2624-2633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piette A, Verschraegen G. Role of coagulase-negative staphylococci in human disease. Vet Microbiol. 2009;134(1–2):45–54. doi: 10.1016/j.vetmic.2008.09.009. [DOI] [PubMed] [Google Scholar]
- Kitao T, Ishimaru M, Nishihara S. Detection of biofilm-producing and methicillin resistance genes in Staphylococcus epidermidis isolated from healthy humans and in blood culture tests. Journal of infection and chemotherapy: official journal of the Japan Society of Chemotherapy. 2010;16(3):170–173. doi: 10.1007/s10156-010-0037-9. [DOI] [PubMed] [Google Scholar]
- Chaudhury A, Kumar AG. In vitro activity of antimicrobial agents against oxacillin resistant staphylococci with special reference to Staphylococcus haemolyticus. Indian J Med Microbiol. 2007;25(1):50–52. doi: 10.4103/0255-0857.31062. [DOI] [PubMed] [Google Scholar]
- Gatermann SG, Koschinski T, Friedrich S. Distribution and expression of macrolide resistance genes in coagulase-negative staphylococci. Clin Microbiol Infect. 2007;13(8):777–781. doi: 10.1111/j.1469-0691.2007.01749.x. [DOI] [PubMed] [Google Scholar]
- Sader HS, Watters AA, Fritsche TR, Jones RN. Daptomycin antimicrobial activity tested against methicillin-resistant staphylococci and vancomycin-resistant enterococci isolated in European medical centers (2005) BMC Infect Dis. 2007;7:29. doi: 10.1186/1471-2334-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner J, Goldmann DA. Coagulase-negative staphylococci: role as pathogens. Annu Rev Med. 1999;50:223–236. doi: 10.1146/annurev.med.50.1.223. [DOI] [PubMed] [Google Scholar]
- El-Mahallawy HA, Loutfy SA, El-Wakil M, El-Al AK, Morcos H. Clinical implications of icaA and icaD genes in coagulase negative staphylococci and Staphylococcus aureus bacteremia in febrile neutropenic pediatric cancer patients. Pediatr Blood Cancer. 2009;52(7):824–828. doi: 10.1002/pbc.21964. [DOI] [PubMed] [Google Scholar]
- Arciola CR, Baldassarri L, Montanaro L. In catheter infections by Staphylococcus epidermidis the intercellular adhesion (ica) locus is a molecular marker of the virulent slime-producing strains. J Biomed Mater Res. 2002;59(3):557–562. doi: 10.1002/jbm.10006. [DOI] [PubMed] [Google Scholar]
- Arciola CR, Baldassarri L, Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J Clin Microbiol. 2001;39(6):2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaieb K, Mahdouani K, Bakhrouf A. Detection of icaA and icaD loci by polymerase chain reaction and biofilm formation by Staphylococcus epidermidis isolated from dialysate and needles in a dialysis unit. J Hosp Infect. 2005;61(3):225–230. doi: 10.1016/j.jhin.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Soboh F, Khoury AE, Zamboni AC, Davidson D, Mittelman MW. Effects of ciprofloxacin and protamine sulfate combinations against catheter-associated Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 1995;39(6):1281–1286. doi: 10.1128/AAC.39.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad GF, El-Feky MA, El-Rehewy MS, Hassan MA, Abolella H, El-Baky RM. Detection of icaA, icaD genes and biofilm production by Staphylococcus aureus and Staphylococcus epidermidis isolated from urinary tract catheterized patients. J Infect Dev Ctries. 2009;3(5):342–351. doi: 10.3855/jidc.241. [DOI] [PubMed] [Google Scholar]
- Virdis S, Scarano C, Cossu F, Spanu V, Spanu C, De Santis EP. Antibiotic resistance in staphylococcus aureus and coagulase negative staphylococci isolated from goats with subclinical mastitis. Veterinary medicine international. 2010;2010:517060. doi: 10.4061/2010/517060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrelli D, Zampaloni C, D’Ercole S, Prenna M, Ballarini P, Ripa S, Vitali LA. Analysis of different genetic traits and their association with biofilm formation in Staphylococcus epidermidis isolates from central venous catheter infections. European journal of clinical microbiology & infectious diseases: official publication of the European Society of Clinical Microbiology. 2006;25(12):773–781. doi: 10.1007/s10096-006-0226-8. [DOI] [PubMed] [Google Scholar]
- Kloos WE, Musselwhite MS. Distribution and persistence of staphylococcus and micrococcus species and other aerobic bacteria on human skin. Appl Microbiol. 1975;30(3):381–385. doi: 10.1128/am.30.3.381-395.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafiso V, Bertuccio T, Santagati M, Campanile F, Amicosante G, Perilli MG, Selan L, Artini M, Nicoletti G, Stefani S. Presence of the ica operon in clinical isolates of staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect. 2004;10(12):1081–1088. doi: 10.1111/j.1469-0691.2004.01024.x. [DOI] [PubMed] [Google Scholar]
- Arslan S, Ozkardes F. Slime production and antibiotic susceptibility in staphylococci isolated from clinical samples. Memorias do Instituto Oswaldo Cruz. 2007;102(1):29–33. doi: 10.1590/S0074-02762007000100004. [DOI] [PubMed] [Google Scholar]
- Foka A, Chini V, Petinaki E, Kolonitsiou F, Anastassiou ED, Dimitracopoulos G, Spiliopoulou I. Clonality of slime-producing methicillin-resistant coagulase-negative staphylococci disseminated in the neonatal intensive care unit of a university hospital. Clin Microbiol Infect. 2006;12(12):1230–1233. doi: 10.1111/j.1469-0691.2006.01565.x. [DOI] [PubMed] [Google Scholar]
- Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, Jabbouri S. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl Microbiol Biotechnol. 2007;75(1):125–132. doi: 10.1007/s00253-006-0790-y. [DOI] [PubMed] [Google Scholar]
- Sadovskaya I, Chaignon P, Kogan G, Chokr A, Vinogradov E, Jabbouri S. Carbohydrate-containing components of biofilms produced in vitro by some staphylococcal strains related to orthopaedic prosthesis infections. FEMS Immunol Med Microbiol. 2006;47(1):75–82. doi: 10.1111/j.1574-695X.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- Gristina AG, Costerton JW. Bacterial adherence to biomaterials and tissue. The significance of its role in clinical sepsis. J Bone Joint Surg Am. 1985;67(2):264–273. [PubMed] [Google Scholar]
- Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. Staphylococcal biofilm exopolysaccharide protects against caenorhabditis elegans immune defenses. PLoS Pathog. 2007;3(4):e57. doi: 10.1371/journal.ppat.0030057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Gara JP. ica and beyond: biofilm mechanisms and regulation in staphylococcus epidermidis and staphylococcus aureus. FEMS Microbiol Lett. 2007;270(2):179–188. doi: 10.1111/j.1574-6968.2007.00688.x. [DOI] [PubMed] [Google Scholar]
- Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F, Hacker J. A novel mechanism of phase variation of virulence in staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32(2):345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- Hennig S, Nyunt Wai S, Ziebuhr W. Spontaneous switch to PIA-independent biofilm formation in an ica-positive staphylococcus epidermidis isolate. International journal of medical microbiology: IJMM. 2007;297(2):117–122. doi: 10.1016/j.ijmm.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Rohde H, Frankenberger S, Zahringer U, Mack D. Structure, function and contribution of polysaccharide intercellular adhesin (PIA) to Staphylococcus epidermidis biofilm formation and pathogenesis of biomaterial-associated infections. Eur J Cell Biol. 2010;89(1):103–111. doi: 10.1016/j.ejcb.2009.10.005. [DOI] [PubMed] [Google Scholar]