Abstract
Throughout the animal kingdom, steroid hormones have been implicated in the defense against microbial infection, but how these systemic signals control immunity is unclear. Here, we show that the steroid hormone ecdysone controls the expression of the pattern recognition receptor PGRP-LC in Drosophila, thereby tightly regulating innate immune recognition and defense against bacterial infection. We identify a group of steroid-regulated transcription factors as well as two GATA transcription factors that act as repressors and activators of the immune response and are required for the proper hormonal control of PGRP-LC expression. Together, our results demonstrate that Drosophila use complex mechanisms to modulate innate immune responses, and identify a transcriptional hierarchy that integrates steroid signalling and immunity in animals.
Keywords: AMPs, ecdysone, IMD pathway, PGRP-LC, Transcription factors
Introduction
Hormones are important regulators of many physiological processes including metabolism, development, reproduction as well as immune responses. In the context of immunity, pharmacologic application of glucocorticoids has multiple potent anti-inflammatory effects on immune cells (Necela and Cidlowski, 2004; Sternberg, 2006). In addition to the well-known immunosuppressive effects of glucocorticoids, many studies have revealed that physiological levels of glucocorticoids actually enhance the immune and inflammatory response (Galon et al, 2002; Shuto et al, 2002; Goulding, 2004; Hermoso et al, 2004; Sakai et al, 2004; Dhabhar, 2009; Busillo et al, 2011). Several other nuclear hormone receptors, including estrogen receptors (ERs), peroxisome proliferator-activated receptors (PPARs), vitamin D receptors (VDRs), retinoid-related orphan receptors (RORs), retinoid X receptors (RXRs) and liver X receptors (LXRs), have also been found to positively regulate innate immunity and proinflammatory cytokine expression (Tontonoz et al, 1998; Hong and Tontonoz, 2008; Jetten, 2009; Baeke et al, 2010; Nunez et al, 2010). All together, these reports exemplify the complex and sometimes contradictory regulation of innate immune and inflammatory responses by steroid/retinoid hormones and their receptors; therefore a more simple and genetically tractable system for studying the interface between steroids and innate immunity would be highly valuable.
Several studies have suggested that the steroid hormone 20-hydroxyecdysone (20E), a central regulator of development, metamorphosis and reproduction in insects (Riddiford, 1993; Kozlova and Thummel, 2000), also positively regulates the innate immune response in the fruit fly, Drosophila melanogaster (Meister and Richards, 1996; Dimarcq et al, 1997; Lanot et al, 2001; Sorrentino et al, 2002; Flatt et al, 2008; Stofanko et al, 2008; Zhang and Palli, 2009). In particular, 20E enhances the pathogen-induced expression of antimicrobial peptide (AMP) genes in both animals and cultured cell lines. These studies suggest that 20E affects AMP expression by regulating the immune deficiency (IMD) pathway, one of two NF-κB signalling pathways in Drosophila, which control the induction of AMP genes in response to DAP-type peptidoglycan (PGN) from the cell wall of Gram-negative and certain Gram-positive bacteria (Kaneko et al, 2006). However, the mechanisms whereby 20E modulates the IMD pathway to affect AMP gene induction remain unclear. In contrast, the mechanisms whereby 20E controls developmental events are well understood.
During development, 20E regulates gene expression through binding to a nuclear hormone receptor heterodimer consisting of the ecdysone receptor (EcR) and ultraspiracle (USP) proteins—orthologs of the vertebrate LXR and RXR receptors, respectively (King-Jones and Thummel, 2005). Ligand binding to the EcR/USP-receptor complex triggers the transcription of ‘early’ response genes, which themselves encode several different transcription factors (e.g., the helix-turn-helix factor Eip93F, the zinc finger factor BR-C, the ETS domain factor Eip74EF and the nuclear hormone receptor Eip75B (Baehrecke and Thummel, 1995; Thummel, 1996; Mugat et al, 2000). Several other genes act as delayed early genes, including the nuclear hormone receptors Eip78C and Hr46 (DHR3), that further contribute to the 20E-triggered transcriptional hierarchy (King-Jones and Thummel, 2005). Here, we identify two novel and distinct mechanisms whereby this ecdysone-inducible transcriptional network controls the IMD immune response in Drosophila. Pattern recognition receptor (PGRP-LC) is a key sensor of DAP-type PGN, and the expression of this receptor, subsequent activation of IMD signal transduction and induction AMP gene expression, is critically dependent on five early ecdysone-inducible transcription factors (BR-C, Eip93F, Eip74EF, Eip78C and HR46) as well as two dGATA factors (Serpent (SRP), and Pannier (PNR)). However, these regulatory connections do not fully explain the 20E control of AMP induction; a second 20E-mediated mechanism, which robustly affects only a subset of AMP genes, is uniquely controlled by BR-C, SRP and PNR. Together, these results demonstrate that 20E functions through multiple, complex and partially overlapping transcriptional circuits to regulate the adult immune response in Drosophila.
Results
20E regulates the expression of PGRP-LC
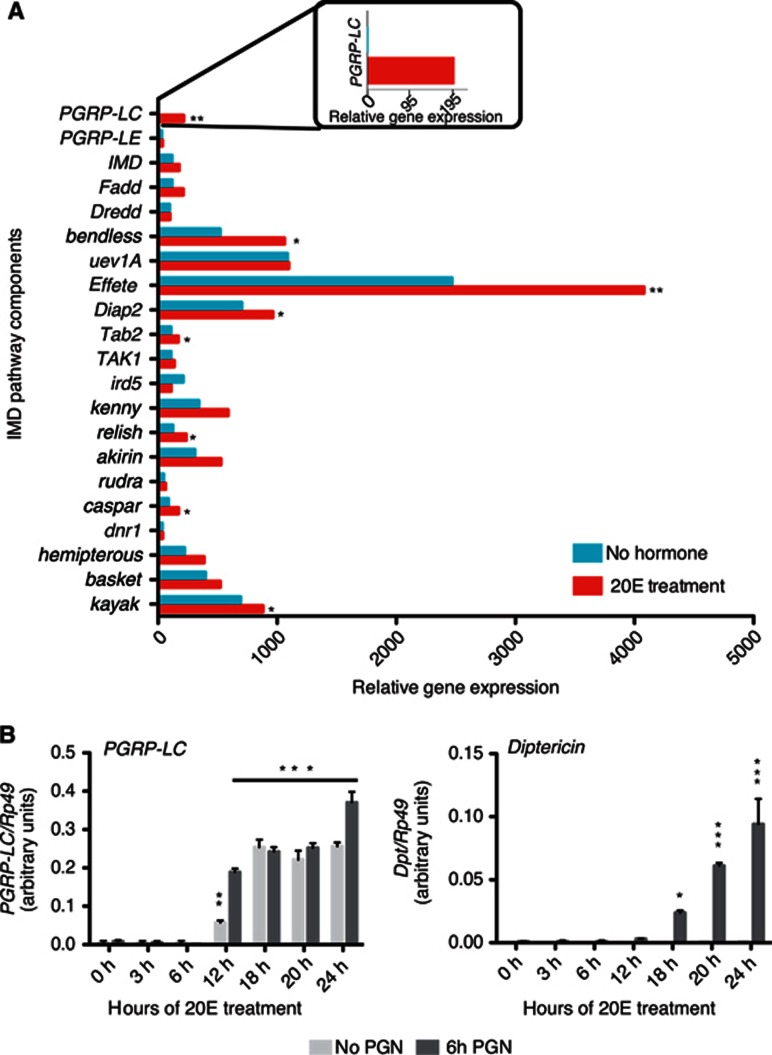
Previously, we and others have demonstrated that the 20E modulates the induction of AMP genes in immune-challenged Drosophila cell culture or whole animals (Meister and Richards, 1996; Dimarcq et al, 1997; Flatt et al, 2008; Zhang and Palli, 2009). In particular, PGN-induced AMP gene induction is nearly undetectable without ∼24 h pretreatment with 20E in S2* cells (Flatt et al, 2008). To identify potential downstream targets of 20E involved in modulating the immune response, microarrays were used to compare the transcriptomes of S2* cells exposed or not to 20E for 24 h. Analysis of the gene expression profiles of all known IMD pathway components identified several signalling factors, such as bendless, effete, Diap2, Tab2, Relish, Caspar and kayak that were modestly but significantly increased upon hormone treatment (Figure 1A), perhaps contributing to the enhancement of PGN-induced AMP gene expression. More striking, PGRP-LC gene expression was undetectable prior to 20E treatment but robustly induced by hormone treatment (P=0.007). PGRP-LC is a critical bacterial sensing receptor required for activating IMD signalling following many bacterial infections (Choe et al, 2002; Gottar et al, 2002; Ramet et al, 2002; Leulier et al, 2003; Werner et al, 2003; Kaneko et al, 2004; Takehana et al, 2004; Kaneko et al, 2006). Validating the array data, qRT–PCR analysis of PGRP-LC expression showed tight hormonal control in S2* cells, with receptor expression preceding the inducibility of the AMP gene Diptericin (Dpt) by approximately 6 h (Figure 1B).
Figure 1.
20E controls PGRP-LC expression. (A) Microarray expression profiles for IMD pathway components. Profiles were generated from triplicate samples of S2* cells before and after 24 h of treatment with 20E. The asterisks represent statistical significance (*P value <0.05; **P<0.01) calculated by unpaired t-test. (B) Real-time qRT–PCR analysis of PGRP-LC and Dpt transcripts from S2* cells that were exposed to 20E for various times, as indicated, and treated with PGN for 6 h or left untreated prior to harvest for RNA isolation. The mean of three independent biological replicates is shown, and error bars represent standard deviation. PGRP-LC levels at 12 h of hormonal treatment with or without PGN stimulation, are significantly increased compared with the untreated samples (0h), while Diptericin levels are significantly increased beginning at 18 h after hormonal treatment, only after PGN stimulation. *P<0.05, **P<0.01, ***P<0.001, as determined by one-way ANOVA with Tukey’s multiple comparison test.
To begin to compare our cell-based results with the intact animal, we examined the modENCODE RNAseq data for the developmental profile of PGRP-LC expression (Graveley et al, 2010). PGRP-LC expression is roughly coincident with the developmental pulses of 20E (Riddiford, 1993; Dubrovsky, 2004; Warren et al, 2006) (Supplementary Figure S1). These observations suggest that 20E plays a major role in controlling PGRP-LC expression, both in vivo and in cell culture.
Ectopic PGRP-LC expression bypasses hormonal control by 20E
To investigate if regulation of PGRP-LC is the primary mechanism by which 20E controls the IMD pathway, a stable cell line expressing C-terminally FLAG-tagged PGRP-LCx from the copper-inducible metallothionein (MT) promoter was established. Fortuitously, we found expression of PGRP-LCx-FLAG to be ‘leaky’ in this cell line, with low-level expression occurring even in the absence of copper (Supplementary Figures S2 and S3, upper panels). As expected, 3 h of treatment with 100 μM CuSO4 further increased the level of PGRP-LCx-FLAG (Supplementary Figures S2 and S3, lower panels). Surprisingly, these data (as well as our unpublished data with other metallothionein promoter-controlled transgenes) show that 20E downregulates the metallothionein promoter (Supplementary Figures S2 and S3); however, this phenomenon is independent of the 20E-control of PGRP-LC expression at its natural locus.
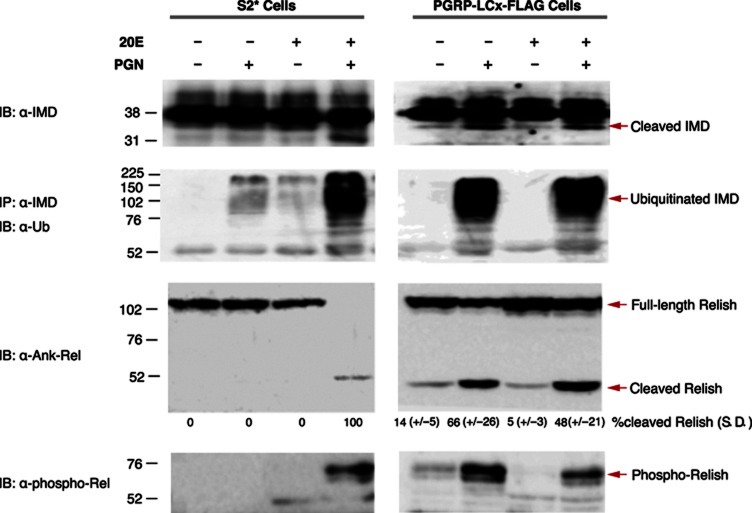
Next, IMD signalling events were evaluated in this PGRP-LCx-FLAG expressing cell line. Previous studies have shown that PGN-stimulation triggers the rapid cleavage and ubiquitination of the IMD protein, while the NF-κB precursor Relish is cleaved, phosphorylated and translocated to the nucleus (Stöven et al, 2000, 2003; Ertürk-Hasdemir et al, 2009; Paquette et al, 2010). Immunoblot analysis of lysates derived from the parental S2* cells showed that 20E pretreatment is required for PGN-induced IMD cleavage and ubiquitination as well as Relish cleavage and phosphorylation (Figure 2, left panels). On the other hand, in the PGRP-LCx expressing cells all of these PGN-triggered events occur upon PGN-stimulation independent of 20E pretreatment (Figure 2, right panels). To examine the nuclear translocation of Relish, the subcellular localization of YFP-tagged Relish was examined by confocal microscopy. In the parental S2* cells, translocation of Relish occurred only when PGN stimulation was preceded by 20E treatment (Supplementary Figure S4A and C), while in the PGRP-LCx-FLAG expressing cells nuclear translation was independent of 20E treatment (Supplementary Figure S4B and D). Together, these biochemical and microscopy analyses demonstrate that ectopic expression of the PGRP-LC receptor is sufficient to support PGN-induced IMD signal transduction, bypassing the need for the steroid hormone 20E.
Figure 2.
20E-independent IMD signalling in PGRP-LCx-FLAG expressing cells. Analysis of whole-cell lysates from parental S2* or PGRP-LCx-FLAG cells with or without 24 h pretreatment with 20E, followed by 10 min stimulation with PGN, as indicated. IMD cleavage was analyzed by immublotting (IB) while IMD ubiquitination was monitored by immunoprecipitation (IP)-IB (upper two panels); Relish cleavage and phosphorylation were analyzed by IB (lower two panels). The percent Relish cleavage was quantified by measuring the intensity of the relevant bands from three different experiments and the mean from three experiments (with s.d.) are shown.
Dual mechanisms of 20E regulation of AMP gene expression
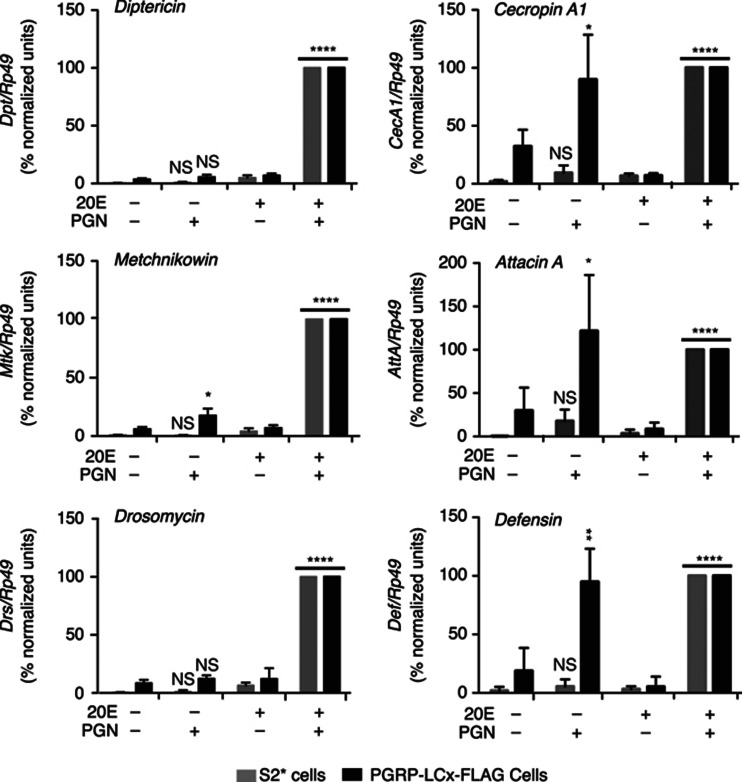
To investigate whether the 20E-independent expression of the PGRP-LC receptor is sufficient to support PGN-induced transcription, AMP gene induction was quantified. In the parental cell line, PGN-triggered AMP gene expression was dependent on pretreatment with 20E (Figure 3, grey bars). Consistent with the biochemical analysis of IMD signalling (shown in Figure 2), the PGN-triggered expression of several AMP genes (Cecropin A1 (CecA1), Attacin A (AttA), and Defensin (Def)) was readily observed without 20E pretreatment in the PGRP-LCx-FLAG expressing cell line (Figure 3, right panels). In fact, ectopic expression of PGRP-LCx in these cells led to modest expression of these AMP genes in the absence of any immune stimulus. On the other hand, the induction of other AMP genes (Diptericin (Dpt), Metchnikowin (Mtk), Drosomycin (Drs)) was still largely dependent on 20E pretreatment, even in the PGRP-LCx expressing cells (Figure 3, left panels, black bars).
Figure 3.
20E-dependent and -independent expression of AMP genes in PGRP-LCx-FLAG cells. qRT–PCR analysis of Dpt, Mtk, Drs, CecA1, AttA and Def expression from parental S2* (grey bars) and PGRP-LCx-FLAG cells (black bars). Cells, with or without 24 h of 20E pretreatment, were stimulated with PGN for 6 h (or not), and then harvested for RNA isolation, as indicated. AMP gene expression was normalized to Rp49 levels, and then normalized between biological replicates and represented as a percentage of the expression level relative to the 20E treated and PGN-stimulated samples from each cell type. For each treatment, the values shown represent the mean of three independent experiments. Error bars represent standard deviations and *P<0.05, **P<0.01, ****P<0.0001 were calculated using unpaired t-test for comparing the corresponding samples with or without hormonal treatment.
Remarkably, these findings suggest two distinct mechanisms for 20E-control of AMP gene induction. The first mechanism involves the 20E-regulated expression of PGRP-LCx. Bypassing this control is sufficient to support PGN-mediated activation of IMD signal transduction and robust induction of a subset of AMP genes (i.e., CecA1, AttA and Def). A second hormone-control mechanism is absolutely required for the immune-induced expression of a distinct subset of AMP genes (Dpt, Mtk and Drs). This second mechanism appears to function downstream of the cleavage and nuclear localization of Relish.
Classic 20E targets are required for the steroid control of IMD signalling
Ecdysone is known to directly regulate a battery of genes through the EcR and ecdysone-response elements found near target genes. Many direct targets of EcR are themselves transcription factors, which initiate a cascade of transcriptional programs downstream of this hormone (Thummel, 2002). Given the 12–18 h required to observe the effect of 20E on PGRP-LC expression and IMD signalling, it seems likely that secondary or tertiary targets of 20E/EcR signalling mediate the IMD-potentiating activity. Therefore, twelve 20E-inducible transcription factors, identified from the literature and our microarray data (Kozlova and Thummel, 2000; Thummel, 2002; King-Jones and Thummel, 2005), were analyzed for their role in 20E-mediated regulation of the IMD immune response (Supplementary Figure S5A and B). RNAi-mediated depletion of seven transcription factors, br-c, Eip78C, Eip93F, Eip74EF, Hr46, srp and pnr, reproducibly blocked 20E-supported PGN-induced Dpt and CecA1 expression in S2* cells (Figures 4A and B). As controls, S2* cells were treated with EcR RNAi, which also prevented 20E immune potentiation (Flatt et al, 2008), or mock RNAi treatment (Figures 4A and B). In contrast, RNAi-mediated depletion of Eip75B robustly enhanced 20E-supported PGN-induced Dpt and CecA1 expression (Figures 4A and B), consistent with an earlier report that Eip75B is a negative regulator of IMD signalling (Kleino et al, 2005). The other four transcription factors analyzed, ERR (estrogen-related receptor), Hsf (heat shock factor), Hnf4 (hepatocyte nuclear factor 4) and luna had no effect on 20E-supported PGN-induced Dpt expression (Supplementary Figure S6).
Figure 4.
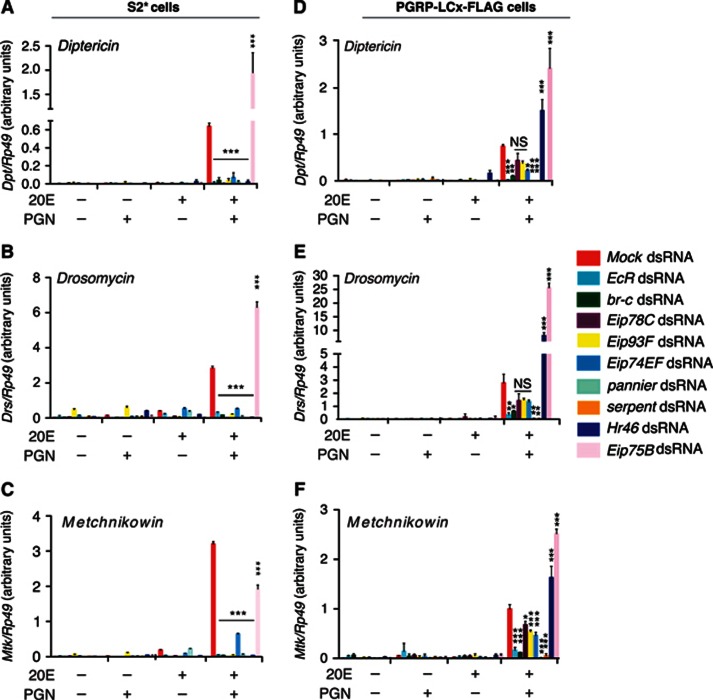
Classic ecdysone signalling pathway components regulate PGRP-LC expression and AMP gene induction. (A and B) qRT–PCR analysis of Dpt (A) or Cecropin A1 (B) induction in S2* cells transfected with RNAi targeting for EcR, br-c, Eip78C, Eip93F, Eip 74EF, Eip75B, Hr46, pnr, srp or mock transfected. Cells with or without exposure to 20E for 24 h were then stimulated (or not) with PGN for an additional 6 h, as indicated. (C) The same RNA was analyzed for the expression of PGRP-LC by qRT–PCR. The mean and s.d. of three independent experiments is shown. P values were calculated by one-way ANOVA with Tukey’s multiple comparison test, ***P<0.001.
Since these results demonstrate that BR-C, Eip78C, Eip93F, Eip74EF, Hr46, SRP and PNR all play a role in 20E-mediated IMD signalling, the role of these transcription factors in the control of PGRP-LC was also quantified. PGRP-LC expression was signficantly reduced in S2* cells when br-c, Eip93F, Eip78C, Eip74EF, srp, pnr, Hr46 or EcR were depleted by RNAi (Figures 4C, ***P<0.001). Depletion of Eip75B, by contrast, increased PGRP-LC levels (***P<0.001), consistent with the higher levels of Dpt and CecA1 expression observed with knockdown of this gene.
A similar RNAi analysis of these transcription factors was also performed in the cell line engineered to express PGRP-LCx-FLAG. Depletion of br-c, srp, pnr or EcR nearly abolished 20E-supported induction of Dpt, Drs and Mtk, even in the presence of PGRP-LCx (***P<0.001, **P<0.01), while targeting the other ecdysone-inducible factors had more modest and variable effects (Figures 5B, D). As expected, the expression of AMP genes was reduced in the parental S2* cells when any of these transcription factors were depleted by RNAi (Figures 5A, B). Together, these data argue that 20E signalling regulates IMD signalling through at least two distinct regulatory mechanisms. First, PGRP-LC expression is controlled by 20E through EcR and Eip78C, Eip93F, Eip74EF, HR46, BR-C, SRP and PNR. Second, the induction of a subset of AMP genes is additionally modulated by hormone, independent of Relish activation, through the activities of EcR, BR-C, SRP and PNR.
Figure 5.
EcR, br-c, srp and pnr are critical for the PGRP-LC-independent hormonal control of AMP gene induction. qRT–PCR analysis of Dpt, Drs and Mtk expression in parental S2* cells (A–C) and PGRP-LCx-FLAG expressing cells (D–F) treated with RNAi targeting for EcR, br-c, Eip78C, Eip93F, Eip74EF, Eip75B, Hr46, pnr, srp or mock transfected. Cells were exposed or not to 20E treatment for 24 h and then stimulated (or not) with PGN for an additional 6 h, as indicated. The results shown represent the mean of three independent experiments and error bars are s.d. Statistical significance was determined by one-way ANOVA with Tukey’s multiple comparison test, *P<0.05, **P<0.01, ***P<0.001.
Ecdysone signalling is required for immune responses in adult flies
To examine the role of the 20E-signalling components in the immune response in vivo, dominant acting mutants affecting ecdysone signalling were exploited (Yoshihara and Ito, 2000; Ishimoto and Kitamoto, 2010). EcRNP5219, which carries P-insertion in an EcR intron, and EcRA483T, which carries a point mutation in the ligand-binding domain of EcR, both exhibit significantly decreased Dpt expression compared to their wild-type controls (cn bw or w1118iso5, respectively) (Supplementary Figure S7A–D). Next, we examined the mutant dominant temperature sensitive 3 (DTS-3), which is deficient for the production of 20E. At the fully restrictive temperature (29°C), DTS-3 mutant displays dominant lethality during development due to a low ecdysone titer (Walker et al, 1987). However, DTS-3/+ females can develop into adults if reared at 25°C, and have a 50% lower 20E titer than wild-type animals (Walker et al, 1987; Ishimoto et al, 2009). Similar to the EcR mutants, DTS-3/+ mutant females reared at 25°C had significantly reduced Dpt expression following E. coli infection as compared to the wild-type control Samarkand (P=0.0001) (Supplementary Figure S7E), while the same strain reared at the permissive temperature (18°C) does not exhibit a significant decrease in Dpt induction (Supplementary Figure S7F) (P=0.1).
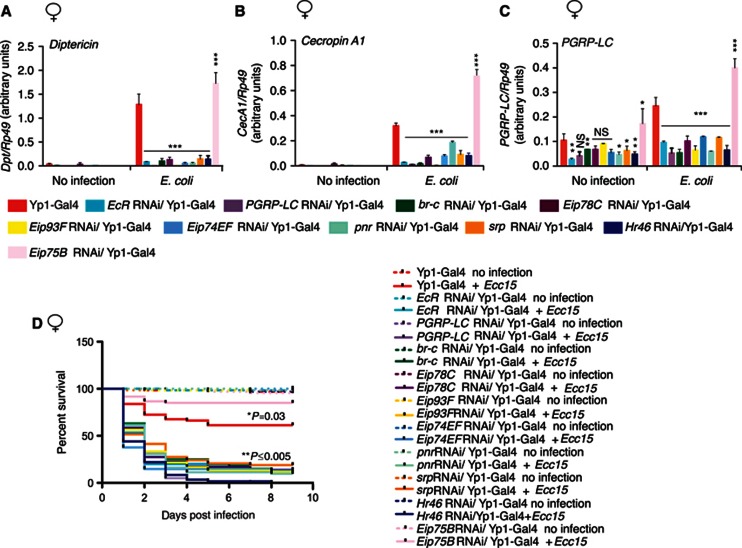
To further investigate the role of the transcription factors functioning downstream of 20E and EcR, an RNAi approach was used. By expressing transgenic hairpin-RNAs with Gal4/UAS system (Brand and Perrimon, 1993; Dietzl et al, 2007), EcR, br-c, Eip78C, Eip93F, Eip74EF, Eip75B, Hr46, srp or pnr were depleted in the adult female fat body, the major site of systemic infection-induced AMP gene expression. Importantly, the Yp1-GAL4 driver used for these studies does not activate gene expression until ∼3 days after eclosion and only in females (Georgel et al, 2001). In contrast to the mutants of EcR and DTS-3 used above, which interfere with ecdysone signalling throughout development, this system allows 20E signalling to be inhibited specifically during adulthood and only in the major immune responsive organ. Experimental Yp1-GAL4>UAS-RNAi animals and controls (Yp1-GAL4 females alone and males with identical genotypes) were infected with E. coli and analyzed for AMP gene expression 24 h later. Similar to the cell-based studies, depletion of EcR, br-c, Eip78C, Eip93F, Eip74EF, Hr46, srp and pnr resulted in significantly reduced induction of Dpt and CecA1 as well as reduced expression of PGRP-LC (Figures 6A–C). Note that males of the identical genotype showed normal AMP and PGRP-LC expression (Supplementary Figure S8A–C). Furthermore, Eip75B RNAi females showed significantly increased levels of the AMP genes as well as the receptor PGRP-LC (Figures 6A–C), thus confirming the role of Eip75B as a negative regulator of IMD signalling in vivo. These results demonstrate that 20E signalling can regulate IMD signalling through br-c, Eip78C, Eip93F, Eip74EF, Hr46, srp and pnr in adults, independent of the developmental properties of 20E.
Figure 6.
EcR, br-c, Eip93F, Eip78C, Eip74EF, Hr46, pnr, and srp knockdown causes immunodeficiency in adult flies. Real-time RT–PCR was used to analyze the expression of Dpt (A), CecA1 (B) and PGRP-LC (C) in EcR, br-c, Eip78C, Eip93F, Eip74EF, Eip75B, Hr46, pnr or srp RNAi expressing flies before or 24 h after infection with E. coli. For all experiments, the yolk protein 1 (Yp1)-GAL4 driver was used to express inverted-repeat RNAs specifically in the adult female fat body, and the Yp1-GAL4 strain is presented as a control. P-values were calculated in comparison to Yp1-GAL4 driver strain by one-way ANOVA with Tukey’s multiple comparison test. *P<0.05, **P<0.01, ***P<0.001, (D) Kaplan–Meier plot showing survival of these same lines after infection with Erwinia carotovora carotovora 15. Survival curves of uninfected animals of all genotypes, overlap and are shown as dashed lines. Statistical significance between the survival of infected RNAi flies and the control Yp1-GAL4 strain were determined by a log-rank test and is equal or less then 0.005 for all comparisons, except for Eip75B RNAi, which survives better with a P=0.03. n=60 for all genotypes, and results are typical of at least three independent experiments.
Fly lines carrying the same UAS-RNAi constructs were also crossed with the C564-GAL4 driver, which expresses GAL4 in the fat body, hemocytes, as well as some male reproductive tissues (Hrdlicka et al, 2002; Buchon et al, 2009). C564-Gal4->UAS-EcR RNAi or UAS-br-C RNAi were not viable to adulthood. However, Dpt expression levels in response to E. coli infection was significantly reduced, compared to control C564-GAL4 driver alone animals, in all other RNAi lines (except Eip75B RNAi), in both males and females (Supplementary Figure S9 A-B). On the other hand, C564-GAL4-driven UAS- Eip75B RNAi significantly enhanced Dpt expression in females, consistent with its role as a negative regulator of IMD signalling. Together, these data demonstrate that ecdysone-signalling pathway has a critical role in regulating IMD signalling and PGRP-LC expression in adult flies.
To determine whether the reduction of infection-induced AMP gene expression observed with depletion of EcR, br-c, Eip78C, Eip93F, Eip74EF, srp or pnr also causes a functional decrease of the realized immune defense, flies were infected with the Gram-negative pathogen Erwinia carotovora carotovora 15 (Ecc15) and their survival monitored. RNAi expressing fly strains were rapidly killed by this infection as compared to Yp1-GAL4 alone females (Figure 6D). Control males did not exhibit any of these phenotypes (Supplementary Figure S8D). On the other hand, when the negative regulator Eip75B was similarly targeted by RNAi, flies showed significantly improved survival, thus loss of Eip75B leads to elevated AMP levels and enhanced immune defense.
Several recent studies have identified genes that affect the ability of flies to survive microbial infections, without altering their ability to kill and clear the infecting pathogen, referred to as tolerance mechanisms rather then resistance mechanisms (Ayres et al, 2008; Schneider and Ayres, 2008). In order to determine whether depletion of EcR, br-c, Eip78C, Eip93F, Eip74EF, Eip75B, Hr46, srp or pnr in the fat body leads to a reduced resistance or tolerance, we measured the number of bacteria present in the fly from 0–48 h post infection with Ecc15 (Supplementary Figure S10). The results show that flies depleted of EcR, br-c, Eip78C, Eip93F, Eip74EF, Hr46, srp or pnr exhibit significantly increased in bacterial loads at 48 h post infection (Supplementary Figure S10A), indicating that interference with ecdysone signalling affects resistance mechanisms in the context of Ecc15 infection. On the other hand, Eip75B RNAi flies more rapidly cleared the infection, with significantly reduced bacterial loads at 24 h consistent with the higher levels of PGRP-LC and AMPs observed in these animals (Supplementary Figure S10A). Control male flies of the same genotypes showed little or no change in Ecc15 clearance, as expected (Supplementary Figure S10B). We also measured bacteria clearance with another Gram-negative bacteria, Enterobacter cloacae, in flies depleted of EcR, br-c, Eip78C and Eip75B (Supplementary Figure S11). Similarly, clearance of this infection was also dependent on 20E signalling, indicating an affect on resistance.
Discussion
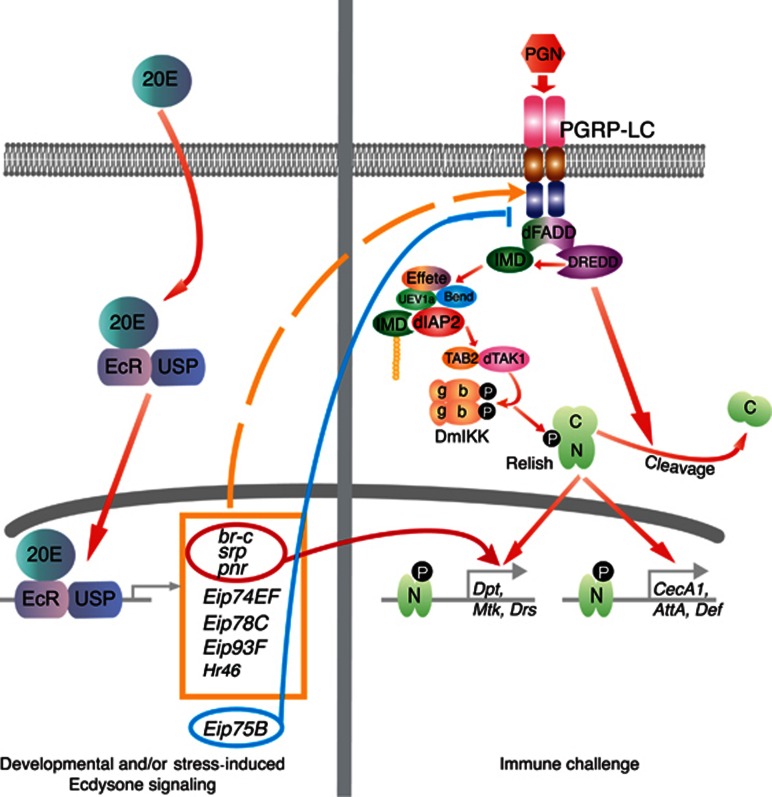
The steroid hormone ecdysone has a critical role in coordinating molting, metamorphosis and reproduction in insects (Riddiford, 1993). Previous studies have also indicated that 20E has profound effects on the Drosophila immune response, especially the IMD-signalling pathway, but the underlying molecular mechanisms have so far remained obscure (Meister and Richards, 1996; Flatt et al, 2008; Zhang and Palli, 2009). In S2* cells, we show here that 20E controls the IMD pathway by at least two distinct mechanisms (Figure 7). First, 20E signalling regulates the expression of the receptor PGRP-LC, thereby affecting all IMD signalling outputs. Ectopic expression of the PGRP-LC receptor is sufficient to support IMD signal transduction and immune-induced expression of some AMP genes, i.e., Cecropin A1, Attacin A and Defensin, in the absence of this steroid hormone. On closer inspection of the data in Figure 3, it is apparent that these AMP genes display weak and variable, but detectable PGN-induced expression in the parental S2* cell line (gray bars) in the absence of hormone pretreatment. This suggests that the Cecropin A1, Attacin A and Defensin loci are primed to respond to PGN stimulation, with AMP expression depending only on the level of the PGRP-LC receptor and the ensuing activity through the IMD signal transduction system. In contrast, the Diptericin, Metchnikowin and Drosomycin loci are nearly unresponsive without hormone pretreatment, regardless of PGRP-LC expression, consistent with the notion that a second, distinct 20E-dependent regulatory circuit directly controls these genes. This second mechanism of hormonal control is absolutely dependent on the EcR, BR-C, SRP and PNR transcription factors, with a lesser contribution from the other ecdysone-inducible transcription factors analyzed.
Figure 7.
Model for 20E regulation of IMD innate immune signalling. 20E controls the IMD innate immune signalling by at least two distinct mechanisms. First, 20E regulates the expression of the peptidoglycan receptor PGRP-LC. This hormonal control involves several ecdysone-inducible transcription factors, including BR-C, Eip78C, Eip93F, Eip74EF, Eip75B, HR46, PNR and SRP. Through this steroid-mediated regulation of the key microbial sensor, immune induction of all AMP genes through the IMD pathway is tightly controlled by prior exposure to this hormone. Through a second mechanism, 20E further regulates the expression of a subset of AMP genes (i.e., Dipt, Mtk and Drs), independent of its control of the receptor PGRP-LC. BR-C, SRP and PNR transcription factors are absolutely required for this PGRP-LC independent hormonal effect. On the other hand, the 20E-inducible nuclear hormone receptor Eip75B negatively regulates the IMD pathway, at least in part, by interfering with PGRP-LC expression.
20E functions through binding to a heterodimeric nuclear hormone-receptor complex consisting of EcR and USP (Yao et al, 1993) that are both required for the effects of 20E on the IMD pathway (Flatt et al, 2008). Many of the immediate transcriptional targets of the EcR/USP complex are themselves transcription factors (Thummel, 1996), driving a complex hierarchy of transcriptional responses downstream of 20E. In addition to activating ‘late’ ecdysone-response genes, which include additional transcription factors, many of the ‘early’ ecdysone-inducible genes also regulate the expression of other ‘early’ response genes (Karim et al, 1993; Thummel, 1996; Lee and Baehrecke, 2001). In this study, we have analyzed 12 of the approximately 64 transcription factors that are induced by 20E at the 24 h time point in our RNA profiling data, while the analysis of the remaining 52 transcription factors await further investigation. Our RNAi-mediated depletion experiments show that seven of these factors (br-c, Eip93F, Eip78C, Ep74EF, Hr46, srp and pnr) have a critical role in the 20E-mediated support of IMD signalling in cells and in adult animals. The in vivo data further suggest that the ecdysone-signalling cascade regulates immune-resistance mechanisms during bacterial infection, in order to limit microbial growth.
Our results demonstrate that reduced 20E signalling can create severe immune deficiency, even in the adult fly where levels of 20E are quite low (Handler, 1982; Schwedes and Carney, 2012), thus underscoring the fundamental role of 20E signalling in promoting immunity in the adult animal.
Interestingly, the dGATAb factor SRP has been previously linked to the Drosophila immune response. In particular, robust induction of Cecropin A1 in the larval fat body requires a GATA element in addition to Relish-binding κB sites (Kadalayil et al, 1997; Petersen et al, 1999; Tingvall et al, 2001). In addition to Cecropin A1, five other AMP genes (Attacin A, Defensin, Drosomycin, Diptericin and Metchnikowin) contain a common organization of regulatory elements: closely linked NF-κB and GATA-binding sites clustered near their transcription start site (Senger et al, 2004, 2006). Interestingly, PGRP-LC also contains a perfect GATA element, in the third exon, that could serve as a binding site for SRP and/or PNR. A direct repeat of NHR-binding elements, which could potentially bind EcR, USP, Hr46, or Eip75B, are also found in the third exon of PGRP-LC. Future studies will be required to evaluate, which of these factors, and/or other factors, directly control hormone-induced PGRP-LC expression.
While five early ecdysone-induced transcription factors (BR-C, Eip93F, Eip78C, Eip74EF and HR46) positively regulate the IMD pathway, Eip75B, another early ecdysone-inducible gene and a nuclear hormone receptor itself, acts as a negative regulator of the innate immune response. Consistent with this observation, Eip75B is known to repress some aspects of the 20E-induced regulatory network through its ability to heterodimerize and interfere with transcriptional activation mediated by another nuclear hormone receptor, HR46 (Thummel, 1997; White et al, 1997; Yamanaka and O'Connor, 2011). Interestingly, HR46 and Eip75B are homologs of ROR and Rev-Erb respectively, that regulate mammalian immune responses. RORα and γ are best known for their critical role in stimulating Th17 development in response to innate immune triggers, such as viral, bacterial, fungal and parasitic infections, promoting production IL-17, IL-21 and IL-22 (Bettelli et al, 2008; Yang et al, 2008; Jetten, 2009; van de Veerdonk et al, 2009), while REV-Erbα has been linked to the circadian control of innate immunity (Ramakrishnan and Muscat, 2006; Gibbs et al, 2012). Interestingly, Eip75B (and REV-Erbβ) utilize heme as a cofactor for the ligand-binding domain. In addition, the heme-E75B complex is repressed by the gases CO or NO, or by oxidation (Caceres et al, 2011; Johnston et al, 2011).
Taken together, these results suggest novel mechanisms for the regulation of insect immunity by both hormonal and environmental factors. Acting through EcR and a set of downstream transcription factors, the steroid 20E potently primes the IMD pathway for a rapid response to infection. In addition, the 20E response also includes a self-limiting component, through the expression of the negative regulator Eip75B. However, Eip75B itself can be further modulated by various environmental factors. These modulators include the heme-mediated activation of E75B, (which is expected to further repress immune responses) or the CO/NO/oxidation-mediated inhibition of the E75B/heme complex, potentiating IMD responses. Interestingly, NO has been implicated in a systemic IMD response following a local oral infection or damage (Foley and O'Farrell, 2003; Wu et al, 2012), although the underlying mechanisms to generate or to respond to this gas remain unclear (Chakrabarti et al, 2012). The findings presented here suggest a potential mechanism whereby NO could enhance an IMD-mediated response by interfering with E75B. In addition, the heme-binding attributes of Eip75B were recently linked to the blood-meal triggered expression of the vitellogenin (Vg) gene in the mosquito Aedes aegypti fat body (Cruz et al, 2012), suggesting that this heme-responsive transcription factor may simultaneously stimulate oogenesis and block IMD signalling, especially in hematophagous insects.
While our current data clearly demonstrate a critical role for 20E signalling in the regulation of the Drosophila immune response, the underlying reasons for this hormonal control of immunity remain opaque. Two possible explanations readily come to mind. In Drosophila and other insects, 20E and the sesquiterpenoid juvenile hormone (JH) have critical roles in orchestrating the major transitions during development, with high levels of 20E and JH driving molting and 20E alone triggering pupation (Dubrovsky, 2004). In an earlier study, we showed that JH counteracts the IMD-potentiating effects of 20E (Flatt et al, 2008). Thus, the ability of 20E and JH to counter-regulate immune function may indicate that the fly sculpts its immune system into the most adaptive configuration for each life stage. The evolutionary pressures that might have created such a regulatory network are unclear, but it may relate to the different microbial threats that are commonly encountered by larvae, pupae or adults.
Another possible reason for tight hormonal control of the Drosophila immune response is inspired by mammalian physiology, where neuroendocrine modulation of the immune response is well-established. The main effectors of this neuroendocrine regulation of mammalian immunity are the glucocorticoids (Webster et al, 2002; Glaser and Kiecolt-Glaser, 2005). Many stressors cause rapid activation of the hypothalamic–pituitary–adrenal axis, thereby initiating a hormonal cascade resulting in the systemic release of glucocorticoids, which in turn regulate the expression of innate immune and inflammatory genes through the glucocorticoid receptor. In adult flies, several studies have demonstrated that stress, for example induced by nutrient restriction, heat treatment or sleep deprivation, leads to increased levels of 20E (Rauschenbach et al, 2000; Terashima et al, 2005; Ishimoto and Kitamoto, 2010, 2011). Thus, it is conceivable that the regulatory network delineated in this study is part of a neuroendocrine-immune axis, whereby stress-induced elevation of the steroid hormone 20E drives elevated PGRP-LC expression and primes all the AMP genes, enabling a more robust immune response during times of stress. Although the direction of this regulation is opposite of that observed in mammals, where stress-induced glucocorticoids are best known to pharmacologically reduce the inflammatory response, several recent reports have clearly shown that glucocorticoids, when produced at physiological levels, actually induce the expression of innate immune receptors like TLR2 and NLRP3 (Shuto et al, 2002; Hermoso et al, 2004; Sakai et al, 2004; Busillo et al, 2011), very similar to the 20E-PGRP-LC axis demonstrated here. Thus, a profound but poorly understood conservation in the neuroendocrine regulation of innate immunity may exist in invertebrates and mammals.
Materials and methods
Microarrays
Affymetrix Drosophila 2.0 Chips were probed in triplicate with RNA isolated from untreated cells or cells treated with 1 μM 20-hydroxyecdysone (Sigma) for 24 h.
cDNA products were hybridized at the Brown University Genomics Core Facility to Affymetrix GeneChip Drosophila_2.0 Genome Arrays (3 replicate chips per treatment) (Li and Wong, 2001). The expression data were analyzed with dCHIP software. (http://biosun1.harvard.edu/complab/dchip/).
Affymetrix microarray data supporting the studies reported here can be found on the GEO database, series record number GSE46020.
Stable cell lines and cell culture
The FLAG-tagged PGRP-LCx construct was cloned into pRmHa3 vector, containing the copper-inducible MT promoter. The construct was then transfected into S2* cells in conjunction with pHs-Neo at a ratio of 50:1 and stable transfectants were selected with G418 (1 mg/ml). The YFP-Relish construct, cloned in pPacPL vector containing the actin promoter, was transfected into the stable cell line expressing the FLAG-tagged PGRP-LCx and into the parental S2* cells in conjunction with BM-IEG-hygromycin at a ratio of 50:1; stable transfectants were then selected with hygromycin (20 U/ml). Drosophila S2* cells were grown in Schneider’s medium (Gibco) with 10% fetal bovine serum, 1% Gluta-MAX (Gibco), and 0.2% penicillin-streptomycin (Gibco) at 27°C.
Co-immunoprecipitation and immunoblotting assays
For immunoblotting analysis, PGRP-LCx-FLAG stable cells and parental S2* cells were split to 1.0 × 106/ml, treated or left untreated with 1 μM 20E (Sigma) for 24 h, and then stimulated with 100 μM copper sulphate for 3 h, when necessary, for increased expression from the metallothionein promoter. For immune stimulation, as indicated, some samples were stimulated for 10 min with 2 μg/ml PGN. The cells were lysed in lysis buffer (20 mM Tris at pH 7.6, 150 mM NaCl, 2 mM EDTA, 10% Glycerol, 1% Triton X-100, 1 mM DTT, NaVO4, glycerol 2-phosphate and protease inhibitors). Total protein extracts were separated by SDS–PAGE and transferred to PVDF membrane (Bio-Rad). Immunoprecipitations were performed with mouse anti-FLAG M2 (Sigma) or anti-IMD (Paquette et al, 2010) antibodies in lysis buffer. For immunoblot analysis, we used anti-FLAG-M2 (Sigma), anti-Ubiquitin (Santa-Cruz) or anti-IMD, anti-Ank-Relish (Stöven et al, 2003) and anti-phospho-Rel (Ertürk-Hasdemir et al, 2009) antibodies. The proteins were detected using HRP-linked anti-mouse IgG (GE Amersham) or anti-rabbit IgG (Bio-Rad) and ECL detection system (Thermo Scientific). Bands were visualized using a Fuji LAS-4000 cooled CCD camera/Dark Box, employing the Image Reader LAS-4000, v 1.1 software. The intensity of Relish protein bands shown in Figure 2 was measured using Multi-Gauge Imaging software and the percentage of Relish cleavage was calculated by dividing the intensity of the cleaved Relish protein by the total Relish intensity.
RNA and qRT–PCR
Total RNA from cultured cells or flies was isolated with the TRIzol reagent (Invitrogen) as previously described (Rutschmann et al, 2000) and then treated with DNase and re-extracted with phenol-chloroform. cDNA was synthesized using iScript cDNA synthesis kit (BioRad) and quantitative PCR analysis was performed on a DNA engine C1000 Thermal Cycler (BioRad), using SYBR Green (BioRad). The specificity of amplification was assessed for each sample by melting curve analysis and relative quantification was performed using a standard curve with dilutions of a standard. Quantified data were normalized to Rp49 levels. To compare AMP gene expression between biological replicates, which always show a robust induction but with variations in the absolute level and amplitude, biological replicates were normalized relative to each other by setting the highest value in any given data set to 100%. Note that qRT–PCR analysis for PGRP-LC examines expression of all three splice-isoforms, with primers hybridizing to the common 5′ exons. In the cell-based experiments, samples were treated with 1 μM 20E for 24 h or left untreated and/or stimulated with 2 μg/ml PGN for additional 6 h prior to harvest for RNA extraction. For in vivo immune stimulation assays, adult flies were infected by pricking in the abdomen with a microsurgery needle dipped into a concentrated pellet of E. coli 1106. RNA was extracted 24 h later and assayed by qRT–PCR.
RNAi
RNAi (dsRNA) to EcR, Eip74EF, Eip75B, Eip78C, Eip93F, br-c, Hr46, ERR, Hnf4, Hsf, luna, srp and pnr, was produced using the T7 RiboMAX Express Large Scale RNA Production System (Promega). S2* cells were split to 1 × 106 cells per ml and incubated for 24 h at 27°C. RNAi (2 μg/ml) was then delivered by calcium phosphate transfection and cells were allowed to recover for ∼24 h at 27°C. Samples were then treated with 1 μM 20E for 24 h or left untreated, and/or stimulated with PGN (2 μg/ml) for additional 6 h before RNA isolation.
Confocal microscopy
For confocal microscopy, double-stable S2* cells expressing FLAG-tagged PGRP-LCx and YFP-Relish and single stables expressing the YFP-Relish were exposed or not to 1 μM 20E for 24 h and then plated on concanavalin A–treated 35 mm glass-bottomed culture. Samples were stimulated with 2 μg/ml PGN for 30 min when required. Cell membranes were stained with Cell Mask 10046 (Invitrogen), while the nuclei were stained with Hoechst 33342 (Invitrogen). The cells were visualized by fluorescence microscopy with a 63X objective on a Leica SP2 AOBS laser-scanning microscope. Images were generated by sequential scanning with 514 nm laser excitation and a 522–599 nm emission window for YFP, 649–666 nm laser excitation for Cell Mask C10046 and a 350–461 nm emission window for Hoechst 33342.
Fly strains and survival experiments
The dominant temperature sensitive 3 (DTS-3) mutant and the EcR mutants: EcRA483T and EcRNP5219, as well as the control strains were obtained from Dr T Kitamoto (University of Iowa, Iowa City, Iowa, USA). DTS-3 was induced in the wild-type strain Samarkand (Holden and Suzuki, 1973); EcRA483T was generated by ethyl methane sulfonate mutagenesis on the cn bw background (Bender et al, 1997), while EcRNP5219 was generated by a P-element insertion in an EcR intron, on an iso(5) background (Yoshihara and Ito, 2000).
EcR, br-c, Eip78C, Eip93F, Eip74EF, Eip75B, srp and pnr RNAi were expressed in the adult female fat body by crossing: w [1118]; P{UAS-EcRGD1428RNAi}v37058, w[1118]; P{UAS-br-cGD4279RNAi}v13705, w[1118]; P{UAS-Eip78CGD4135RNAi}v10396, w[1118]; P{UAS-Eip93FGD4449RNAi}v45857, P{UAS-Eip74EFKK109288RNAi} VIE-260B, P{UAS-Eip75BKK108982RNAi}VIE-260B, w[1118]; P{srpGD12779RNAi}v35578, P{pnrKK108962RNAi}VIE-260B lines from the Vienna Drosophila RNAi Center (VDRC) (Dietzl et al, 2007) to the Yp1-GAL4 driver line (w;P{GAL4-Yp1.JMR}20(yolkGAL4)) (Georgel et al, 2001) or in the fat body and hemocytes, using C564-GAL4 driver line (Hrdlicka et al, 2002; Buchon et al, 2009). In all experiments 3–5 days old mated female flies were used and, as control, male flies with identical genotypes and/or GAL4 driver alone strains were included. Survival experiments were performed with 60 flies, following infection by pricking in the abdomen with a microsurgery needle dipped into a concentrated pellet of Erwinia carotovora carotovora 15 (Basset et al, 2000; Zaidman-Remy et al, 2006). Surviving flies were transferred to fresh vials and counted daily for 9 days. Kaplan–Meier plots are presented and P-values were determined using unpaired t-test analysis.
Determination of CFU counts
To determine the bacterial load in flies at 0 h, 24 h and 48 h after infection with Erwinia carotovora carotovora 15 or Enterobacter cloacae, individual flies were homogenized in 200 μl and of phosphate-buffered saline. The homogenates were diluted in series (usually 10−1 to 10−3), and the dilutions were plated on LB-Ampicilin or LB-nalidixic acid plates, as appropriate, and incubated overnight at 37°C for CFU counting.
Statistical analysis
Throughout, data are presented as mean values and error bars represent standard deviation. The variance and statistical significance were assessed using unpaired t-test or one-way ANOVA, with Tukey's post-test for multiple comparisons, as appropriate. For analysis of bacterial clearance assays, data were first normalized by logarithmic transformation and then the statistical significance was calculated by two-way ANOVA with gender and genotype as main effects, and paired comparisons to driver-alone control using Bonferroni post-test. Throughout, significance level is indicated as **** for P<0.0001, *** for P<0.001, ** for P<0.01, * for P<0.05 and NS for P>0.05.
Supplementary Material
Acknowledgments
We are grateful to all the members of the Silverman Laboratory, especially to Deniz Ertürk-Hasdemir and Nicholas Paquette, for technical and intellectual contributions throughout this work. We also thank K Adelman for her insightful suggestions; B Graveley for providing the modENCODE RNAseq data; M Brodsky and S Wolfe for sharing transcription factor binding site data; T Ip, T Kitamoto, JM Reichhart for bacterial strains, fly strains and/or antibodies; and the Vienna Drosophila RNAi Center (VDRC) for the various RNAi fly strains. NS was supported by NIH grants (AI060025, AI099708 and AG033561) and MT was supported by NIH grants (AG024360, AG031152 and AG033561).
Author contributions: FR, TF, MT and NS designed research; FR, TF, KA, KA, MT, TO, KO and EY performed research; FR, TF and NS analyzed the data; and FR and NS wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ayres JS, Freitag N, Schneider DS (2008) Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178: 1807–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehrecke EH, Thummel CS (1995) The Drosophila E93 gene from the 93F early puff displays stage- and tissue-specific regulation by 20-hydroxyecdysone. Dev Biol 171: 85–97 [DOI] [PubMed] [Google Scholar]
- Baeke F, Takiishi T, Korf H, Gysemans C, Mathieu C (2010) Vitamin D: modulator of the immune system. Curr Opin Pharmacol 10: 482–496 [DOI] [PubMed] [Google Scholar]
- Basset A, Khush RS, Braun A, Gardan L, Boccard F, Hoffmann JA, Lemaitre B (2000) The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci USA 97: 3376–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender M, Imam FB, Talbot WS, Ganetzky B, Hogness DS (1997) Drosophila ecdysone receptor mutations reveal functional differences among receptor isoforms. Cell 91: 777–788 [DOI] [PubMed] [Google Scholar]
- Bettelli E, Korn T, Oukka M, Kuchroo VK (2008) Induction and effector functions of T(H)17 cells. Nature 453: 1051–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, Lemaitre B (2009) A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci USA 106: 12442–12447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busillo JM, Azzam KM, Cidlowski JA (2011) Glucocorticoids sensitize the innate immune system through regulation of the NLRP3 inflammasome. J Biol Chem 286: 38703–38713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres L, Necakov AS, Schwartz C, Kimber S, Roberts IJ, Krause HM (2011) Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev 25: 1476–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S, Liehl P, Buchon N, Lemaitre B (2012) Infection-induced host translational blockage inhibits immune responses and epithelial renewal in the Drosophila gut. Cell Host Microbe 12: 60–70 [DOI] [PubMed] [Google Scholar]
- Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV (2002) Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296: 359–362 [DOI] [PubMed] [Google Scholar]
- Cruz J, Mane-Padros D, Zou Z, Raikhel AS (2012) Distinct roles of isoforms of the heme-liganded nuclear receptor E75, an insect ortholog of the vertebrate Rev-erb, in mosquito reproduction. Mol Cell Endocrinol 349: 262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhabhar FS (2009) Enhancing versus suppressive effects of stress on immune function: implications for immunoprotection and immunopathology. Neuroimmunomodulation 16: 300–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Dimarcq JL, Imler JL, Lanot R, Ezekowitz RA, Hoffmann JA, Janeway CA, Lagueux M (1997) Treatment of l(2)mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect Biochem Mol Biol 27: 877–886 [DOI] [PubMed] [Google Scholar]
- Dubrovsky EB (2004) Hormonal cross talk in insect development. Trends Endocrinol Metab 16: 6–11 [DOI] [PubMed] [Google Scholar]
- Ertürk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stoven S, Meier P, Silverman N (2009) Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA 106: 9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Heyland A, Rus F, Porpiglia E, Sherlock C, Yamamoto R, Garbuzov A, Palli SR, Tatar M, Silverman N (2008) Hormonal regulation of the humoral innate immune response in Drosophila melanogaster. J Exp Biol 211: 2712–2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley E, O'Farrell PH (2003) Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev 17: 115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galon J, Franchimont D, Hiroi N, Frey G, Boettner A, Ehrhart-Bornstein M, O'Shea JJ, Chrousos GP, Bornstein SR (2002) Gene profiling reveals unknown enhancing and suppressive actions of glucocorticoids on immune cells. FASEB J 16: 61–71 [DOI] [PubMed] [Google Scholar]
- Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, Kopczynski C, Duyk G, Reichhart JM, Hoffmann JA (2001) Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell 1: 503–514 [DOI] [PubMed] [Google Scholar]
- Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, Loudon AS (2012) The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci USA 109: 582–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R, Kiecolt-Glaser JK (2005) Stress-induced immune dysfunction: implications for health. Nat Rev Immunol 5: 243–251 [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J (2002) The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416: 640–644 [DOI] [PubMed] [Google Scholar]
- Goulding NJ (2004) The molecular complexity of glucocorticoid actions in inflammation - a four-ring circus. Curr Opin Pharmacol 4: 629–636 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo RN, Miller D, Sturgill D et al. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler AM (1982) Ecdysteroid titers during pupal and adult development in Drosophila melanogaster. Dev Biol 93: 73–82 [DOI] [PubMed] [Google Scholar]
- Hermoso MA, Matsuguchi T, Smoak K, Cidlowski JA (2004) Glucocorticoids and tumor necrosis factor alpha cooperatively regulate toll-like receptor 2 gene expression. Mol Cell Biol 24: 4743–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden JJ, Suzuki DT (1973) Temperature-sensitive mutations in Drosophila melanogaster. XII. The genetic and developmental effects of dominant lethals on chromosome 3. Genetics 73: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Tontonoz P (2008) Coordination of inflammation and metabolism by PPAR and LXR nuclear receptors. Curr Opin Genet Dev 18: 461–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdlicka L, Gibson M, Kiger A, Micchelli C, Schober M, Schock F, Perrimon N (2002) Analysis of twenty-four Gal4 lines in Drosophila melanogaster. Genesis 34: 51–57 [DOI] [PubMed] [Google Scholar]
- Ishimoto H, Kitamoto T (2010) The steroid molting hormone Ecdysone regulates sleep in adult Drosophila melanogaster. Genetics 185: 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto H, Kitamoto T (2011) Beyond molting-roles of the steroid molting hormone ecdysone in regulation of memory and sleep in adult Drosophila. Fly (Austin) 5: 215–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimoto H, Sakai T, Kitamoto T (2009) Ecdysone signaling regulates the formation of long-term courtship memory in adult Drosophila melanogaster. Proc Natl Acad Sci USA 106: 6381–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten AM (2009) Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal 7: e003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DM, Sedkov Y, Petruk S, Riley KM, Fujioka M, Jaynes JB, Mazo A (2011) Ecdysone- and NO-mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Mol Cell 44: 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadalayil L, Petersen UM, Engstrom Y (1997) Adjacent GATA and kappa B-like motifs regulate the expression of a Drosophila immune gene. Nucleic Acids Res 25: 1233–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T, Goldman WE, Mellroth P, Steiner H, Fukase K, Kusumoto S, Harley W, Fox A, Golenbock D, Silverman N (2004) Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20: 637–649 [DOI] [PubMed] [Google Scholar]
- Kaneko T, Yano T, Aggarwal K, Lim JH, Ueda K, Oshima Y, Peach C, Erturk-Hasdemir D, Goldman WE, Oh BH, Kurata S, Silverman N (2006) PGRP-LC and PGRP-LE have essential yet distinct functions in the Drosophila immune response to monomeric DAP-type peptidoglycan. Nat Immunol 7: 715–723 [DOI] [PubMed] [Google Scholar]
- Karim FD, Guild GM, Thummel CS (1993) The Drosophila Broad-Complex plays a key role in controlling ecdysone-regulated gene expression at the onset of metamorphosis. Development 118: 977–988 [DOI] [PubMed] [Google Scholar]
- King-Jones K, Thummel CS (2005) Nuclear receptors--a perspective from Drosophila. Nat Rev Genet 6: 311–323 [DOI] [PubMed] [Google Scholar]
- Kleino A, Valanne S, Ulvila J, Kallio J, Myllymaki H, Enwald H, Stoven S, Poidevin M, Ueda R, Hultmark D, Lemaitre B, Rämet M (2005) Inhibitor of apoptosis 2 and TAK1-binding protein are components of the Drosophila Imd pathway. EMBO J 24: 3423–3434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlova T, Thummel CS (2000) Steroid regulation of postembryonic development and reproduction in Drosophila. Trends Endocrinol Metab 11: 276–280 [DOI] [PubMed] [Google Scholar]
- Lanot R, Zachary D, Holder F, Meister M (2001) Postembryonic hematopoiesis in Drosophila. Dev Biol 230: 243–257 [DOI] [PubMed] [Google Scholar]
- Lee CY, Baehrecke EH (2001) Steroid regulation of autophagic programmed cell death during development. Development 128: 1443–1455 [DOI] [PubMed] [Google Scholar]
- Leulier F, Parquet C, Pili-Floury S, Ryu JH, Caroff M, Lee WJ, Mengin-Lecreulx D, Lemaitre B (2003) The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat Immunol 4: 478–484 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98: 31–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister M, Richards G (1996) Ecdysone and insect immunity: the maturation of the inducibility of the diptericin gene in Drosophila larvae. Insect Biochem Mol Biol 26: 155–160 [DOI] [PubMed] [Google Scholar]
- Mugat B, Brodu V, Kejzlarova-Lepesant J, Antoniewski C, Bayer CA, Fristrom JW, Lepesant JA (2000) Dynamic expression of broad-complex isoforms mediates temporal control of an ecdysteroid target gene at the onset of Drosophila metamorphosis. Dev Biol 227: 104–117 [DOI] [PubMed] [Google Scholar]
- Necela BM, Cidlowski JA (2004) Mechanisms of glucocorticoid receptor action in noninflammatory and inflammatory cells. Proc Am Thorac Soc 1: 239–246 [DOI] [PubMed] [Google Scholar]
- Nunez V, Alameda D, Rico D, Mota R, Gonzalo P, Cedenilla M, Fischer T, Bosca L, Glass CK, Arroyo AG, Ricote M (2010) Retinoid X receptor alpha controls innate inflammatory responses through the up-regulation of chemokine expression. Proc Natl Acad Sci USA 107: 10626–10631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Ertürk-Hasdemir D, Reichhart JM, Meier P, Silverman N (2010) Caspase-mediated cleavage, IAP binding, and ubiquitination: linking three mechanisms crucial for Drosophila NF-kappaB signaling. Mol Cell 37: 172–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen UM, Kadalayil L, Rehorn KP, Hoshizaki DK, Reuter R, Engstrom Y (1999) Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J 18: 4013–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan SN, Muscat GE (2006) The orphan Rev-erb nuclear receptors: a link between metabolism, circadian rhythm and inflammation? Nucl Recept Signal 4: e009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA (2002) Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416: 644–648 [DOI] [PubMed] [Google Scholar]
- Rauschenbach IY, Sukhanova MZ, Hirashima A, Sutsugu E, Kuano E (2000) Role of the ecdysteroid system in the regulation of Drosophila reproduction under environmental stress. Dokl Biol Sci 375: 641–643 [DOI] [PubMed] [Google Scholar]
- Riddiford LM (1993) Hormone receptors and the regulation of insect metamorphosis. Receptor 3: 203–209 [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D (2000) Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol 1: 342–347 [DOI] [PubMed] [Google Scholar]
- Sakai A, Han J, Cato AC, Akira S, Li JD (2004) Glucocorticoids synergize with IL-1beta to induce TLR2 expression via MAP kinase phosphatase-1-dependent dual inhibition of MAPK JNK and p38 in epithelial cells. BMC Mol Biol 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DS, Ayres JS (2008) Two ways to survive infection: what resistance and tolerance can teach us about treating infectious diseases. Nat Rev Immunol 8: 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwedes CC, Carney GE (2012) Ecdysone signaling in adult Drosophila melanogaster. J Insect Physiol. 58: 293–302 [DOI] [PubMed] [Google Scholar]
- Senger K, Armstrong GW, Rowell WJ, Kwan JM, Markstein M, Levine M (2004) Immunity regulatory DNAs share common organizational features in Drosophila. Mol Cell 13: 19–32 [DOI] [PubMed] [Google Scholar]
- Senger K, Harris K, Levine M (2006) GATA factors participate in tissue-specific immune responses in Drosophila larvae. Proc Natl Acad Sci USA 103: 15957–15962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuto T, Imasato A, Jono H, Sakai A, Xu H, Watanabe T, Rixter DD, Kai H, Andalibi A, Linthicum F, Guan YL, Han J, Cato AC, Lim DJ, Akira S, Li JD (2002) Glucocorticoids synergistically enhance nontypeable Haemophilus influenzae-induced Toll-like receptor 2 expression via a negative cross-talk with p38 MAP kinase. J Biol Chem 277: 17263–17270 [DOI] [PubMed] [Google Scholar]
- Sorrentino RP, Carton Y, Govind S (2002) Cellular immune response to parasite infection in the Drosophila lymph gland is developmentally regulated. Dev Biol 243: 65–80 [DOI] [PubMed] [Google Scholar]
- Sternberg EM (2006) Neural regulation of innate immunity: a coordinated nonspecific host response to pathogens. Nat Rev Immunol 6: 318–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stofanko M, Kwon SY, Badenhorst P (2008) A misexpression screen to identify regulators of Drosophila larval hemocyte development. Genetics 180: 253–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D (2000) Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep 1: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, Maniatis T, Hultmark D (2003) Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci USA 100: 5991–5996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehana A, Yano T, Mita S, Kotani A, Oshima Y, Kurata S (2004) Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J 23: 4690–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima J, Takaki K, Sakurai S, Bownes M (2005) Nutritional status affects 20-hydroxyecdysone concentration and progression of oogenesis in Drosophila melanogaster. J Endocrinol 187: 69–79 [DOI] [PubMed] [Google Scholar]
- Thummel CS (1996) Flies on steroids--Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet 12: 306–310 [DOI] [PubMed] [Google Scholar]
- Thummel CS (1997) Dueling orphans--interacting nuclear receptors coordinate Drosophila metamorphosis. Bioessays 19: 669–672 [DOI] [PubMed] [Google Scholar]
- Thummel CS (2002) Ecdysone-regulated puff genes 2000. Insect Biochem Mol Biol 32: 113–120 [DOI] [PubMed] [Google Scholar]
- Tingvall TO, Roos E, Engstrom Y (2001) The GATA factor serpent is required for the onset of the humoral immune response in Drosophila embryos. Proc Natl Acad Sci USA 98: 3884–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM (1998) PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell 93: 241–252 [DOI] [PubMed] [Google Scholar]
- van de Veerdonk FL, Gresnigt MS, Kullberg BJ, van der Meer JW, Joosten LA, Netea MG (2009) Th17 responses and host defense against microorganisms: an overview. BMB Rep 42: 776–787 [DOI] [PubMed] [Google Scholar]
- Walker VK, Watson KL, Holden JJA, Steel CGH (1987) Vitellogenesis and fertility in Drosophila females with low ecdysteroid titres; the L(3)3DTS mutation. J Insect Physiol 33: 137–142 [Google Scholar]
- Warren JT, Yerushalmi Y, Shimell MJ, O'Connor MB, Restifo LL, Gilbert LI (2006) Discrete pulses of molting hormone, 20-hydroxyecdysone, during late larval development of Drosophila melanogaster: correlations with changes in gene activity. Dev Dyn 235: 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JI, Tonelli L, Sternberg EM (2002) Neuroendocrine regulation of immunity. Annu Rev Immunol 20: 125–163 [DOI] [PubMed] [Google Scholar]
- Werner T, Borge-Renberg K, Mellroth P, Steiner H, Hultmark D (2003) Functional diversity of the Drosophila PGRP-LC gene cluster in the response to lipopolysaccharide and peptidoglycan. J Biol Chem 278: 26319–26322 [DOI] [PubMed] [Google Scholar]
- White KP, Hurban P, Watanabe T, Hogness DS (1997) Coordination of Drosophila metamorphosis by two ecdysone-induced nuclear receptors. Science 276: 114–117 [DOI] [PubMed] [Google Scholar]
- Wu SC, Liao CW, Pan RL, Juang JL (2012) Infection-induced intestinal oxidative stress triggers organ-to-organ immunological communication in Drosophila. Cell Host Microbe 11: 410–417 [DOI] [PubMed] [Google Scholar]
- Yamanaka N, O'Connor MB (2011) Nitric oxide directly regulates gene expression during Drosophila development: need some gas to drive into metamorphosis? Genes Dev 25: 1459–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, Watowich SS, Tian Q, Jetten AM, Dong C (2008) T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity 28: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao TP, Forman BM, Jiang Z, Cherbas L, Chen JD, McKeown M, Cherbas P, Evans RM (1993) Functional ecdysone receptor is the product of EcR and Ultraspiracle genes. Nature 366: 476–479 [DOI] [PubMed] [Google Scholar]
- Yoshihara M, Ito K (2000) Improved Gal4 screening kit for large-scale generation of enhancer-trap strains. Dros. Inf. Serv 83: 199–202 [Google Scholar]
- Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, Oh BH, Ueda R, Mengin-Lecreulx D, Lemaitre B (2006) The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24: 463–473 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Palli SR (2009) Identification of a cis-regulatory element required for 20-hydroxyecdysone enhancement of antimicrobial peptide gene expression in Drosophila melanogaster. Insect Mol Biol 18: 595–605 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.