Abstract
Global increases in small ubiquitin-like modifier (SUMO)-2/3 conjugation are a neuroprotective response to severe stress but the mechanisms and specific target proteins that determine cell survival have not been identified. Here, we demonstrate that the SUMO-2/3-specific protease SENP3 is degraded during oxygen/glucose deprivation (OGD), an in vitro model of ischaemia, via a pathway involving the unfolded protein response (UPR) kinase PERK and the lysosomal enzyme cathepsin B. A key target for SENP3-mediated deSUMOylation is the GTPase Drp1, which plays a major role in regulating mitochondrial fission. We show that depletion of SENP3 prolongs Drp1 SUMOylation, which suppresses Drp1-mediated cytochrome c release and caspase-mediated cell death. SENP3 levels recover following reoxygenation after OGD allowing deSUMOylation of Drp1, which facilitates Drp1 localization at mitochondria and promotes fragmentation and cytochrome c release. RNAi knockdown of SENP3 protects cells from reoxygenation-induced cell death via a mechanism that requires Drp1 SUMOylation. Thus, we identify a novel adaptive pathway to extreme cell stress in which dynamic changes in SENP3 stability and regulation of Drp1 SUMOylation are crucial determinants of cell fate.
Keywords: apoptosis, Drp1, mitochondria, SENP3, SUMO
Introduction
Post-translational modification of substrate proteins by the covalent attachment of small ubiquitin-like modifier (SUMO) proteins regulates multiple cellular pathways, ranging from nuclear organization and transcriptional regulation to membrane protein trafficking and stability (Wilkinson and Henley, 2010). There are three SUMO paralogues (SUMO-1–3) in mammals, which are conjugated to lysine residues in target proteins by an enzymatic pathway comprising an E1 complex, SAE1/SAE2, a single E2, Ubc9 and various E3 enzymes (Wilkinson and Henley, 2010). SUMO-2 and SUMO-3 differ by just three N-terminal amino acids, but share only ∼50% sequence identity to SUMO-1. SUMO proteins are deconjugated from substrates via the actions of the SENP family of SUMO-specific isopeptidases. There are six mammalian SENPs: SENP1–3 and SENP5–7. Of these, SENP1 and 2 show a broad specificity against SUMO-1 and SUMO-2/3, SENP3 and SENP5 favour removal of SUMO-2/3 over SUMO-1 and SENP6–7 edit SUMO-2/3 chains on substrates (Yeh, 2009). Thus, the dynamic balance between Ubc9-mediated SUMO conjugation and SENP-mediated SUMO removal determines the SUMOylation status of substrate proteins.
Brain ischaemia is a major cause of death and disability. It occurs when the blood supply to a part of the brain is interrupted by trauma, occlusion following a stroke or by heart failure. The oxygen and glucose deprivation (OGD) during ischaemia, and the reperfusion damage that occurs when the blood supply is restored and oxygen and glucose become available, exposes cells to extreme metabolic stress.
At the cellular level, ischaemia causes ATP depletion, glutamate excitotoxicity, calcium overload, mitochondrial dysfunction and oxidative damage. To counteract these stressors, cells utilize a variety of adaptive responses to decrease energy expenditure, increase nutrient availability and promote cell survival. The endoplasmic reticulum unfolded protein response (UPR) is activated when misfolded proteins accumulate in the ER lumen as a result of oxidative stress (Hetz, 2012). The protein kinase RNA (PKR)-like ER kinase (PERK) is a critical initiator of UPR signalling, which attempts to restore normal ER function by inhibiting general protein synthesis while promoting transcription of ER chaperones and folding enzymes to enhance ER processing and alleviate protein aggregation (Yang and Paschen, 2009). The UPR initially promotes cell survival; however, when overwhelmed, it initiates pro-apoptotic pathways involving signalling to B-cell lymphoma protein 2 (Bcl-2) family proteins located at mitochondria (Szegezdi et al, 2009). Thus, the UPR is fundamental in dictating whether cells survive post-ischaemia through signalling to mitochondria.
SUMOylation is strongly implicated in a range of neurodegenerative disorders, suggesting a critical role for protein SUMOylation in regulating neuronal function (Wilkinson et al, 2010) and dysfunction (Dorval and Fraser, 2007; Anderson et al, 2009). In response to ischaemic stress, global levels of SUMO-2/3 conjugation are massively enhanced in neurons (Cimarosti et al, 2008, 2012; Yang et al, 2008a, 2008b). Intriguingly, protein SUMO-2/3-ylation is also increased in the brains of hibernating animals leading to the proposal that it constitutes a cytoprotective pathway for ischaemic preconditioning (Lee et al, 2007). This is supported by observations that overexpression of SUMO-1 or SUMO-2 can increase resistance to ischaemia (Lee et al, 2009), and silencing SUMO-2/3 using microRNA makes cells more vulnerable to ischaemic stress (Datwyler et al, 2011). However, the mechanisms that regulate protein SUMOylation and the identity of many of the substrate proteins involved in the adaptive response to stress have remained elusive.
Severe ischaemia causes cell death through cytochrome c release into the cytosol, which leads to caspase cleavage and apoptosis. Cytochrome c can be released by mitochondrial fission (Wilson et al, 2013) and/or via mitochondrial outer membrane permeabilization (MOMP) through the formation of channels by oligomerization of the pro-apoptotic Bcl-2 family members on the outer mitochondrial membrane (Montessuit et al, 2010). The mitochondrial GTPase dynamin-related protein 1 (Drp1) plays key roles in both fission and MOMP. Under basal conditions, Drp1 is primarily localized in the cytosol but when recruited to the mitochondrial membrane it can assemble into spiral structures that wrap around the mitochondria to mediate fission (Frank et al, 2001; Cassidy-Stone et al, 2008; Jourdain and Martinou, 2009; Wilson et al, 2013). Additionally, in response to apoptotic stimuli Drp1 is recruited to the mitochondrial membrane to initiate oligomerization of the pro-apoptotic Bcl-2 family proteins Bax and Bak and activate MOMP (Montessuit et al, 2010). Inhibition of Drp1 is neuroprotective against glutamate excitotoxicity and OGD in cultured cells, and ischaemic brain damage in vivo (Grohm et al, 2012). Furthermore, depending on the conditions, either upregulation or downregulation of Drp1 has been reported to protect against apoptosis (Lee et al, 2004; Szabadkai et al, 2004). Drp1 has been reported to be a substrate for both SUMO-1 and SUMO-2/3 conjugation, and it has been suggested that SUMOylation may regulate the partitioning of Drp1 between mitochondria and the cytosol (Wasiak et al, 2007; Figueroa-Romero et al, 2009).
We investigated the molecular mechanisms of SUMOylation in the cellular response to OGD. We show that levels of the SUMO-2/3-specific deSUMOylating enzyme SENP3 are greatly reduced during OGD via a pathway that requires activation of the UPR-associated kinase PERK. This downregulation of SENP3 increases Drp1 SUMO-2/3-ylation and decreases its mitochondrial association, which reduces Drp1-dependent cytochrome c release via reduced mitochondrial fission and fragmentation. Thus, the loss of SENP3 during ischaemia enhances cell survival. However, SENP3 levels recover during reoxygenation after OGD leading to Drp1 dSUMOylation, which is instrumental in initiating apoptotic cell death. Taken together, our results reveal a novel pro-survival cross-talk pathway between the UPR and mitochondria, mediated through loss of the deSUMOylating enzyme SENP3. Furthermore, our results demonstrate that SUMO-2/3-ylation of Drp1 is a crucial determinant of cell survival after ischaemic insult.
Results
OGD enhances SUMO-2/3 conjugation by degrading SENP3
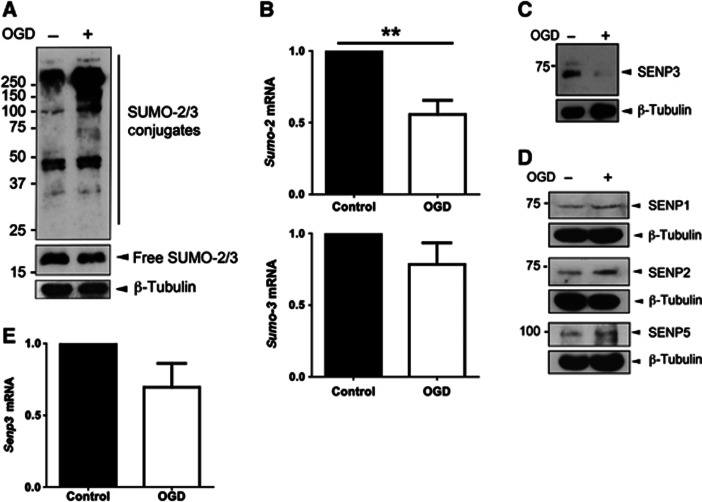
OGD in dissociated primary cultured rat cortical neurons caused a substantial increase in SUMO-2/3 conjugation (Figure 1A) but not in SUMO-1 conjugation (Supplementary Figure 1A). Levels of sumo-3 and sumo-2 mRNA were unchanged and significantly reduced respectively, indicating that increased SUMO-2/3-ylation was not due to increased transcription (Figure 1B). We therefore investigated the effects of OGD on the deSUMOylating SENP enzymes in cultured neurons. Levels of the SUMO-2/3-specific protease SENP3 were dramatically reduced immediately following OGD (Figure 1C), whereas levels of SENP1, 2 and 5 remained relatively unchanged (Figure 1D). Levels of senp3 mRNA were not significantly altered by OGD (Figure 1E). We also observed a substantial increase in SUMO-2/3 conjugation in HEK293 cells and a corresponding marked decrease in SENP3 levels immediately after OGD (Supplementary Figure 1B). After termination of OGD followed by reoxygenation, levels of endogenous SENP3 recovered with a time course that mirrored the decrease in global SUMO-2/3 conjugation (Supplementary Figure 1C). Indeed, consistent with the inverse correlation between SENP3 levels and SUMO-2/3-ylation, SENP3 knockdown significantly increased SUMO-2/3 conjugation in all cell types investigated (Supplementary Figure 1D). Furthermore, SENP3 knockdown occluded the OGD-induced increase in SUMO-2/3 conjugation in HEK293 cells (Supplementary Figure 1E). Taken together, these findings suggest that levels of SENP3 are tightly controlled during and after OGD to regulate SUMO-2/3 conjugation.
Figure 1.
OGD increases global SUMO-2/3 conjugation and decreases SENP3 in neurons. (A) Lysates of primary cortical neurons were blotted for SUMO-2/3 and β-tubulin after OGD (1 h). (B) sumo-2 mRNA levels are decreased and sumo-3 mRNA levels remain unchanged after OGD (1 h) in primary cortical neurons. (C) Levels of SENP3 are reduced in neurons after OGD (30 min). (D) OGD (30 min) does not alter SENP1, SENP2 or SENP5 levels in neurons. (E) senp3 mRNA levels are not reduced by OGD (1 h). For (B) and (E) significance was determined using a two-tailed Paired t-test; for sumo-2 mRNA (n=9; **P<0.01), for sumo-3 mRNA (n=7) and for senp3 mRNA (n=8).
Source data for this figure is available on the online supplementary information page.
PERK activation is required for SENP3 degradation
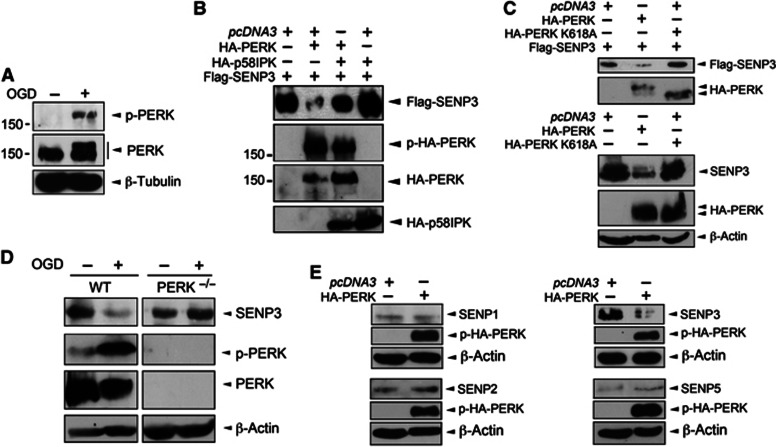
The ER kinase PERK is activated by ischaemia and reperfusion (Kumar et al, 2001). We detected increased levels of phosphorylated, active PERK (p-PERK) in cultured neurons following OGD (Figure 2A). To explore if PERK activation is required for SENP3 degradation during OGD, we co-expressed HA-PERK and Flag-SENP3 in HEK293 cells. PERK expression dramatically decreased SENP3 levels via a mechanism dependent on its kinase activity since co-expression of the PERK-specific inhibitor p58IPK (Yan et al, 2002; Figure 2B) or expression of a kinase-dead PERK mutant (K618A; Harding et al, 1999; Figure 2C) did not reduce SENP3 levels. To further confirm a role for PERK in regulating SENP3 stability, we subjected wild-type (WT) and PERK−/− mouse embryonic fibroblasts (MEFs) (Harding et al, 2001) to severe OGD. In MEFs from WT mice OGD induced PERK phosphorylation and dramatically reduced SENP3 levels. In stark contrast, OGD had no affect on SENP3 in PERK−/− MEFs, indicating that PERK activation is directly responsible for regulating SENP3 levels during ischaemic stress (Figure 2D).
Figure 2.
PERK activation is required for decreased SENP3. (A) PERK is phosphorylated (p-PERK) in neurons by OGD (30 min). Blots were probed with phospho-PERK, PERK and β-tubulin antibodies. (B) Overexpression of HA-PERK causes loss of Flag-SENP3 in HEK293 cells, which is blocked by overexpression of the PERK inhibitor p58IPK. Lysates were blotted with Flag, phospho-PERK and HA antibodies. (C) The kinase activity of PERK is required for SENP3 removal in HEK293 cells. HA-PERK, but not kinase-dead HA-PERK K618A decreases SENP3. The top panel shows the effect of PERK on overexpressed Flag-SENP3, while the lower panel shows endogenous SENP3. (D) PERK is required for OGD-induced loss of SENP3. Confluent wild-type and PERK−/− MEFs were subjected to OGD (2 h), harvested and the lysates probed as indicated. (E) Overexpression of HA-PERK in HEK293 cells reduces levels of SENP3 but not of SENP1, SENP2 or SENP5.
Source data for this figure is available on the online supplementary information page.
Although proteolysis of SENP3 requires PERK kinase activity we were unable to detect direct PERK phosphorylation of SENP3 in in vitro [32P]ATP kinase assays or pulldown experiments from HEK293 cells (Supplementary Figure 2A and B). We individually mutated 11 serine and 3 threonine residues in SENP3 identified as potential phosphorylation sites to alanine and tested the resistance of the SENP3 mutants to PERK-mediated degradation. All of these mutants were degraded when co-expressed with PERK (Supplementary Figure 2C).
An important PERK target is the eukaryotic initiation factor eIF2α (Van Der Kelen et al, 2009), which under stress conditions is phosphorylated by PERK to inhibit protein synthesis (Koumenis et al, 2002). We used MEF cells expressing a non-activatable form of eIF2α (eIF2α S51A) (Scheuner et al, 2001) to test if PERK phosphorylation of eIF2α plays a role in SENP3 degradation. OGD reduced SENP3 levels in both WT and eIF2α S51A MEF cells, indicating that phosphorylation of eIF2α and inhibition of translation are not involved in the OGD-induced loss of SENP3 (Supplementary Figure 2D).
SENP3 degradation does not involve ubiquitination but requires cathepsin B
We next tested whether SENP3 degradation was dependent on ubiquitination. Co-expression of SENP3 with ubiquitin (Ub) or the chain-forming deficient mutant Ub K7R indicated that SENP3 was efficiently mono-ubiquitinated (Supplementary Figure 3A). However, mutation of K231, the major ubiquitination site, did not prevent PERK-mediated SENP3 degradation (Supplementary Figure 3B and C). Furthermore, we individually mutated each of the 28 lysines in SENP3 to arginine but observed no affect on PERK-mediated degradation (Supplementary Figure 3D). Similarly, a series of truncation mutants spanning the full length of SENP3 (amino acids 1–200, 1–400 and 370–574) were each susceptible to PERK-mediated degradation (Supplementary Figure 3E). Taken together, these results suggest that canonical ubiquitin-mediated degradation is not responsible for SENP3 loss during OGD.
Despite this apparent independence of ubiquitination, OGD-induced SENP3 degradation was largely blocked by co-application of the lysosome inhibitors leupeptin and chloroquine, but was unaffected by application of the proteasome inhibitor MG132, suggesting that SENP3 is susceptible to the actions of lysosomal proteases (Supplementary Figure 3F). Lysosomal rupture and release of cathepsin proteases can trigger caspase-dependent cell death (Boya et al, 2003; Boya and Kroemer, 2008) and cathepsin B is implicated in the UPR stress response (Friedlander et al, 2000; Kim et al, 2006). Since PERK-mediated SENP3 degradation can be blocked by lysosomal inhibition, we investigated the effects of OGD on cathepsin B in cortical neurons. The precursor protein pro-cathepsin B was cleaved to active cathepsin B by OGD in neurons (Supplementary Figure 4A) and PERK overexpression in HEK293 cells (Supplementary Figure 4B and C). Importantly, selective inhibition of cathepsin B prevented SENP3 degradation invoked by overexpression of PERK (Supplementary Figure 4D) or OGD in HEK293 cells (Supplementary Figure 4E). Additionally, cathepsin B-mediated degradation of SENP3 was assessed in in vitro assays. While GST was only slightly affected by incubation with purified cathepsin B, GST-SENP3 was extensively degraded (Supplementary Figure 4F) suggesting that cathepsin B directly degrades SENP3. We next measured PERK activation and SENP3 levels in cathepsin B−/− MEF cells subjected to OGD. Consistent with the inhibitor data, in cathepsin B−/− MEF cells, OGD still led to PERK activation, but there was no decrease in SENP3 levels (Supplementary Figure 4G). Thus, PERK-mediated maturation and release of cathepsin B contribute to SENP3 proteolysis during OGD. However, since cathepsin B inhibitor II did not entirely prevent OGD-induced SENP3 loss in primary neuronal cultures, we conclude that there are also other pathways present in neurons that can degrade SENP3 during OGD.
SUMOylation of Drp1 regulates cytochrome c release
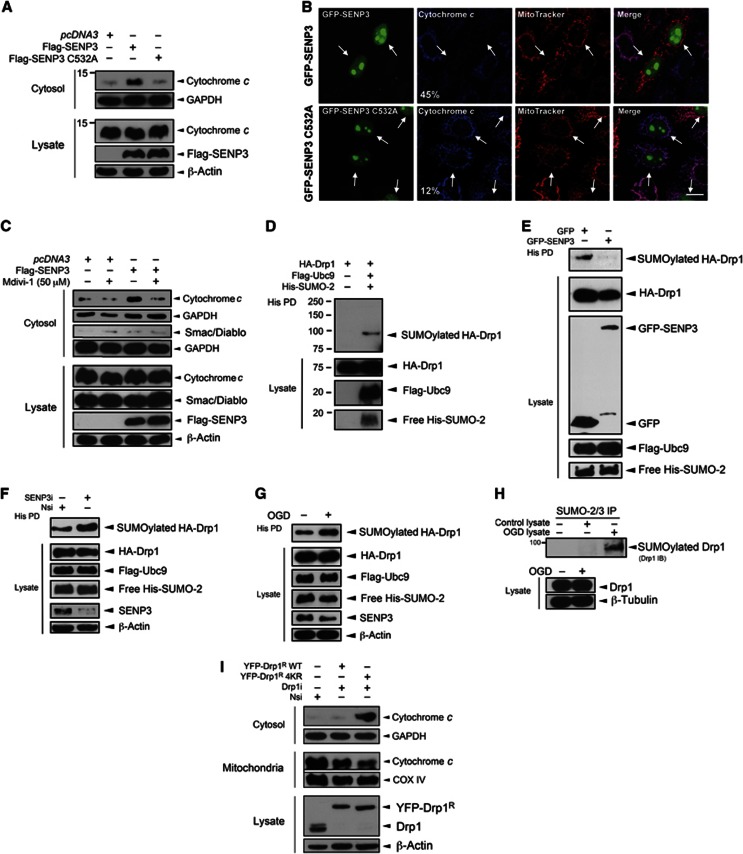
Pro-apoptotic stimuli cause cytochrome c release from mitochondria (Tait and Green, 2011) and, once in the cytosol, cytochrome c initiates a caspase pathway that culminates in cell death (Riedl and Salvesen, 2007). We therefore investigated if SENP3 regulates cytochrome c release in HEK293 cells. Overexpression of SENP3, but not the inactive mutant SENP3 C532A, caused a marked increase in cytosolic cytochrome c under basal conditions (Figure 3A). In confocal imaging experiments, overexpression of SENP3 but not of SENP3 C532A consistently reduced the localization of cytochrome c in mitochondria in HeLa cells (Figure 3B). Although the majority of SENP3 is found in the nucleus, there is robust cytosolic SENP3 immunostaining (Supplementary Figure 5A). This raises the possibility that extranuclear SENP3 regulates cytochrome c release through deSUMOylation of cytosolic or mitochondrial SUMO substrate(s). The mitochondrial GTPase Drp1 can regulate cytochrome c release through mitochondrial fission (Wilson et al, 2013) and/or via stimulation of Bax/Bak oligomerization and MOMP (Montessuit et al, 2010). Drp1 is a SUMO substrate (Harder et al, 2004) so we tested if SENP3-mediated cytochrome c release is dependent on Drp1 activity. Consistent with this, the selective Drp1 inhibitor mitochondrial division inhibitor-1 (mdivi-1) (Cassidy-Stone et al, 2008) markedly reduced SENP3-mediated cytochrome c release in HEK293 cells (Figure 3C). Interestingly, however, in contrast to cytochrome c, overexpression of SENP3 did not increase the cytosolic localization of another intramembrane space (IMS) protein Smac/Diablo (Figure 3C), suggesting that Drp1 release is independent of MOMP.
Figure 3.
SENP3 regulates cytochrome c release via deSUMOylation of Drp1. (A) Overexpression of Flag-SENP3, but not inactive C532A mutant causes cytochrome c release in HEK293 cells. The cytosol fraction or whole cell lysate was blotted for cytochrome c, Flag, and the cytosolic marker GAPDH, or β-actin. (B) Overexpressing SENP3 decreases mitochondrial cytochrome c in HeLa cells. Fixed cells were immunostained to assess the localization of cytochrome c in mitochondria labelled with MitoTracker. Representative images showing GFP-SENP3 (top panel) and GFP-SENP3 C532A expressing cells (lower panel, Green: SENP3; Red: MitoTracker; Blue: cytochrome c; scale bar: 10 μm). White arrows indicate transfected cells with percentages denoting the relative proportions of transfected cells with significantly reduced cytochrome c in mitochondria for wild-type versus inactive mutant overexpression (n=75 cells for GFP-SENP and n=113 for GFP-SENP3 C532A). (C) SENP3-mediated cytochrome c release is Drp1 dependent. HEK293 cells expressing Flag-SENP3 were treated with Mdivi-1 (50 μM) for 4 h. The cytosol fraction or whole cell lysate was blotted for cytochrome c, Smac/Diablo, Flag, and GAPDH, or β-actin. (D) Drp1 is SUMO-2-ylated in HEK293 cells expressing Flag-Ubc9, His-SUMO-2 and HA-Drp1. Lysate was incubated with Ni2+ beads to precipitate His-SUMO-2-ylated proteins (His PD). (E) Overexpression of SENP3 decreases Drp1 SUMOylation. Constructs expressing HA-Drp1, Flag-Ubc9, His-SUMO-2 and either GFP or GFP-SENP3 were transfected into HEK293 cells. (F) SENP3 knockdown enhances Drp1 SUMOylation. Either non-specific siRNA (Nsi) or SENP3 siRNA (SENP3i) together with constructs expressing HA-Drp1, Flag-Ubc9 and His-SUMO-2 were co-expressed in HEK293 cells. (G) OGD enhances Drp1 SUMOylation. Two days post transfection, HEK293 cells were exposed to OGD (2 h). In (C–F), His-pulldown and lysate samples were blotted as indicated with antibodies against HA, Flag, His, GFP, SENP3 and β-actin. (H) OGD (2 h) for primary cortical neurons increases SUMOylation of endogenous Drp1. (I) Preventing Drp1 SUMOylation results in cytochrome c release. YFP-Drp1R WT or Drp1R 4KR were expressed in HEK293 cells after knockdown of endogenous Drp1.
Source data for this figure is available on the online supplementary information page.
Drp1 is efficiently conjugated by SUMO-2 in HEK293 cells (Figure 3D; Supplementary Figure 5B) and this Drp1 SUMO-2-ylation is abolished by SENP3 overexpression (Figure 3E). Furthermore, Drp1 SUMOylation is promoted by SENP3 knockdown (Figure 3F) and, consistent with the OGD-mediated loss of SENP3, OGD enhances Drp1 SUMO-2/3-ylation in both HEK293 cells and neurons (Figure 3G and H).
To fully confirm the critical role for Drp1 SUMO-2/3-ylation in cytochrome c release, we made a non-SUMOylatable Drp1 by mutating four acceptor lysines to arginines (Drp1 4KR; Supplementary Figure 5C; Figueroa-Romero et al, 2009) and used this in knockdown rescue experiments. Following siRNA knockdown of endogenous Drp1, we expressed RNAi-resistant YFP-Drp1R WT or a non-SUMOylatable mutant Drp1R 4KR. As shown in Figure 3I, rescue with YFP-Drp1R 4KR, but not Drp1R WT, caused a substantial increase in cytochrome c levels in the cytosol with a corresponding decrease in the mitochondrial fraction. Thus, SUMOylation of Drp1 reduces its ability to evoke cytochrome c release. This is consistent with our observations that SENP3 overexpression decreases Drp1 SUMOylation and enhances the release of cytochrome c.
SUMOylation regulates mitochondrial association of Drp1
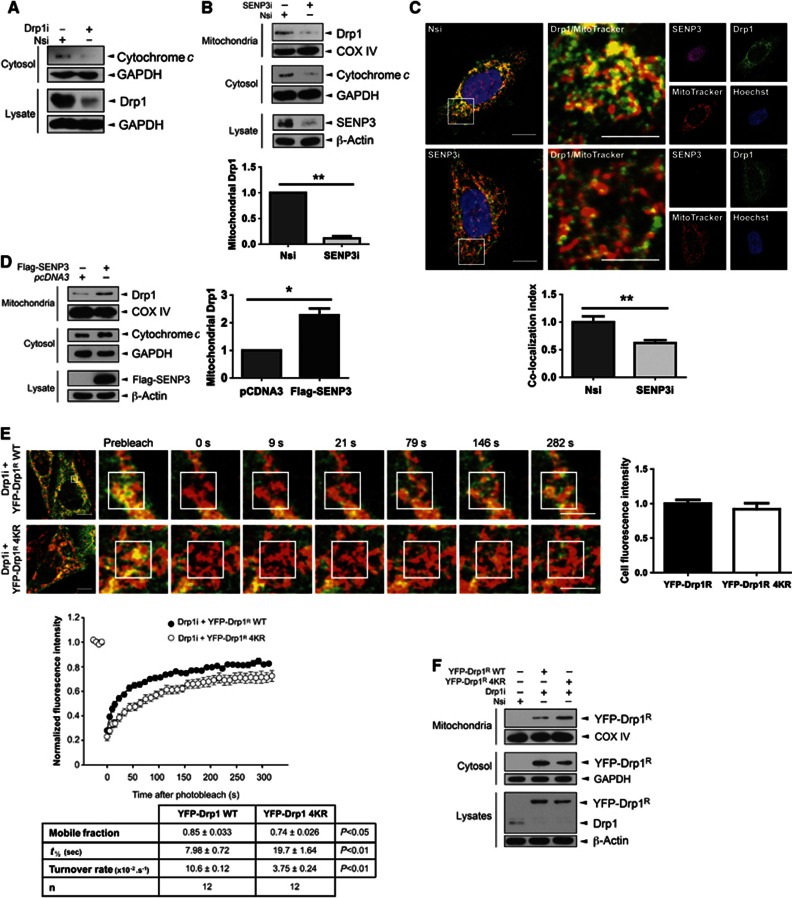
We next explored how SENP3-mediated deSUMOylation regulates Drp1 function and localization. As expected, knockdown of Drp1 decreased basal levels of cytochrome c release in HEK293 cells (Figure 4A). Similarly, under basal conditions SENP3 knockdown decreased the amount of Drp1 localized at mitochondria and reduced cytochrome c release (Figure 4B). Localization of endogenous Drp1 following SENP3 knockdown was determined in HeLa cells by immunocytochemistry. Consistent with SENP3 removal decreasing Drp1 association with the mitochondria, we observed reduced co-localization of Drp1 with MitoTracker following SENP3 knockdown (Figure 4C). Conversely, SENP3 overexpression promoted mitochondrial localization of Drp1 and increased cytochrome c release (Figure 4D). Taken together, these data indicate that enhanced Drp1 SUMOylation, facilitated by reduced levels of SENP3, reduces mitochondrial localization and downregulates cytochrome c release.
Figure 4.
SENP3 regulates mitochondrial association of Drp1. (A) Knockdown of Drp1 decreases cytosolic cytochrome c. (B) Knockdown of SENP3 decreases the mitochondrial association of endogenous Drp1 and reduces levels of cytosolic cytochrome c in HEK293 cells (n=3; **P<0.01). (C) SENP3 knockdown decreases mitochondrial localization of Drp1 in HeLa cells. Fixed cells were immunostained to assess the co-localization of endogenous Drp1 with mitochondria. Representative images showing control (top panel, Nsi) and SENP3 knockdown cells (lower panel, SENP3i; Green: Drp1; Red: MitoTracker; Yellow: co-localization of Drp1 and MitoTracker; Magenta: SENP3; Blue: Hoechst; scale bar: 10 μm). The region of interest defined by the white box is enlarged in the middle panels (scale bar: 5 μm). Right hand panels illustrate the individual channel data. The histogram shows the comparative levels of Drp1 localization at mitochondria (n=36 cells for Nsi and n=27 cells for SENP3i; **P<0.01). (D) SENP3 overexpression increases localization of Drp1 at mitochondria in HEK293 cells (n=3; *P<0.05). (E) SUMOylation affects Drp1 dynamics. YFP-Drp1R WT or non-SUMOylatable Drp1R 4KR was expressed in HeLa cells after knockdown of endogenous Drp1 and the cells subjected to FRAP analysis. The left hand images show the representative cells sampled and the subsequent image panels are enlargements of frames at the specified time points (Green: YFP-DRP1R; Red: MitoTracker; Yellow: co-localization. Scale bar: 10 μm for first frame, 5 μm for enlarged frames). The area defined by the white box is the photobleached region of interest. Recovery curves for Drp1R WT (black) and Drp1R 4KR (white) are shown and the values presented in the table. Values=mean±s.e.m. (n=12 cells per condition). (F) Non-SUMOylatable Drp1 shows enhanced mitochondrial association. YFP-Drp1R WT or Drp1R 4KR was expressed in HeLa cells after knockdown of endogenous Drp1, and cytosolic fraction, mitochondrial fraction, or whole cell lysates were blotted as indicated.
Source data for this figure is available on the online supplementary information page.
We next used knockdown of endogenous Drp1 and investigated the mitochondrial association of RNAi-resistant YFP-Drp1R WT and non-SUMOylatable YFP-Drp1R 4KR using live-cell imaging and fluorescence recovery after photobleach (FRAP) analysis in HeLa cells and neurons (Figure 4E; Supplementary Figure 5D). YFP-Drp1R WT rapidly recycled between the cytoplasm and mitochondria. YFP-Drp1R 4KR, however, displayed significantly slower rates of recovery (t1/2 values) and turnover at mitochondrial loci. Moreover, there was a greater immobile fraction YFP-Drp1R 4KR compared to YFP-Drp1R WT, indicative of longer residence times and more stable association of the non-SUMOylatable Drp1 with mitochondria. Consistent with this, subcellular fractionation and immunoblotting revealed greater mitochondrial association of YFP-Drp1R 4KR compared to YFP-Drp1R WT (Figure 4F). Taken together, our data show that SUMOylation regulates Drp1 mitochondrial association. However, it has also been reported that phosphorylation of Drp1 at S637 controls its mitochondrial localization (Chang and Blackstone, 2007). Given the growing number of examples of interplay between SUMOylation and phosphorylation (Hietakangas et al, 2006; Wilkinson and Henley, 2010; Konopacki et al, 2011), we tested if OGD also regulated Drp1 phosphorylation. However, although we observed strong phosphorylation following forskolin application, we did not detect any S637 phosphorylation under control or OGD conditions in our system (Supplementary Figure 6A). To further examine any potential cross-talk between Drp1 SUMOylation and phosphorylation at S637, we investigated whether SUMOylation of Drp1 is affected by mutation of S637 to a non-phosphorylatable alanine. Drp1 SUMOylation was not altered by the S637A mutation, further suggesting that SUMOylation of Drp1 occurs independently of phosphorylation (Supplementary Figure 6B).
DeSUMOylation of Drp1 increases mitochondrial fragmentation
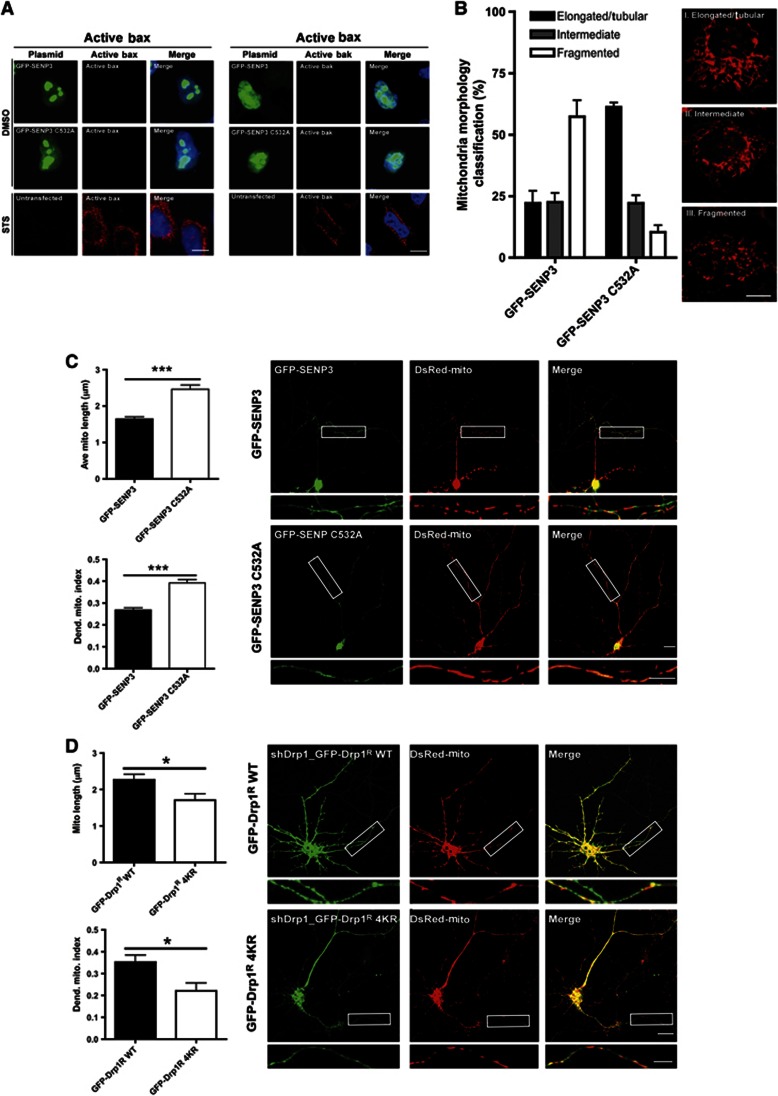
Overexpression of SENP3 significantly increases cytochrome c release but does not release the IMS protein Smac/Diablo, a marker for MOMP (Figure 3C; Tait and Green, 2011). Consistent with this, SENP3 overexpression did not lead to Bax/Bak activation in HeLa cells whereas the protein kinase inhibitor staurosporine (STS) caused robust activation of Bax/Bak (Figure 5A). Cytochrome c is also released during mitochondrial fission, a process driven by Drp1 but not dependent on MOMP (Youle and van der Bliek, 2012).To examine whether SENP3 regulates Drp1-dependent mitochondrial fission, we used MitoTracker to monitor mitochondrial morphology in cells overexpressing GFP-SENP3 or inactive GFP-SENP3 C532A. We observed increased mitochondrial fragmentation in GFP-SENP3 expressing compared to inactive GFP-SENP3 C532A expressing HeLa cells (Figure 5B), resulting in altered mitochondrial morphology from predominantly elongated/tubular to more rounded and fragmented. Similarly, overexpression of SENP3 in primary cortical neurons had little effect on the number of mitochondria in dendrite processes (1.7 mitochondria/10 μm in both GFP-SENP3 and GFP-SENP3 C532A expressing cells), although the mean length of mitochondria was significantly reduced in cells expressing WT SENP3 (Figure 5C). This resulted in a decreased dendritic mitochondrial index (Li et al, 2004), indicative of increased fission.
Figure 5.
SENP3 regulates Drp1-mediated mitochondrial fission. (A) Overexpression of SENP3 does not cause Bax/Bak activation in HeLa cells whereas staurosporine (STS; 1 μM; 2 h) potently activates Bax/Bak. Representative images show immunostaining for active Bax (left panels) or active Bak (right panels) in GFP-SENP3 or GFP-SENP C532A transfected cells or untransfected staurosporine-treated cells (Green: GFP-SENP3, Red: active Bax/Bak; Blue: Hoechst; Scale bar: 10 μm). (B) Overexpression of SENP3 promotes mitochondrial fission in HeLa cells. Cells overexpressing GFP-SENP3 or inactive C532A mutant were stained with MitoTracker and mitochondrial morphology classified into one of three categories: (I) Elongated/Tubular, (II) Intermediate or (III) Fragmented. There was a significantly increased proportion of cells with fragmented mitochondria in GFP-SENP3 compared to C532A overexpressing cells (P<0.0001, chi square test; n=99 cells for GFP-SENP3 and n=86 cells for GFP-SENP3 C532A). (C) Representative images showing GFP-SENP3 expressing (upper panels) and GFP-SENP3 C532A expressing neurons (lower panels) co-transfected with Mito-DsRed (scale: 10 μm, white box ROI magnified below). Data were quantified and expressed as a dendritic mitochondrial index (total mitochondrial length/dendrite length). Values bar charts are shown as mean±s.e.m. n=29 cells for GFP-SENP3 and n=27 for C532A, ***P<0.0001). (D) Representative images showing knockdown of endogenous Drp1 and rescue with GFP-Drp1R WT or non-SUMOylatable Drp1R 4KR in neurons. Non-SUMOylatable Drp1R 4KR increases mitochondrial fission visualized using Mito-DsRed (scale bar: 10 μm, white box ROI magnified below). Mitochondria morphology analysis was performed as in (C). Values in bar charts are shown as mean±s.e.m. (n=8 cells for GFP-Drp1R WT and n=7 cells for GFP-Drp1R 4KR), *P<0.05.
In addition, knockdown of endogenous Drp1 and rescue with non-SUMOylatable GFP-Drp1R 4KR also significantly increased mitochondrial fission in cultured neurons (Figure 5D), suggesting that SENP3 regulates mitochondrial fission through the SUMOylation status of Drp1. Taken together, these results indicate that SENP3-mediated deSUMOylation of Drp1 promotes cytochrome c release via mitochondrial fission.
SENP3 recovery during reoxygenation after OGD promotes cell death
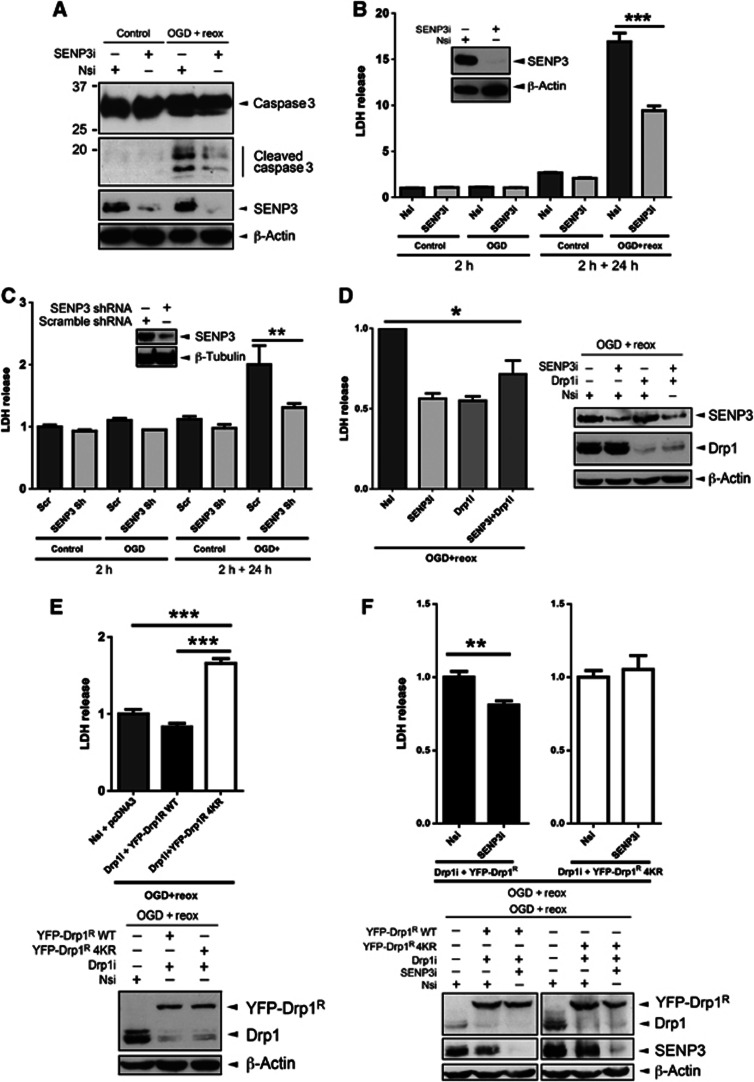
Most neuronal damage occurs during reperfusion after ischaemia rather than during the initial insult (Saito et al, 2005). We therefore investigated the effects of OGD or OGD plus reoxygenation in HEK293 cells and cortical neurons in which SENP3 was knocked down by RNAi. We monitored activation of the executioner caspase 3 and measured lactate dehydrogenase (LDH) release as markers for cell death. No caspase 3 activation was detected during OGD but marked cleavage occurred during reoxygenation in HEK293 cells. SENP3 knockdown greatly attenuated caspase 3 cleavage, indicating that SENP3 acts upstream caspase 3 activation (Figure 6A). Similarly, LDH release from HEK293 cells or cortical neurons was not increased by OGD alone but it was significantly increased after OGD plus reoxygenation and this affect was reduced or abolished, respectively, in SENP3 knockdown cells (Figure 6B and C). These effects were specific to SENP3 since knockdown of SENP1 failed to reduce LDH release from HEK293 cells after OGD plus reoxygenation (Supplementary Figure 7A). These results demonstrate that preventing the return of SENP3 can protect cells from the otherwise lethal effects of reoxygenation after OGD.
Figure 6.
SENP3 regulation of Drp1 SUMOylation plays a critical role in cell death following reoxygenation. (A) SENP3 knockdown decreases OGD plus reoxygenation-induced caspase 3 cleavage in HEK293 cells. Two days post transfection with Nsi or SENP3i HEK293 cells were subjected to OGD (2 h) and then reoxygenation (24 h). Lysates were blotted as indicated. (B) SENP3 knockdown decreases LDH release from HEK293 cells after OGD plus reoxygenation. Cells were treated as in (A) and culture media was sampled after OGD (2 h) and after 24 h reoxygenation (2 h+24 h). (C) SENP3 knockdown decreases LDH release in neurons following OGD plus reoxygenation. Neurons were infected with retrovirus containing either scrambled shRNA (Scr) or SENP3 shRNA (SENP3 sh), subjected to OGD (2 h) and then reoxygenation (24 h), and media sampled for LDH as in (B). Inset panels in (B) and (C) show immunoblots of cell lysates confirming SENP3 knockdown. (D) Knockdown of both SENP3 and Drp1 is not additive on OGD plus reoxygenation-evoked LDH release from HEK293 cells. Immunoblots (right panel) confirm knockdown of SENP3 and/or Drp1. (E) Drp1 knockdown and rescue with non-SUMOylatable Drp1R 4KR increases LDH release in response to OGD plus reoxygenation in HEK293 cells (upper panel). Immunoblots (lower panel) confirm knockdown and rescue of Drp1. (F) SENP3 knockdown does not reduce LDH release induced by OGD plus reoxygenation in Drp1R 4KR-rescued cells. Immunoblots (lower panels) confirm knockdown of Drp1/SENP3 and rescue of Drp1. In (B–F), values are presented as mean±s.e.m. (n?5 replicates for each group; *P<0.05; **P<0.01; ***P<0.001).
Source data for this figure is available on the online supplementary information page.
In a complimentary approach, we enhanced SUMO-2/3 conjugation by overexpression of YFP-SUMO-2 Q90P and YFP-SUMO-2 ΔGG in HEK293 cells. SUMO-2 Q90P is resistant to SENP3-mediated deSUMOylation (Bekes et al, 2011) and should provide maximal SUMO-2 conjugation whereas SUMO-2 ΔGG is unable to conjugate (Li et al, 2006). As predicted, YFP-SUMO-2 Q90P reduced LDH release compared to SUMO-2 ΔGG in cells subjected to OGD plus reoxygenation (Supplementary Figure 7B) further highlighting the crucial role of SUMO-2/3 conjugation in cell survival after OGD plus reoxygenation.
Similar to knocking down SENP3, Drp1 knockdown also reduced LDH release from HEK293 cells subjected to OGD plus reoxygenation. Interestingly, there was no additive protective effect in cells in which both Drp1 and SENP3 were ablated (Figure 6D), strongly suggesting that the two proteins act in the same pathway. We then knocked down endogenous Drp1 in HEK293 cells and rescued with either RNAi-resistant YFP-Drp1R WT or YFP-Drp1R 4KR (Figure 6E). LDH release after OGD plus reoxygenation was significantly increased in cells rescued with Drp1R 4KR compared to Drp1R WT.
Finally, we confirmed that the protective effect of SENP3 knockdown occurs through regulation of Drp1 SUMOylation. We monitored LDH release after OGD plus reoxygenation from cells in which both Drp1 and SENP3 were knocked down and endogenous Drp1 was rescued with either RNAi-resistant YFP-Drp1R WT or YFP-Drp1R 4KR. Knockdown of SENP3 reduced LDH release in cells rescued with YFP-Drp1R WT. However, in cells where endogenous Drp1 was replaced by YFP-Drp1R 4KR, removal of SENP3 did not reduce the OGD plus reoxygenation induced LDH release (Figure 6F). Since SENP3 removal failed to protect cells expressing non-SUMOylatable Drp1 these results demonstrate that SUMO-2/3-ylation of Drp1 is a critical step in this previously uncharacterized protective pathway.
Discussion
The UPR is a critical arbiter of cell survival in response to stress. If the UPR fails to restore proper function then insurmountable ER stress leads to cell death, but the transition from pro-survival to pro-apoptotic UPR signalling and the pathways involved are poorly understood. Protein SUMOylation is dramatically upregulated by cell stress and this has been proposed to be a protective cellular response (Datwyler et al, 2011; Lee et al, 2011; Renner et al, 2011). However, the mechanisms that regulate SUMOylation and the key target substrate proteins involved in cytoprotection are not known. Our results demonstrate that ER stress-induced degradation of SENP3 modulates protein SUMOylation. This represents a new aspect of the UPR signalling network in which enhanced SUMO-2/3-ylation of the GTPase Drp1 provides a pro-survival signal from the UPR to the mitochondria (Figure 7).
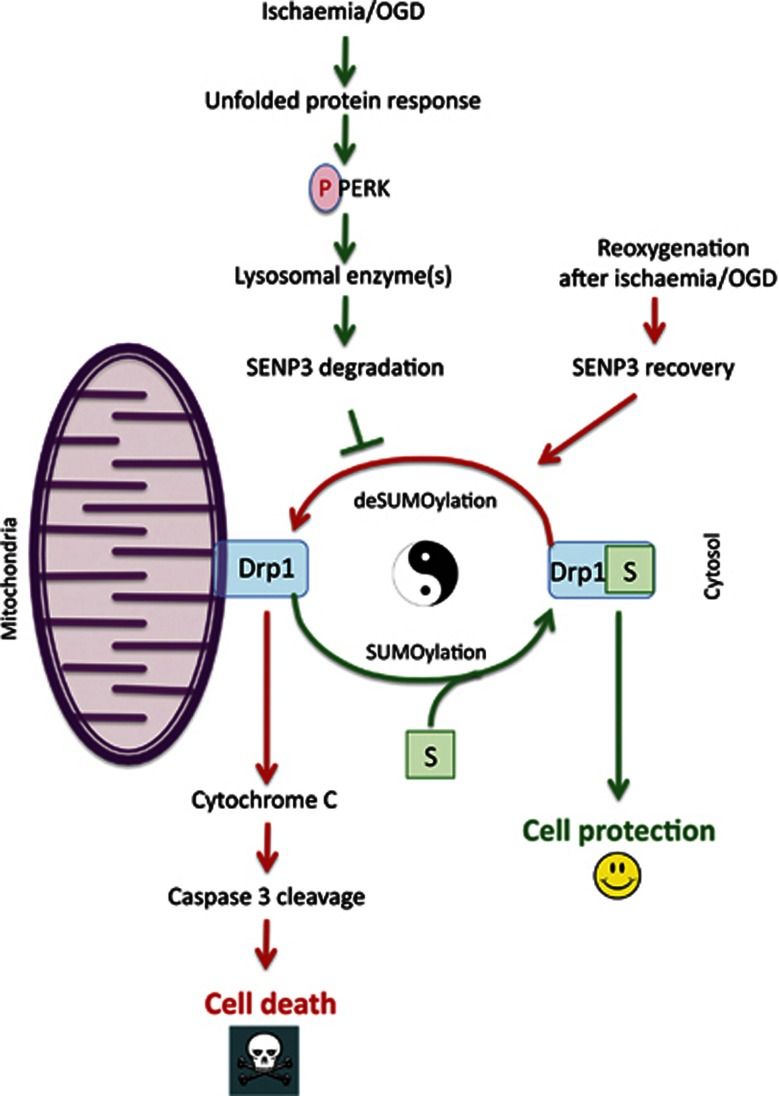
Figure 7.
Schematic of proposed cell death/survival pathway. During ischaemic stress, the UPR kinase PERK is activated, which leads to lysosome-mediated degradation of the SUMO-2/3-specific deSUMOylating enzyme SENP3. The absence of SENP3 prolongs Drp1 SUMOylation, favouring localization in the cytosol and reducing Drp1-mediated cytochrome c release. Following reoxygenation, however, SENP3 levels recover, promoting mitochondrial association of Drp1 and cell death.
A pivotal finding of this study is that selective degradation of a SUMO-2/3-specific deSUMOylating enzyme, SENP3, is a crucial mediator of the cellular response during OGD. SENP3 is selectively degraded during ischaemia leading to increased total levels of SUMO-2/3 conjugation, which contributes to cell survival. Levels of SUMO-1 conjugation are not altered. During reoxygenation after OGD, on the other hand, SENP3 levels are restored with a corresponding decrease in global SUMO-2/3 conjugation and cells die by apoptosis. Consistent with this, RNAi-mediated SENP3 knockdown to prevent the recovery after OGD significantly increases cell survival during reoxygenation. Importantly, we identify Drp1 as a key SUMO-2/3 substrate regulated by changes in SENP3 availability. Knockdown of endogenous Drp1 and rescue with a non-SUMOylatable mutant abolishes the protective effects of SENP3 knockdown. Thus, our data demonstrate that enhanced degradation and decreased availability of SENP3 during OGD is part of a protective cellular response. Furthermore, we propose that SENP3 reappearance during reoxygenation is a major factor in cell death due to reperfusion injury.
Nuclear localized SENP3 is continuously degraded through the ubiquitin-proteasome pathway under basal conditions. The rate of proteasomal degradation of SENP3 in the nucleus can be increased by phosphorylation (Kuo et al, 2008) and decreased by relatively small increases in reactive oxygen species (ROS), which can act to enhance SENP3 stability (Huang et al, 2009). By contrast, our results describe a distinct pathway activated by extreme cell stress in which cytoplasmic SENP3 is rapidly degraded by a non-proteasomal mechanism that does not rely on direct canonical ubiquitination or phosphorylation. Rather we show that cathepsin B inhibitor II prevents the OGD-induced loss of SENP3 in HEK293 cells and that SENP3 is not degraded in MEFs from cathepsin B knockout mice. In cultured cortical neurons, however, we were unable to reliably prevent OGD-induced SENP3 degradation with cathepsin B inhibitor II. The reasons for this are unclear but we speculate that either the cathepsin B inhibitor does not work well in primary cultured neurons, or other lysosomal enzymes compensate for its inhibition. Nonetheless, taken together our data strongly suggest that cathepsin B plays a major role in SENP3 degradation.
Our results indicate that regulation of SENP stability is an important mechanism for controlling cellular SUMOylation. We show that PERK overexpression or subjecting cells to OGD for 2 h activates cathepsin B. We attribute the fact that this cathepsin B activation does not cause significant cell death to the intensity of the response. We propose that a likely explanation is that exposure time is the major factor for cell viability. Prolonged OGD can trigger necroptosis via lysosome permeabilization whereas shorter OGD triggers cellular survival mechanisms that may contribute to the phenomenon of ischaemic pre-conditioning. Further, the loss of SENP3 during ischaemia and its recovery during reoxygenation provide an explanation for the widely observed phenomenon that cells, particularly neurons, can survive extraordinarily well during the oxygen deprivation phase of ischaemic insult but undergo massive delayed cell death following reoxygenation.
SENP3 specifically cleaves SUMO-2/3 from target proteins and we show that SENP3 knockdown causes a dramatic increase in total SUMO-2/3-ylation of multiple target proteins. Although changes in global SUMOylation clearly have far-reaching effects, to investigate the mechanisms underlying the cytoprotective actions of SUMO-2/3 it is necessary to identify specific target proteins. In an attempt to define pivotal SUMO substrates we reasoned that proteins involved in the pathway leading to apoptosis would be likely candidates.
Drp1 plays a crucial role in mediating cytochrome c release during apoptosis (Frank et al, 2001; Taguchi et al, 2007; Ishihara et al, 2009) so how Drp1 is recruited to the mitochondrial outer membrane is likely to be fundamental in regulating the cellular response to stress. Phosphorylation of Ser637 has been reported to regulate the mitochondrial association of Drp1 (Chang and Blackstone, 2007; Cribbs and Strack, 2007) but in our system we did not observe any Drp1 Ser637 phosphorylation in neurons.
In addition, SUMO-1-ylation of Drp1 during apoptotic cell death has been proposed to enhance stable association of Drp1 at the mitochondrial membrane (Wasiak et al, 2007), suggesting that SUMO-1-ylation of Drp1 could be pro-apoptotic. Consistent with this model, overexpression of SENP5 rescues SUMO-1-induced mitochondrial fragmentation whereas silencing of SENP5 caused mitochondrial fragmentation (Zunino et al, 2007).
Here, we show that Drp1 is a SUMO-2/3 substrate and that it is an important target for SENP3-mediated deSUMOylation. In contrast to the apoptotic affects reported for SUMO-1 conjugation, our data using OGD to stress cells strongly support a cytoprotective role for SUMO-2/3 modification of Drp1. We demonstrate that SUMO-2/3-ylation modification acts to partition Drp1 away from mitochondria and that it is deSUMOylation that enhances mitochondrial localization and promotes cytochrome c release. It is important to note, however, that the SENP3 pathway we describe is specific for SUMO-2/3 and does not impact on SUMO-1. Furthermore, our OGD protocols, which dramatically reduced SENP3, did not alter levels of SENP5 protein.
Combining previously reported data with our results it appears that SUMO-1-ylation of Drp1 enhances mitochondrial association and promotes fragmentation and apoptosis (Harder et al, 2004; Wasiak et al, 2007; Zunino et al, 2007), whereas Drp1 SUMO-2/3-ylation decreases mitochondrial localization and reduces apoptosis. Our findings are consistent with the overall picture that is emerging of SUMOylation, particularly SUMO-2/3-ylation, as an adaptive protective response to stress. Nonetheless, depending on the state of the cell, SUMO-1 and SUMO-2/3 conjugation to Drp1 may exert different effects and exhibit distinct conjugation dynamics. Although exactly how the balance between SUMO-1 and/or SUMO-2/3 conjugation to Drp1 is regulated is not known, our results strongly suggest that the targeted degradation of a paralogue-specific SENP may provide a mechanism. Similarly, there is no information on the interrelationship between conjugation by SUMO-1 and SUMO-2/3 and further work will be needed to resolve these issues.
The small proportion of Drp1 that is modified at any given time argues against a static model in which SUMO-2/3-ylated Drp1 is cytosolic and deSUMOylated Drp1 associated with the mitochondrial membrane. Rather, our FRAP data suggest that SUMO-2/3-ylation can act as a ‘mobilization factor’ in a dynamic model of constitutive cycling of Drp1 between the mitochondrial membrane and the cytosol. Thus, SUMOylation regulates Drp1 residence time at mitochondria through a balance of Ubc9-mediated SUMO-2/3 modification and SENP3-mediated deSUMOylation. The downregulation of SENP3-mediated deSUMOylation during OGD shifts the balance to decrease the mitochondrial residence time of Drp1, consequently reducing Drp1-mediated cytochrome c release, caspase cleavage and apoptosis. Exactly how deSUMOylation of Drp1 enhances mitochondrial localization is currently unclear but we speculate that SUMOylation may regulate the interaction between Drp1 and its putative receptors on the mitochondrial outer membrane such as Mff, Mid49 and Mid51 (Youle and van der Bliek, 2012).
We show that SENP3 enhances cytochrome c, but not Smac/Diablo, release via a mechanism that requires Drp1 activity, but occurs independently of Bax activation and MOMP. Further, we show that replacement of endogenous Drp1 with non-SUMOylatable Drp1 4KR enhances mitochondrial fragmentation demonstrating that SENP3 activity and Drp1 SUMOylation regulate mitochondrial fission, which leads to cytochrome c release. We do not yet have a clear idea of the mechanism that allows this to be selective for cytochrome c compared to Smac/Diablo. This will be the focus of future work but one possibility is that SUMOylation of Drp1 may regulate its interaction with the mitochondrial anionic phospholipid cardiolipin (Montessuit et al, 2010).
Since SENP3 levels correlate with cytochrome c release, and SENP3 recovers during reoxygenation, we reasoned that removing SENP3 or promoting SUMO-2/3-ylation during reoxygenation might reduce cell death post-OGD. Consistent with this, SENP3 knockdown or overexpression of a non-deconjugatable SUMO-2 afforded significant protection against cell death during reoxygenation. Enhancing general levels of SUMOylation has been reported to be protective after OGD (Lee et al, 2007; Datwyler et al, 2011; Cimarosti et al, 2012) but here we provide the first example of the protective effects of SENP3 depletion. Furthermore, the degree of protection from SENP3 knockdown was greater than that observed by overexpressing SUMO-2, suggesting that exploiting the substrate specificity of SENP3-mediated deSUMOylation could provide a novel therapeutic strategy.
In conclusion, we define a novel cytoprotective pathway in which SENP3 availability during ischaemia regulates SUMO-2/3-ylation of Drp1, which, in turn, controls the dynamics of Drp1 cycling between the mitochondrial outer membrane and the cytosol. Drp1 SUMOylation promotes partitioning into the cytosol, which reduces cytochrome c release and spares the cell from apoptosis. This pathway represents a novel example of a substrate-specific protective action of SUMO-2/3-ylation during cell stress and reveals a potential therapeutic target for increasing resistance to apoptosis after ischaemia and other insults.
Materials and methods
Plasmids
Constructs were generated using standard cloning procedures and PCR-based mutagenesis. Details of the plasmids used, their source and how they were modified are provided in Supplementary data.
Cell culture
Cortical cultures were prepared from E18 Wistar rat embryos by dissection, trypsin treatment and mechanical dissociation. Cells were then plated on poly-L-lysine-coated culture dishes at a density of 3 million per 10 cm dish, 600 K per well for 6-well plates or 25 mm glass coverslips (250 K per 3.5 mm dish) in Neurobasal medium (Gibco) containing 10% horse serum, B27 (Gibco), and 5 mM glutamine and incubated at 37°C in humidified air supplemented with 5% CO2. On the second day, the media was changed for Neurobasal medium containing B27 and 2 mM glutamine. Cortical neurons were then fed each week with this serum-free medium until experimental usage at 14 DIV.
HEK293 and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Lonza) containing 10% fetal bovine serum (FBS), 5 mM glutamine, and 100 units/ml penicillin/streptomycin at 37°C in humidified air supplemented with 5% CO2.
MEF cell lines were cultured in DMEM containing 10% FBS, 5 mM glutamine, 100 units/ml penicillin/streptomycin, 0.1 mM non-essential amino acids (Invitrogen), and 0.05 mM β-mercaptoethanol (Invitrogen) at 37°C in humidified air supplemented with 5% CO2. PERK−/− MEFs and their WT controls were a gift from D Ron. eIF2α S51A MEFs and their WT controls were a gift from R Kaufman. Cathepsin B−/− primary MEFs and their WT controls were a gift from C Peters and were cultured in DMEM containing 10% FBS, 5 mM glutamine, 100 units/ml penicillin/streptomycin.
OGD and reoxygenation
For cortical neurons, OGD was performed within a MACS-VA500 anaerobic workstation (Don Whitley Scientific Limited) supplemented with 95% N2 and 5% CO2 at 37°C for 30–120 min. Details of the protocols used are provided in Supplementary data.
DNA and siRNA transfections
For DNA transfection into HEK293 cells and HeLa cells, we used jetPEI reagents (Polyplus Transfection) according to manufacturer’s instructions, and cultured cells until use within 48 h. INTERFERin reagent (Polyplus Transfection) was used for siRNA transfection and cells were used within 72 h. For DNA and siRNA co-transfection, we used jetPRIME reagents (Polyplus Transfection) and used the cells within 72 h. MEFs were transfected with siRNAs by nucleofector electroporation (program A-23, and MEF solution 2; Amaxa Biosystems). Primary rat cortical neurons were transfected using Lipofectamine 2000 reagent (Invitrogen) according to manufacturer’s instructions. siRNA duplexes were used as follows: mouse SENP3 siRNA (ON-TARGET plus SMART pool L-057149-01-005 set of 4, Thermo Scientific), human SENP3 siRNA (sc-44451, Santa Cruz) and human Drp1 siRNA (synthesized by Eurofins MWG Operon).
Retroviral shRNA infection
The shRNA constructs containing gene-specific shRNA expression vectors in pGFP-V-RS plasmid were purchased from Origene Technologies, Inc. The following sequence was chosen to silence SENP3: 5′-ACTGGCTCAATGACCAGGTGATGAACATG-3′. A construct of scrambled non-effective shRNA cassette in the same plasmid was used as negative control. In all, 1 μg plasmids were used to transfect am12 cells, a murine retroviral packaging system (Patience et al, 1998) with Lipofectamine 2000 transfection reagent (Invitrogen). In all, 46 h post transfection the culture media was collected and centrifuged at 2000 g for 5 min. Supernatant was aliquoted and kept at −20°C as virus stock. In all, 15–30 μl virus stock was used to infect ∼1 million primary cultured cortical neurons at DIV7, supplemented with a 4 μg/ml concentration of Polybrene (hexadimethrine bromide) for 6 h. Culture media was then replaced with fresh media and the cells left until DIV14.
Quantitative RT–PCR
RNA was extracted from 14 DIV primary cortical neuronal cultures using an RNeasy kit (Qiagen). RNA was DNAse treated and reverse transcribed (Ambion). SUMO2, SUMO3 and SENP3 mRNAs were measured using a TaqMan assay-on-demand (Applied Biosystems) and normalized to 18srRNA (Applied Biosystems) using multiplexing on a SMx3000P system (Stratagene). The probes used were Rn00821719_g1* for Rat SUMO2, Rn01429214_g1 for Rat SUMO3 and Rn01496196_g1 for Rat SENP3 (Applied Biosystems).
LDH assay
LDH levels in conditioned culture media were assessed using the In Vitro Toxicology Assay Kit (Lactic Dehydrogenase Based; Sigma). Briefly, HEK293 cells cultured on 24-well plates or dissociated cortical neurons cultured on 12-well plates, were subjected to 2 h OGD or 2 h OGD plus 24 h reoxygenation. In all, 30 μl of conditioned culture medium was taken from each well of the 24-well plates or 12-well plates and spun down to pellet cells or cell debris and 10 μl of the medium was transferred to each well of a 96-well plate. LDH assay mixture containing substrate, cofactor and dye solutions was prepared and 50 μl of the mixture was added to each well of the 96-well plate. Following 15 min incubation, the absorbance at 492 nm and at reference 620 nm was measured using a Tecan program on a plate reader. In some initial experiments using HEK293 cells, absorbance was measured only at a wavelength of 492 nm. Results shown in each histogram are representative of at least three experiments carried out using different cell populations.
Subcellular fractionation
Nuclear and non-nuclear fractions from HEK293 cells were prepared using a Nuclear Extract Kit (Active Motif). Cytosol and mitochondria fractions were prepared using a ProteoExtract Cytosol/Mitochondria Fractionation Kit (Calbiochem).
In vitro cleavage assay of SENP3 by cathepsin B
Technical difficulties in expressing the catalytic domain of SENP3 in E. coli have previously been reported previously (Mikolajczyk et al, 2007; Kolli et al, 2010). In agreement with those reports, we were unable to detect expression of full-length SENP3 in E. coli. To overcome this, GST-SENP3 or Flag-SENP3 was purified from HEK293 cells transfected with pEBG-SENP3 or Flag-SENP3 by GST-pulldown or immunoprecipitation, respectively. In vitro cleavage by Cathepsin B was performed as previously described (Taha et al, 2005). Briefly, for each reaction, 10 μl of GST/GST-SENP3-bound glutathione-Sepharose 4B beads was added to 7 μl of 3 × Cathepsin B reaction buffer containing 150 mM sodium acetate, pH 6.0, 12 mM EDTA, and 24 mM dithiothreitol, 0.5 μl of Cathepsin B (∼0.01 units; ENZO Life Sciences), and 3.5 μl of ddH2O. The cleavage reaction was carried out by incubation at 37°C for 30 min. Following incubation, 5 μl 6 × SDS loading buffer was added to each sample. All the samples were subjected to SDS–PAGE and immunoblotting under denaturing and reducing conditions.
In vitro kinase assay
In vitro kinase assays were performed as reported previously (Yan et al, 2002). Briefly, GST or GST-SENP3 was purified from transfected HEK293 cells by GST-pulldown. HA-PERK, HA-PERK K618A mutant, or HA-p58IPK was purified from transfected HEK293 cells by immunoprecipitation. Purified proteins were mixed in a kinase reaction buffer containing 25 mM HEPES pH 7.4, 25 mM glycerophosphate, 25 mM MgCl2, 2 mM DTT, 0.1 mM orthovanadate, and 50 M [32P]ATP (10 Ci/mmol), and incubated at 30°C for 30 min. The reaction was terminated by adding 10 μl 6 × SDS loading buffer. All the samples were subjected to SDS–PAGE and the gels were stained with Coomasssie blue, destained and dried before exposure to X-ray autoradiography film.
Preparation of cell lysate, immunoprecipitation and GST-pulldown
Transfected HEK293 cells were washed once with ice-cold phosphate-buffered saline and then lysed in buffer containing 20 mM Tris, pH 7.4, 137 mM NaCl, 25 mM β-glycerophosphate, 2 mM sodium pyrophosphate, 2 mM EDTA, 1% Triton X-100, 10% glycerol, 1 mM phenylmethylsulphonyl fluoride, 1 mM sodium orthovanadate, 10 μg/ml aprotinin and 10 μg/ml leupeptin. Following sonication, insoluble material was removed from lysates by centrifugation for 15 min at 16 000 g. In experiments to detect cellular levels of SUMOylation, 20 mM N-ethylmaleimide (NEM) was also added to the lysis buffer. HA or FLAG-tagged proteins were immunoprecipitated from HEK293 cell lysates using HA antibody (Roche) or M2 antibody (Sigma). SUMO-2/3 conjugates were immunoprecipitated from whole cell lysates prepared in the above-mentioned lysis buffer plus 0.5% SDS using SUMO-2/3 antibody (Clone 1E7, MBL). GST-tagged proteins were isolated by incubating lysates with glutathione-Sepharose 4B (GE Healthcare).
Nickel affinity pulldown
His-SUMO-2 or His-Ub conjugates were purified by Nickel affinity pulldowns. Briefly, transfected HEK293 cells were immediately lysed in denaturing buffer containing 6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris–HCl, pH 8.0, and incubated with Ni2+—NTA beads (Qiagen) for 2–3 h at room temperature. The beads were washed twice with the denaturing buffer, two times with wash buffer containing 8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris–HCl, pH 6.3, and once with PBS. Bound proteins were eluted in 6 × SDS loading buffer and resolved by SDS–PAGE, His-SUMO-2 or His-Ub conjugates were detected by immunoblotting using HA or Flag antibody.
Immunoblotting
Samples were resolved by SDS–PAGE (7.5–15% gels) and transferred onto Immobilon-P membranes (Millipore Inc.), which were immunoblotted with the antibodies listed in Supplementary data.
Immunocytochemistry
Cells were fixed for 12 min at RT in 4% paraformladehyde/PBS with 5% sucrose. Following 5 min incubation with 50 mM NH4Cl to quench residual PFA, cells were permeabilized in 0.1% Triton X-100/PBS, blocked for 20 min with 10% horse serum/PBS and then incubated with antibodies. Primary and secondary antibody incubations were performed in 5% horse serum/PBS for 1 h at RT, followed by 15 min incubation with Hoechst 33542 to stain the cell nuclei. Coverslips were mounted on glass slides in fluoromount medium (Sigma). Immunostaining was performed using antibodies against SENP3 (D20A10, Cell Signalling, 1:400), Drp1 (Clone 8, BD Biosciences, 1:100), cytochrome c (6H2.B4, BD Biosciences, 1:100), active Bax (Clone 3, BD Biosciences, 1:100), and active Bak (Ab-1/TC-100, Millipore, 1:100).
Microscopy, image acquisition and analysis
Detailed methods used for the live and fixed cell confocal imaging, FRAP, mitochondrial morphology and statistical analysis are provided in Supplementary data.
Supplementary Material
Acknowledgments
The European Research Council, the MRC, the Wellcome Trust and the BBSRC supported this work. We thank AD Sharrocks, AJ Whitmarsh, AM van der Bliek, BM Burgering, M Matsushita, RT Hay, S Strack and M Wambach for supplying plasmids; D Ron, D Cavener, RJ Kaufman and C Peters for supplying knockout/knock-in MEFs; H Cimarosti for help with initial experiments; and P Rubin for excellent technical assistance.
Author contributions: CG and JMH conceived the project with valuable input from KAW. CG designed and performed most biochemical and molecular biological experiments, KLH performed all cell imaging assays in cell culture, JL performed experiments of SENP3 knockdown and LDH assay in dissociated primary cultured rat cortical neurons, LD performed quantitative RT–PCR, and KAW performed some molecular biological experiments. JMH provided project management and wrote the manuscript with hypothesis development, experimental design and data interpretation contributed by all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson DB, Wilkinson KA, Henley JM (2009) Protein SUMOylation in neuropathological conditions. Drug News Perspect 22: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekes M, Prudden J, Srikumar T, Raught B, Boddy MN, Salvesen GS (2011) The dynamics and mechanism of SUMO chain deconjugation by SUMO-specific proteases. J Biol Chem 286: 10238–10247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Andreau K, Poncet D, Zamzami N, Perfettini JL, Metivier D, Ojcius DM, Jaattela M, Kroemer G (2003) Lysosomal membrane permeabilization induces cell death in a mitochondrion-dependent fashion. J Exp Med 197: 1323–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boya P, Kroemer G (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27: 6434–6451 [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J (2008) Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 14: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CR, Blackstone C (2007) Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem 282: 21583–21587 [DOI] [PubMed] [Google Scholar]
- Cimarosti H, Ashikaga E, Jaafari N, Dearden L, Rubin P, Wilkinson KA, Henley JM (2012) Enhanced SUMOylation and SENP-1 protein levels following oxygen and glucose deprivation in neurones. J Cereb Blood Flow Metab 32: 17–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarosti H, Lindberg C, Bomholt SF, Ronn LC, Henley JM (2008) Increased protein SUMOylation following focal cerebral ischemia. Neuropharmacology 54: 280–289 [DOI] [PubMed] [Google Scholar]
- Cribbs JT, Strack S (2007) Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep 8: 939–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datwyler AL, Lattig-Tunnemann G, Yang W, Paschen W, Lee SL, Dirnagl U, Endres M, Harms C (2011) SUMO2/3 conjugation is an endogenous neuroprotective mechanism. J Cereb Blood Flow Metab 31: 2152–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorval V, Fraser PE (2007) SUMO on the road to neurodegeneration. Biochim Biophys Acta 1773: 694–706 [DOI] [PubMed] [Google Scholar]
- Figueroa-Romero C, Iniguez-Lluh JA, Stadler J, Chang CR, Arnoult D, Keller PJ, Hong Y, Blackstone C, Feldman EL (2009) SUMOylation of the mitochondrial fission protein Drp1 occurs at multiple nonconsensus sites within the B domain and is linked to its activity cycle. FASEB J 23: 3917–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ (2001) The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell 1: 515–525 [DOI] [PubMed] [Google Scholar]
- Friedlander R, Jarosch E, Urban J, Volkwein C, Sommer T (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol 2: 379–384 [DOI] [PubMed] [Google Scholar]
- Grohm J, Kim SW, Mamrak U, Tobaben S, Cassidy-Stone A, Nunnari J, Plesnila N, Culmsee C (2012) Inhibition of Drp1 provides neuroprotection in vitro and in vivo. Cell Death Differ 19: 1446–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder Z, Zunino R, McBride H (2004) Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol 14: 340–345 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D (2001) Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell 7: 1153–1163 [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397: 271–274 [DOI] [PubMed] [Google Scholar]
- Hetz C (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13: 89–102 [DOI] [PubMed] [Google Scholar]
- Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA 103: 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Han Y, Wang Y, Sun X, Yan S, Yeh ET, Chen Y, Cang H, Li H, Shi G, Cheng J, Tang X, Yi J (2009) SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J 28: 2748–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Nomura M, Jofuku A, Kato H, Suzuki SO, Masuda K, Otera H, Nakanishi Y, Nonaka I, Goto Y, Taguchi N, Morinaga H, Maeda M, Takayanagi R, Yokota S, Mihara K (2009) Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat Cell Biol 11: 958–966 [DOI] [PubMed] [Google Scholar]
- Jourdain A, Martinou JC (2009) Mitochondrial outer-membrane permeabilization and remodelling in apoptosis. Int J Biochem Cell Biol 41: 1884–1889 [DOI] [PubMed] [Google Scholar]
- Kim R, Emi M, Tanabe K (2006) Role of mitochondria as the gardens of cell death. Cancer Chemother Pharmacol 57: 545–553 [DOI] [PubMed] [Google Scholar]
- Kolli N, Mikolajczyk J, Drag M, Mukhopadhyay D, Moffatt N, Dasso M, Salvesen G, Wilkinson KD (2010) Distribution and paralogue specificity of mammalian deSUMOylating enzymes. Biochem J 430: 335–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopacki FA, Jaafari N, Rocca DL, Wilkinson KA, Chamberlain S, Rubin P, Kantamneni S, Mellor JR, Henley JM (2011) Agonist-induced PKC phosphorylation regulates GluK2 SUMOylation and kainate receptor endocytosis. Proc Natl Acad Sci USA 108: 19772–19777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumenis C, Naczki C, Koritzinsky M, Rastani S, Diehl A, Sonenberg N, Koromilas A, Wouters BG (2002) Regulation of protein synthesis by hypoxia via activation of the endoplasmic reticulum kinase PERK and phosphorylation of the translation initiation factor eIF2alpha. Mol Cell Biol 22: 7405–7416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Azam S, Sullivan JM, Owen C, Cavener DR, Zhang P, Ron D, Harding HP, Chen JJ, Han A, White BC, Krause GS, DeGracia DJ (2001) Brain ischemia and reperfusion activates the eukaryotic initiation factor 2alpha kinase, PERK. J Neurochem 77: 1418–1421 [DOI] [PubMed] [Google Scholar]
- Kuo ML, den Besten W, Thomas MC, Sherr CJ (2008) Arf-induced turnover of the nucleolar nucleophosmin-associated SUMO-2/3 protease Senp3. Cell Cycle 7: 3378–3387 [DOI] [PubMed] [Google Scholar]
- Lee YJ, Castri P, Bembry J, Maric D, Auh S, Hallenbeck JM (2009) SUMOylation participates in induction of ischemic tolerance. J Neurochem 109: 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ (2004) Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell 15: 5001–5011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Miyake S, Wakita H, McMullen DC, Azuma Y, Auh S, Hallenbeck JM (2007) Protein SUMOylation is massively increased in hibernation torpor and is critical for the cytoprotection provided by ischemic preconditioning and hypothermia in SHSY5Y cells. J Cereb Blood Flow Metab 27: 950–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, Mou Y, Maric D, Klimanis D, Auh S, Hallenbeck JM (2011) Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PLoS ONE 6: e25852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Santockyte R, Shen RF, Tekle E, Wang G, Yang DC, Chock PB (2006) Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J Biol Chem 281: 36221–36227 [DOI] [PubMed] [Google Scholar]
- Li Z, Okamoto K, Hayashi Y, Sheng M (2004) The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell 119: 873–887 [DOI] [PubMed] [Google Scholar]
- Mikolajczyk J, Drag M, Bekes M, Cao JT, Ronai Z, Salvesen GS (2007) Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. J Biol Chem 282: 26217–26224 [DOI] [PubMed] [Google Scholar]
- Montessuit S, Somasekharan SP, Terrones O, Lucken-Ardjomande S, Herzig S, Schwarzenbacher R, Manstein DJ, Bossy-Wetzel E, Basanez G, Meda P, Martinou JC (2010) Membrane remodeling induced by the dynamin-related protein Drp1 stimulates Bax oligomerization. Cell 142: 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patience C, Takeuchi Y, Cosset FL, Weiss RA (1998) Packaging of endogenous retroviral sequences in retroviral vectors produced by murine and human packaging cells. J Virol 72: 2671–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner F, Moreno R, Schmitz ML (2011) SUMOylation-dependent localization of IKKepsilon in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol Cell 37: 503–515 [DOI] [PubMed] [Google Scholar]
- Riedl SJ, Salvesen GS (2007) The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol 8: 405–413 [DOI] [PubMed] [Google Scholar]
- Saito A, Maier CM, Narasimhan P, Nishi T, Song YS, Yu F, Liu J, Lee YS, Nito C, Kamada H, Dodd RL, Hsieh LB, Hassid B, Kim EE, Gonzalez M, Chan PH (2005) Oxidative stress and neuronal death/survival signaling in cerebral ischemia. Mol Neurobiol 31: 105–116 [DOI] [PubMed] [Google Scholar]
- Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Szabadkai G, Simoni AM, Chami M, Wieckowski MR, Youle RJ, Rizzuto R (2004) Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell 16: 59–68 [DOI] [PubMed] [Google Scholar]
- Szegezdi E, Macdonald DC, Ni Chonghaile T, Gupta S, Samali A (2009) Bcl-2 family on guard at the ER. Am J Physiol Cell Physiol 296: C941–C953 [DOI] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K (2007) Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem 282: 11521–11529 [DOI] [PubMed] [Google Scholar]
- Taha TA, Kitatani K, Bielawski J, Cho W, Hannun YA, Obeid LM (2005) Tumor necrosis factor induces the loss of sphingosine kinase-1 by a cathepsin B-dependent mechanism. J Biol Chem 280: 17196–17202 [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR (2011) Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 11: 621–632 [DOI] [PubMed] [Google Scholar]
- Van Der Kelen K, Beyaert R, Inze D, De Veylder L (2009) Translational control of eukaryotic gene expression. Crit Rev Biochem Mol Biol 44: 143–168 [DOI] [PubMed] [Google Scholar]
- Wasiak S, Zunino R, McBride HM (2007) Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol 177: 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Henley JM (2010) Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428: 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Nakamura Y, Henley JM (2010) Targets and consequences of protein SUMOylation in neurons. Brain Res Rev 64: 195–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson TJ, Slupe AM, Strack S (2013) Cell signaling and mitochondrial dynamics: Implications for neuronal function and neurodegenerative disease. Neurobiol Dis 51: 13–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Frank CL, Korth MJ, Sopher BL, Novoa I, Ron D, Katze MG (2002) Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc Natl Acad Sci USA 99: 15920–15925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Paschen W (2009) The endoplasmic reticulum and neurological diseases. Exp Neurol 219: 376–381 [DOI] [PubMed] [Google Scholar]
- Yang W, Sheng H, Warner DS, Paschen W (2008a) Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J Cereb Blood Flow Metab 28: 892–896 [DOI] [PubMed] [Google Scholar]
- Yang W, Sheng H, Warner DS, Paschen W (2008b) Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab 28: 269–279 [DOI] [PubMed] [Google Scholar]
- Yeh ET (2009) SUMOylation and De-SUMOylation: wrestling with life's processes. J Biol Chem 284: 8223–8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM (2012) Mitochondrial fission, fusion, and stress. Science 337: 1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM (2007) The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci 120: 1178–1188 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.