Abstract
Lifespan of C. elegans is affected by the nervous system; however, the underlying neural integration still remains unclear. In this work, we targeted an antagonistic neural system consisting of low-oxygen sensing BAG neurons and high-oxygen sensing URX neurons. While ablation of BAG neurons increases lifespan of C. elegans, ablation of URX neurons decreases lifespan. Genetic analysis revealed that BAG and URX neurons counterbalance each other via different guanylate cyclases (GCYs) to control lifespan balance. Lifespan-modulating effects of GCYs in these neurons are independent of the actions from insulin/IGF-1 signalling, germline signalling, sensory perception, or dietary restriction. Given the known gas-sensing property of these neurons, we profiled that lifespan of C. elegans is promoted under moderately low oxygen (4–12%) or moderately high carbon dioxide (5%) but inhibited under high-level oxygen (40%); however, these pro-longevity and anti-longevity effects are counteracted, respectively, by BAG and URX neurons via different GCYs. In conclusion, BAG and URX neurons work as a neural-regulatory system to counterbalance each other via different GCYs to control lifespan homeostasis.
Keywords: C. elegans , lifespan, neuron
Introduction
What controls the lifespan of a given species is an intriguing question for centuries. Based on C. elegans, lifespan is impacted by environmental conditions and related physiological activities such as feeding, sensory perception, and reproduction (Finch and Ruvkun, 2001; Antebi, 2007; Bishop and Guarente, 2007a; Mair and Dillin, 2008; Kenyon, 2010). Indeed, feeding control through dietary restriction (DR) is known as an environmental manipulation for longevity across many species, and interestingly this effect was associated with the nervous system in C. elegans (Bishop and Guarente, 2007b). In molecular biology, metabolic pathway consisting of DAF-2 (a homologue of mammalian insulin and IGF-1 receptor) and downstream DAF-16 has been studied and found to control lifespan in C. elegans (Kenyon et al, 1993; Kimura et al, 1997; Ogg et al, 1997; Hsin and Kenyon, 1999), and the underlying basis has been related to the gonad and the nervous system (Wolkow et al, 2000; Lin et al, 2001; Arantes-Oliveira et al, 2002). Recently, it was shown that microRNA mir-71 can act in neurons to promote germline-mediated longevity through DAF-16 (Boulias and Horvitz, 2012). In addition, sensory perception has been shown to contribute to lifespan control, for example, lifespan increase was seen in C. elegans with genetic defects of sensory perception (Apfeld and Kenyon, 1999; Lans and Jansen, 2007) or with functional loss of gustatory neurons ASI and ASG or olfactory neurons AWA and AWC (Alcedo and Kenyon, 2004), and the effect of sensory perception could be partially mediated by DAF-16 (Apfeld and Kenyon, 1999). Hinted by this background, we hypothesized that a neural system may exist to work through antagonistic neuronal subgroups that balance lifespan upregulation versus downregulation and thus mediate lifespan balance. To identify this potential neural pathway, we focused our strategy on neurons that respond to important environmental cues, and our attention was directed to the structurally and functionally interlinked BAG and URX neurons—which were recently found to differentially sense low- versus high-level oxygen (O2) (Coates and de Bono, 2002; Cheung et al, 2004; Gray et al, 2004; Chang et al, 2006; Rogers et al, 2006; Zimmer et al, 2009). Excitingly, we discovered that BAG and URX neurons counteractively control lifespan via different guanylate cyclases (GCYs) in a manner that is independent of DAF-2/DAF-16 or other canonical pathways involved in the lifespan-modulating effects of feeding, sensory perception, or reproduction. These findings lead us to propose a model of BAG-URX neuronal network that controls lifespan homeostasis.
Results
BAG and URX neurons opposingly control lifespan
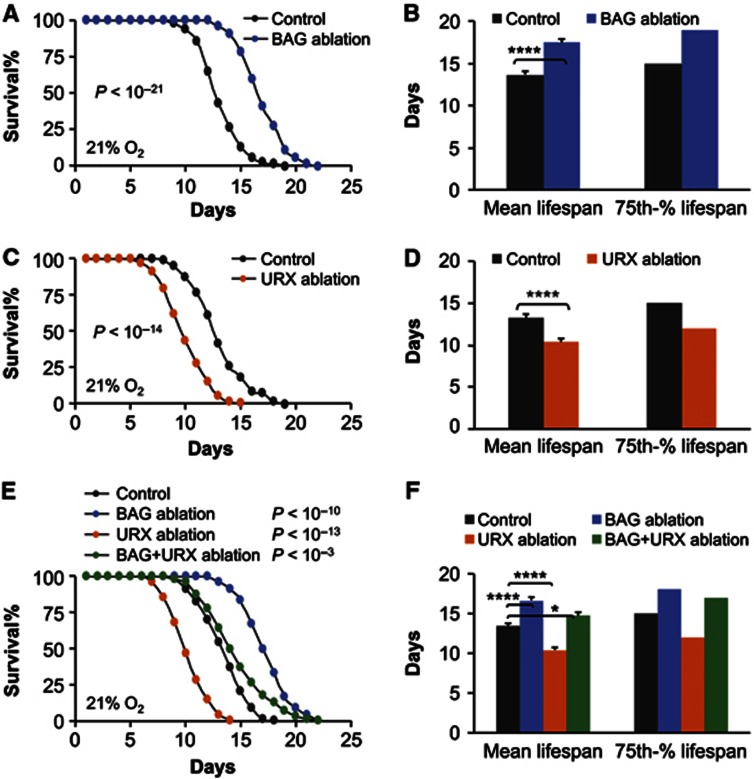
As established, C. elegans sense low-level O2 via BAG neurons (Zimmer et al, 2009), and sense high-level O2 via URX neurons and related AQR and PQR neurons among which URX neurons are most important (Coates and de Bono, 2002; Cheung et al, 2004, 2005; Chang et al, 2006; Busch et al, 2012). In this study, we first employed laser ablation approach to selectively kill BAG versus URX neurons and examined the outcomes on lifespan. We found that BAG-ablated animals had significantly extended lifespan compared to ablation controls. As shown in Figure 1A and B, BAG ablation increased mean lifespan from 13.7±0.4 days to 17.5±0.4 days, 75th percentile lifespan from 15 days to 19 days, and maximal lifespan from 19 days to 22 days. These results indicate that BAG neurons act to limit lifespan extension in C. elegans. In contrast, URX-ablated animals showed pronounced lifespan decrease compared to ablation control. As shown in Figure 1C and D, URX ablation led to decreases in mean lifespan from 13.3±0.4 days to 10.4±0.4 days, 75th percentile lifespan from 15 days to 12 days, and maximal lifespan from 18 days to 14 days. The short lifespan induced by URX ablation was not a result of developmental defects or sickness, as URX-ablated animals and controls had similar developmental profiles, appearances, reproduction, and behaviours (data not shown). Altogether, these data indicate that URX neurons are involved in maintaining lifespan in C. elegans. Subsequently, we examined whether BAG and URX neurons interact with each other to control lifespan, and to do so, we simultaneously ablated both types of neurons in C. elegans. Results showed that double-ablated animals showed an intermediate lifespan, compared to long-lived phenotype of BAG-ablated animals and short-lived phenotype of URX-ablated animals. As shown in Figure 1E and F, BAG ablation completely prevented URX ablation-induced lifespan shortening, whereas URX ablation partially reduced the pro-longevity effect of BAG ablation, indicating that BAG and URX neurons substantially antagonize each other to control the lifespan of C. elegans. Of note, animals with simultaneous BAG and URX ablations still did better than control animals in terms of both mean lifespan and 75th percentile lifespan, and their maximal lifespan remained extended (Figure 1E and F), suggesting that BAG neurons have a more predominant role in the BAG-URX neural network. In our study, we also considered whether the lifespan phenotypes of BAG- or URX-ablated animals involved feeding behavioural changes. Using a method adapted from the literature (Shtonda and Avery, 2006; Bendesky et al, 2011), we measured the amount of time that worms spent on food, and found that animals with either ablation were similar to controls (Supplementary Figure S1A and B). Taken together, all these results demonstrate that lifespan of C. elegans can be inhibited and promoted by BAG and URX neurons, respectively.
Figure 1.
Effects of ablating BAG versus URX neurons on C. elegans lifespan. Survival curves (A, C and E) and mean lifespan and 75th percentile lifespan (B, D and F) of C. elegans following laser ablation of BAG neurons versus ablation controls (n=74–94) (A, B), laser ablation of URX neurons versus ablation controls (n=76–94) (C, D), or simultaneous ablation of BAG and URX neurons versus ablation controls (n=103–120) (E, F). For comparison, single ablation of BAG neurons or URX neurons (n=22–28) was also included in (E) and (F). (A, C and E) show animals pooled from multiple independent experiments; (B, D and F) show analysis from a representative single experiment (n=22–48 per group). *P<0.05, ****P<0.0001, statistics for curve comparisons are shown in the figure. Error bars represent mean±s.e.m.
GCY-31 and GCY-33 negatively control lifespan
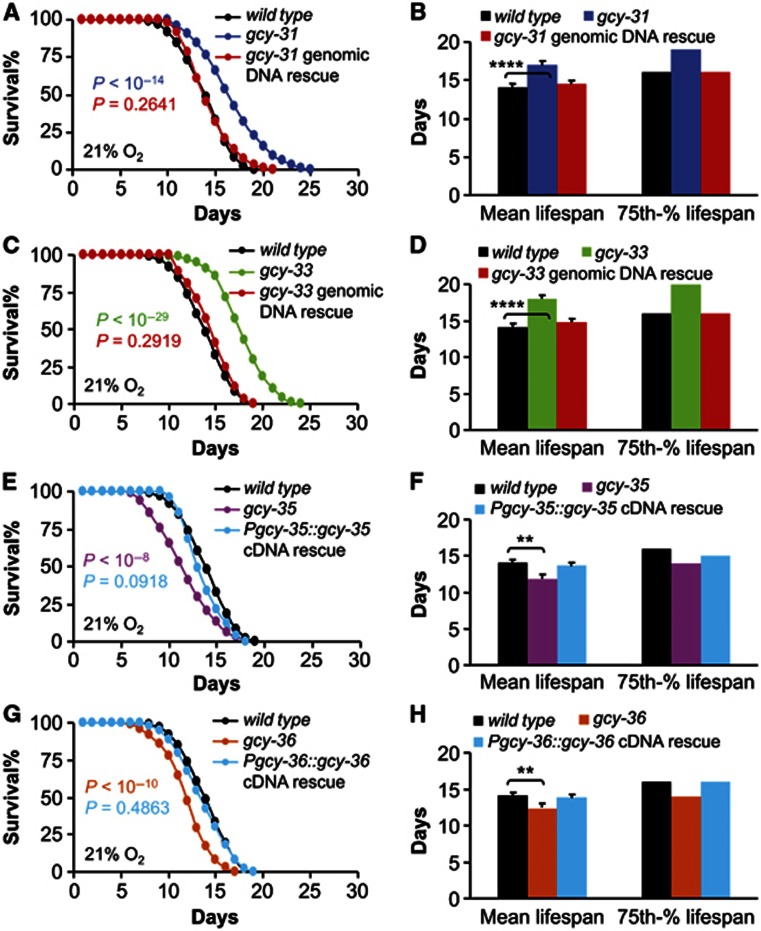
Following the ablation studies above, we continued our investigation using molecular approaches that targeted GCYs which are a class of soluble GCYs that mediate O2 sensing in C. elegans (Morton et al, 1999; Cheung et al, 2004, 2005; Gray et al, 2004; Chang et al, 2006; Rogers et al, 2006; Zimmer et al, 2009). Among GCYs, GCY-31 and GCY-33 are the only two that are expressed in BAG neurons (Yu et al, 1997; Zimmer et al, 2009), and genetic studies have shown that GCY-31 and GCY-33 are required non-redundantly by BAG neurons to sense low-level O2 (Zimmer et al, 2009). Therefore, we studied the potential effects of GCY-31 and GCY-33 on lifespan, using gcy-31(ok296) and gcy-33(ok232) mutants. Both of these mutants were generated from exonic deletions that led to the loss of the key enzymatic domains and thus abolishment of gene function (Zimmer et al, 2009), and have been established as functional null mutations in the literature (Hallem and Sternberg, 2008; Zimmer et al, 2009). We performed our experiments under atmospheric normoxic condition, which helped dissociate the actions of these molecules from any inputs of O2 changes. Data showed that both gcy-31(ok296) (Figure 2A and B) and gcy-33(ok232) (Figure 2C and D) mutants had significantly increased mean lifespan and 75th percentile lifespan. Mean lifespan of gcy-31(ok296) mutants increased by 20.4% compared to wildtype, while gcy-33(ok232) mutants showed a larger increase by 27.3% compared to wildtype. Transgenic expression of C. elegans wildtype genomic copies of gcy-31 gene in gcy-31(ok296) mutants (Figure 2A and B) or of gcy-33 gene in gcy-33(ok232) mutants (Figure 2C and D) rescued the lifespan changes in these mutants. Consistent with the longevity phenotypes, ageing process in gcy-31 and gcy-33 mutants was slower than wildtype, for example, ageing-related loss of muscular functions delayed in gcy-31 and gcy-33 mutants according to head movement, sinusoidal body posture, and locomotion at the time point of population median lifespan (Day 17 for gcy-31 mutants, Day 18 for gcy-33 mutants, and Day 14 for wildtype) (Supplementary Figure S2; Supplementary Movies S1–S3). Also, gcy-31 and gcy-33 mutants spent similar time on food as did wildtype controls (Supplementary Figure S1C). It should be mentioned that gcy-31 is expressed exclusively in BAG neurons (Zimmer et al, 2009), and thus the longevity phenotype of gcy-31 mutants can be attributed specifically to BAG neurons. Altogether, these data indicate that GCY-31 and GCY-33 act to suppress lifespan in C. elegans.
Figure 2.
Lifespan phenotypes of gcy-31, gcy-33, gcy-35, and gcy-36 mutants. Survival curves (A, C, E and G) and mean lifespan and 75th percentile lifespan (B, D, F and H) of gcy-31(ok296) mutants and gcy-31(ok296) mutants expressing wildtype gcy-31 genomic DNA (n=93–113 per genotype) (A, B), gcy-33(ok232) mutants and gcy-33(ok232) mutants expressing wildtype gcy-33 genomic DNA (n=37–138 per genotype) (C, D), gcy-35(ok769) mutants and gcy-35(ok769) mutants expressing gcy-35 cDNA from its endogenous promoter (n=96–140 per genotype) (E, F), or gcy-36(db66) mutants and gcy-36(db66) mutants expressing gcy-36 cDNA from its endogenous promoter (n=116–142 per genotype) (G, H) versus shared wildtype controls (n=154). (A, C, E and G) show animals pooled from multiple independent experiments, and the same pool of wildtype controls was used in these curves. (B, D, F and H) show analysis from a representative single experiment (n=28–39 per group). **P<0.01, ****P<0.0001, statistics for curve comparisons are shown in the figure. Error bars represent mean±s.e.m.
GCY-35 and GCY-36 positively control lifespan
Together with our studies on GCY-31/33, we simultaneously investigated GCY-35 and GCY-36, two GCY isoforms that can mediate high-level O2 sensing in URX neurons and related AQR and PQR neurons (Cheung et al, 2004, 2005; Gray et al, 2004; Chang et al, 2006; Rogers et al, 2006; Zimmer et al, 2009). Our experiments were based on functional loss of GCY-35 and GCY-36, using the established gcy-35(ok769) and gcy-36(db66) mutants (Cheung et al, 2004, 2005; Gray et al, 2004; Chang et al, 2006; Rogers et al, 2006; Zimmer et al, 2009). We found that gcy-35(ok769) (Figure 2E and F) and gcy-36(db66) (Figure 2G and H) mutants had significantly decreased mean lifespan by 16.1 and 13.9%, respectively. Rescue experiments were further performed through transgenic expression of gcy-35 cDNA under gcy-35 promoter in gcy-35(ok769) mutants (Figure 2E and F) or transgenic expression of gcy-36 cDNA under gcy-36 promoter in gcy-36(db66) mutants (Figure 2G and H). As shown in these figures, the short lifespan phenotypes of these mutant strains were normalized by restoration of gcy-35 or gcy-36 genes. All these data indicate that GCY-35 and GCY-36 work to promote lifespan, and URX neurons are probably most important for this effect, since GCY-36 is present restrictedly in URX/AQR/PQR neurons (Cheung et al, 2004; Gray et al, 2004), and consistently, our ablation experiment has revealed the comparable lifespan phenotype in URX-ablated animals (Figure 1C and D). We also examined feeding behaviour of gcy-35 and gcy-36 mutants, and found that they spent normal amount of time on food as did wildtype (Supplementary Figure S1D), which agrees with the similar observation from the URX ablation study. In summary, our data have shown that GCY-35 and GCY-36 can work to increase lifespan in C. elegans.
BAG versusURX neurons-specific actions of GCYs opposingly control lifespan
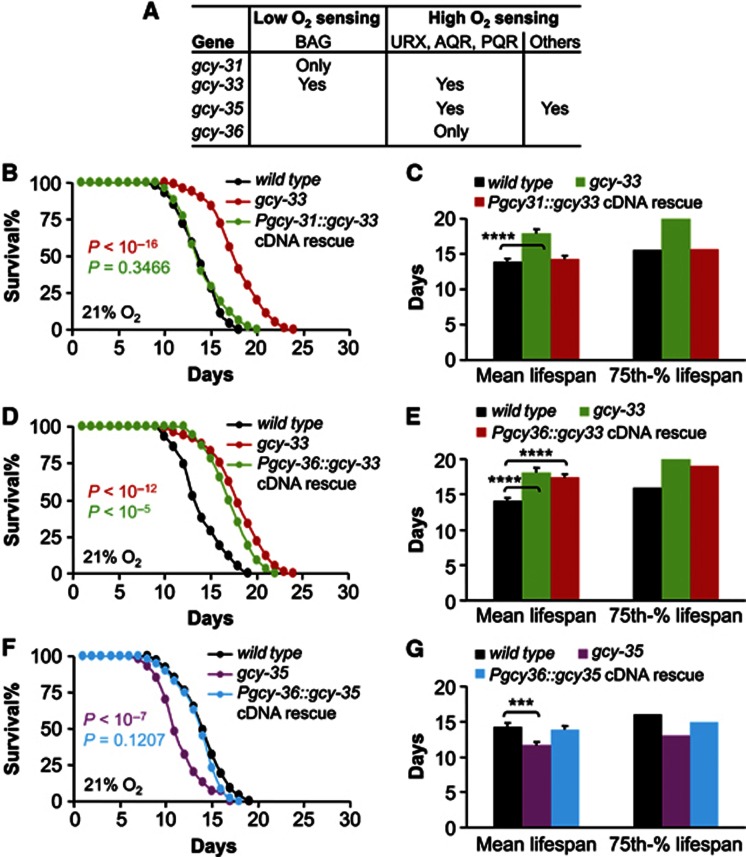
Next, we performed cell-specific rescue to dissect the types of neurons that are responsible for the revealed lifespan phenotypes of gcy mutants. As elucidated in Figure 3A, gcy-33 is expressed in BAG neurons as well as in URX/AQR/PQR neurons (Yu et al, 1997; Zimmer et al, 2009). Since these two groups of neurons have antagonizing roles in lifespan control, we questioned how gcy-33 acts in these neurons to regulate lifespan. In experiments, we used gcy-31 promoter (Zimmer et al, 2009) to direct BAG neuron-specific gcy-33 expression in gcy-33 mutants. Expression of Pgcy-31::gcy-33 cDNA rescued the longevity phenotype of gcy-33(ok232) mutants (Figure 3B and C), indicating that gcy-33 in BAG neurons per se is sufficient to suppress lifespan. Further, we used gcy-36 promoter (Cheung et al, 2004; Gray et al, 2004) to direct gcy-33 expression in URX/AQR/PQR neurons of gcy-33 mutants. However, expression of Pgcy-36::gcy-33 cDNA did not alter the longevity of gcy-33(ok232) mutants (Figure 3D and E), suggesting that the lifespan-regulating action of gcy-33 does not reside in URX/AQR/PQR neurons. This finding also agrees with the literature showing that BAG neurons rather than URX/AQR/PQR neurons are important for gcy-33 to induce aerotaxis (Zimmer et al, 2009). In parallel, we applied tissue-specific rescue to gcy-35(ok769) mutants, since gcy-35 is expressed not only in URX/AQR/PQR neurons but also in SDQ/ALN/PLN neurons (Cheung et al, 2004; Gray et al, 2004). SDQ group neurons additionally regulate O2-sensing behaviours in C. elegans (Chang et al, 2006; Rogers et al, 2006), but they do not have structural connections with BAG or URX/AQR/PQR neurons (White et al, 1986; Altun et al, 2012). We used gcy-36 promoter (Cheung et al, 2004; Gray et al, 2004) to drive gcy-35 expression specifically in URX/AQR/PQR neurons of gcy-35 mutants. Data showed that expression of Pgcy-36::gcy-35 cDNA rescued the short lifespan phenotype of gcy-35(ok769) mutants (Figure 3F and G), indicating that gcy-35 in URX/AQR/PQR neurons per se is sufficient to promote longevity in C. elegans.
Figure 3.
Tissue-specific transgenic rescue of lifespan phenotypes of gcy-33 and gcy-35 mutants. (A) Expression profiles of individual gcy genes in O2-sensing neurons. (B–G) Survival curves (B, D and F) and mean lifespan and 75th percentile lifespan (C, E and G) of wildtype controls versus gcy-33(ok232) mutants and gcy-33(ok232) mutants expressing gcy-33 cDNA from gcy-31 promoter (n=52–80 per genotype) (B, C), of wildtype controls versus gcy-33(ok232) mutants (n=84) and gcy-33(ok232) mutants expressing gcy-33 cDNA from gcy-36 promoter (n=32–84 per genotype) (D, E), and of wildtype controls versus gcy-35(ok769) mutants and gcy-35(ok769) mutants expressing gcy-35 cDNA from gcy-36 promoter (n=60–74 per genotype) (F, G). (B, D and F) show animals pooled from multiple independent experiments; (C, E and G) show analysis from a representative single experiment (n=25–42 per group). ***P<0.001, ****P<0.0001, statistics for curve comparisons are shown in the figure. Error bars represent mean±s.e.m.
Counterbalance of different GCYs controls lifespan homeostasis
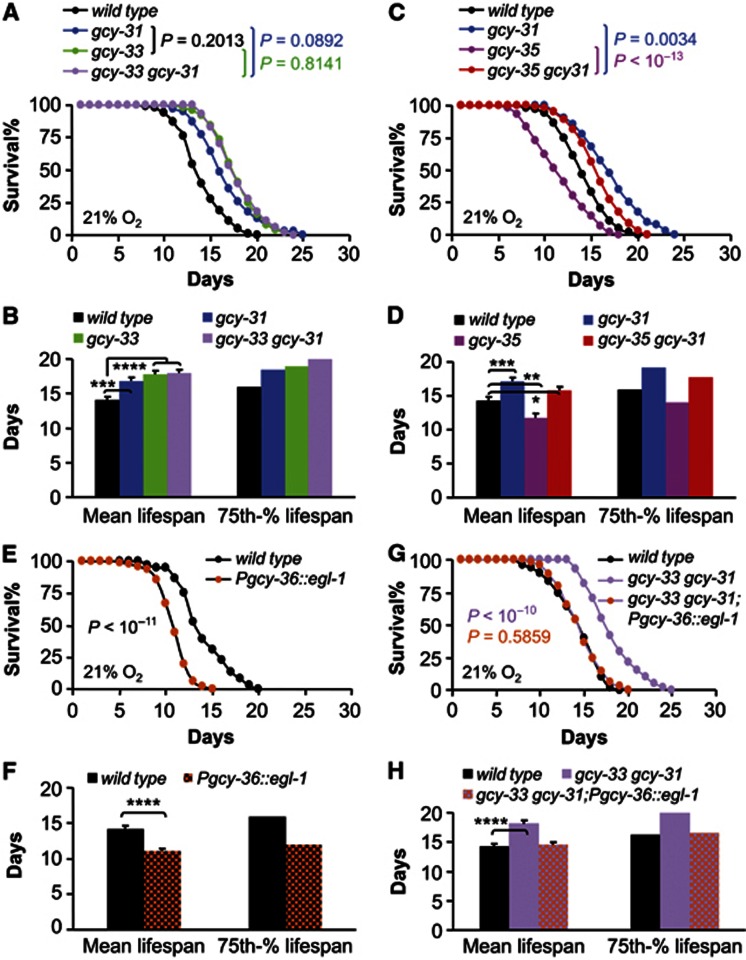
To gain further insight into the interplay among different GCYs in lifespan control, we examined the genetic epistatic relationship among these GCYs. We first tested whether GCY-31 and GCY-33 may have synergistic effects to suppress lifespan, given that increase in lifespan was seen in both gcy-31 mutants (Figure 2A and B) and gcy-33 mutants (Figure 2C and D). To do this, we constructed gcy-33 gcy-31 double mutants, and found that the double mutants did not further increase lifespan compared to gcy-31(ok296) or gcy-33(ok232) single mutants (Figure 4A and B). Thus, these data may suggest that GCY-31 and GCY-33 act in the same pathway to suppress lifespan, which is in line with our revealed importance of BAG neurons for both GCY-31 and GCY-33 to affect lifespan. Next, we asked whether the pro-longevity phenotype of gcy mutations in BAG neurons can be affected by gcy mutations in URX/AQR/PQR neurons. Since gcy-31 is specifically expressed in BAG neurons, we tested if the pro-longevity effect of gcy-31 loss-of-function could be affected by gcy-35 loss-of-function. To do so, double mutants with gcy-35(ok769) and gcy-31(ok296) were compared to single mutants, and results showed that the pro-longevity effect in gcy-31 mutants was significantly although partially reduced (Figure 4C and D). This result indicates that GCY-35 in URX/AQR/PQR neurons can counteract the lifespan-suppressing action of GCY-31 in BAG neurons. In addition to gcy-35 mutants, we alternatively inhibited URX/AQR/PQR neurons through transgene qaIs2241 which expressed gcy-36 promoter-controlled cell-death activator egl-1 (Chang et al, 2006; Styer et al, 2008). Indeed, this qaIs2241 mutant strain showed lifespan shortening (Figure 4E and F)—which resembled the effect of URX ablation (Figure 1C and D), and importantly, functional loss of GCY-31/33 via gcy-33 gcy-31 double mutants completely abolished the lifespan-shortening effect of transgene qaIs2241 (Figure 4G and H). These findings further proved that URX/AQR/PQR neurons are counteractively involved in the lifespan-inhibiting actions of GCY-31/33 in BAG neurons.
Figure 4.
Interaction of low O2-sensing and high O2-sensing pathways in lifespan control. Survival curves (A, C, E and G) and mean lifespan and 75th percentile lifespan (B, D, F and H) of wildtype controls versus gcy-31(ok296) mutants (Mantel-Cox P<10−5), gcy-33(ok232) mutants (Mantel-Cox P<10−13), and gcy-33(ok232); gcy-31(ok296) double mutants (Mantel-Cox P<10−25) (n=39–140 per genotype) (A, B), of wildtype controls versus gcy-31(ok296) mutants (Mantel-Cox P<10−5), gcy-35(ok769) mutants (Mantel-Cox P<10−4), and gcy-35(ok769); gcy-31(ok296) double mutants (Mantel-Cox P<10−3) (n=40–103 per genotype) (C, D), of wildtype controls versus animals with genetic ablation of URX, AQR, and PQR neurons (n=36–140 per genotype) (E, F), and of wildtype controls versus gcy-33(ok232); gcy-31(ok296) mutants, and gcy-33(ok232); gcy-31(ok296) mutants with genetic ablation of URX, AQR, and PQR neurons (Mantel-Cox P<10−12 compared to gcy-33 gcy-31 double mutants) (n=48–115 per genotype) (G, H). (A, C, E and G) show animals pooled from multiple independent experiments; (B, D, F and H) show analysis from a representative single experiment (n=26–48 per group). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001, statistics for curve comparisons are shown in the figure. Error bars represent mean±s.e.m.
GCYs modulate lifespan independently of canonical pathways
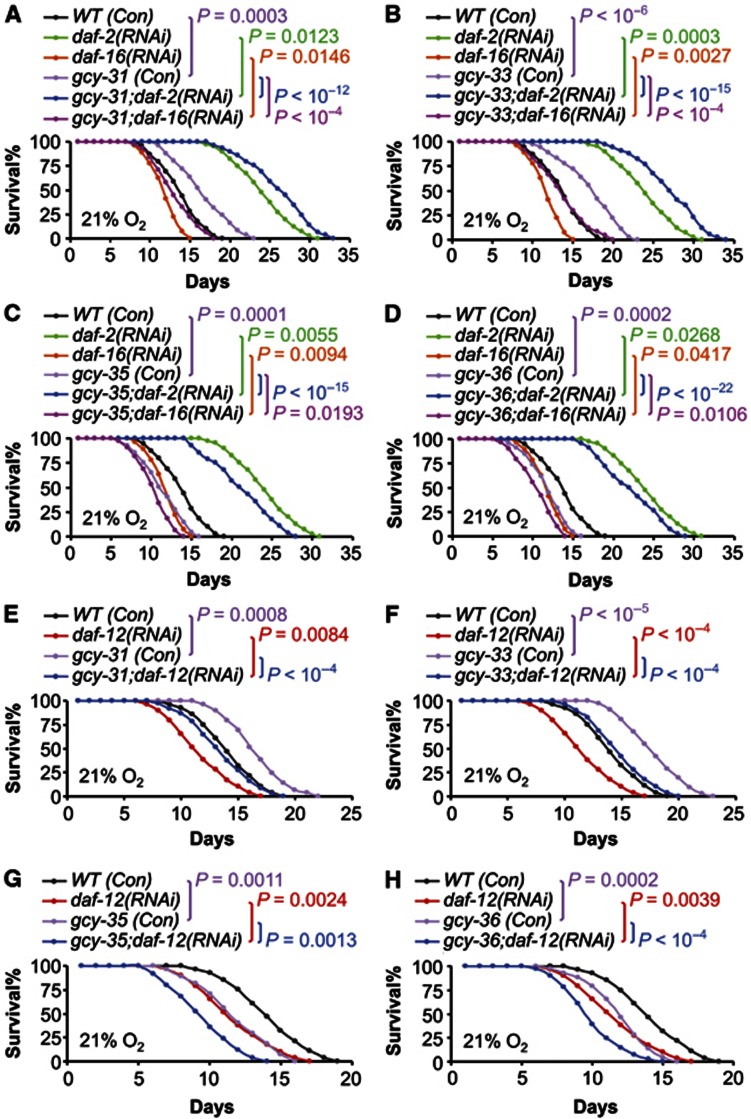
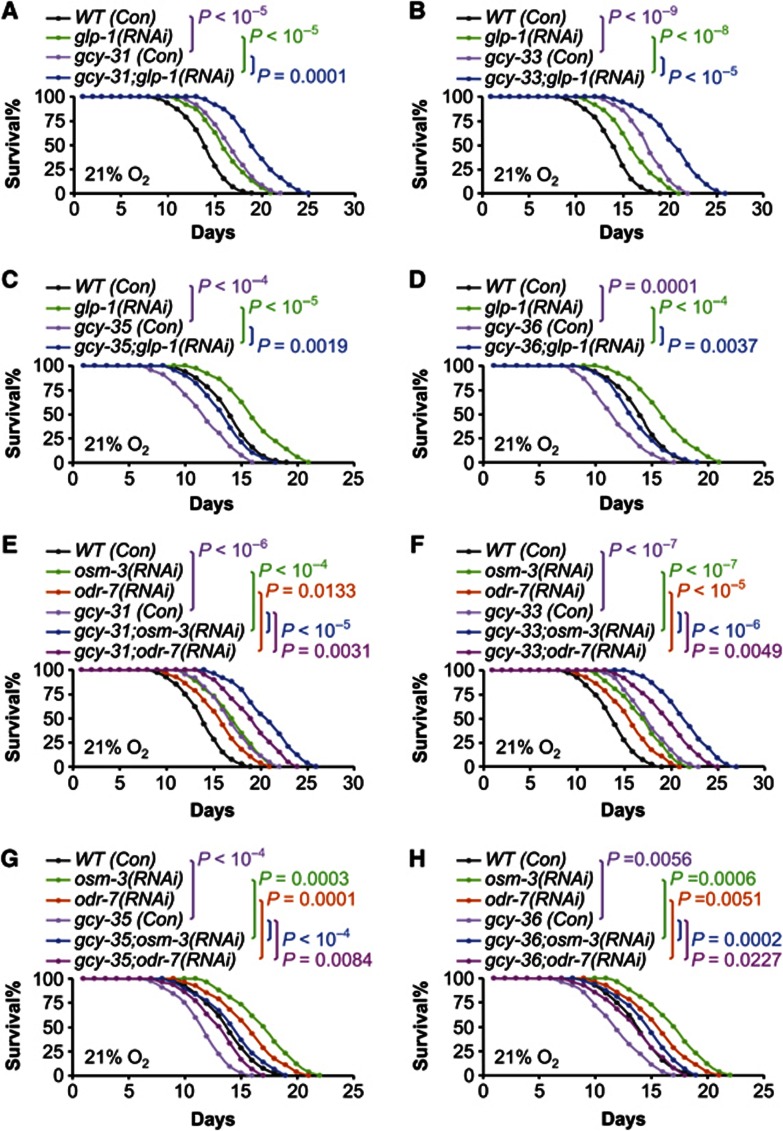
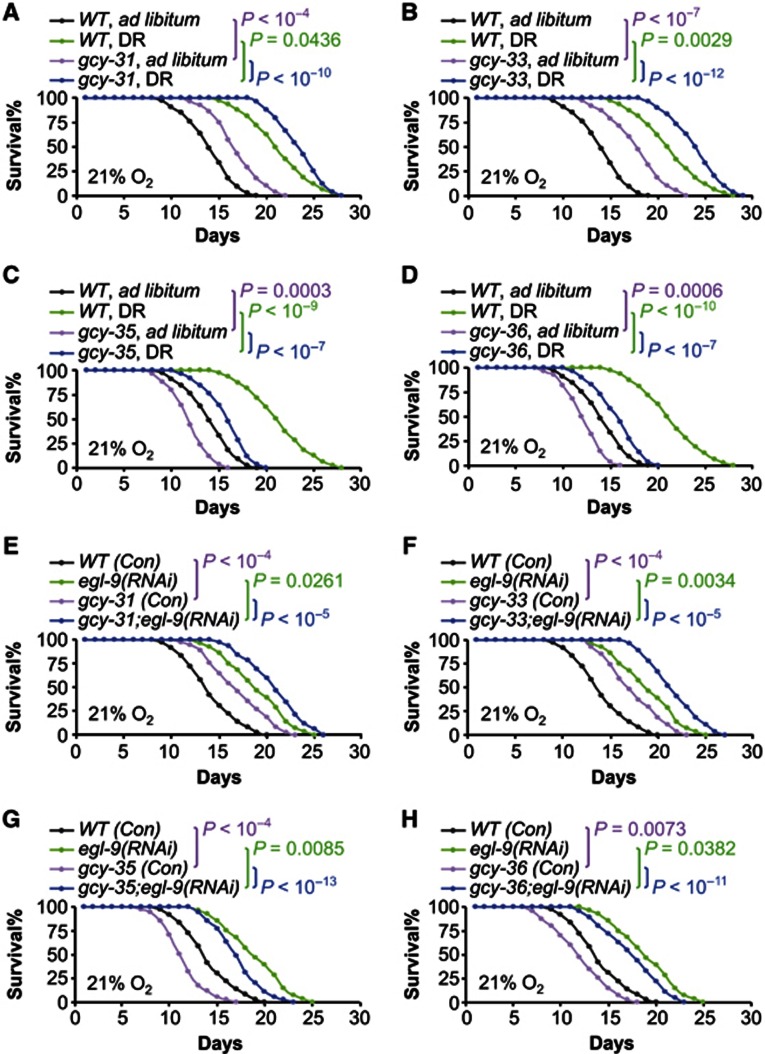
In C. elegans research, a few canonical pathways are known to regulate lifespan including DAF-2/DAF-16 signalling, DAF-12 signalling, germline signalling, and sensory perception (Kenyon et al, 1993; Sengupta et al, 1994; Tabish et al, 1995; Kimura et al, 1997; Ogg et al, 1997; Hsin and Kenyon, 1999; Gerisch et al, 2001; Arantes-Oliveira et al, 2002; Broue et al, 2007; Gerisch et al, 2007; Lee and Kenyon, 2009). We examined if inhibition of these canonical pathways could affect the lifespan phenotypes in our models. Using RNAi approach, gcy mutants were treated with daf-2 or daf-16 knockdown to alter DAF-2/DAF-16 signalling, daf-12 knockdown to inhibit DAF-12 signalling, glp-1 knockdown to inhibit germline signalling, or osm-3 or odr-7 knockdown to inhibit sensory perception. Overall, through all these experiments, we consistently found that the patterns of lifespan phenotypes in gcy mutants were not altered by RNAi manipulation of DAF-2/DAF-16 (Figure 5A–D; Supplementary Figure S3), DAF-12 (Figures 5E–H; Supplementary Figure S4), germline signalling (Figures 6A–D; Supplementary Figure S5), or sensory perception (Figures 6E–H; Supplementary Figure S6). Further, we tested if DR and GCYs might work in an overlapping pathway to affect lifespan. Our study employed a modified solid DR method—which was shown to induce lifespan extension independently of DAF-16 (Chen et al, 2009), and it elicited ∼50% lifespan extension in N2 worms in our experimental conditions. Of interest, we found that DR-induced longevity was additive to the lifespan phenotype of individual gcy mutants (Figure 7A–D; Supplementary Figure S7). Finally, we considered hypoxia-inducible factor-1 (HIF-1), given the literature context showing that HIF-1 activation increases lifespan (Chen et al, 2009; Mehta et al, 2009; Muller et al, 2009; Zhang et al, 2009; Leiser et al, 2011). We tested HIF-1 gain-of-function using RNAi knockdown of egl-9 (a gene encoding an HIF-1-degrading enzyme), and this model has been shown to induce lifespan extension in C. elegans (Mehta et al, 2009). Our data showed that egl-9 knockdown extended lifespan in wildtype animals, and this effect was additive to lifespan phenotypes of gcy mutants (Figure 7E–H; Supplementary Figure S8). These data suggest that HIF-1 and GCYs may work in parallel pathways to modulate lifespan, but possibility also exists that our experimental system may be inadequate to discern an interconnection between HIF-1 and GCYs. To summarize, the lifespan-modulating effects of GCYs are not mediated by these canonical regulatory pathways in C. elegans. Also importantly, GCY-31/33 inhibition can further enhance the longevity outcomes from suppressing insulin/IGF-1 signalling, germline pathway, sensory perception or feeding, and on the other hand, GCY-35/36 inhibition can exacerbate lifespan shortening induced by DAF-16 or DAF-12 inhibition. Therefore, different GCYs can work to broadly counterbalance the pro- and anti-longevity actions of endocrine signalling, reproduction, sensory perception, and feeding.
Figure 5.
GCYs modulate lifespan independently of DAF-2/DAF-16 or DAF-12 signalling. (A–D) Survival curves of N2 wildtype (WT) strain versus gcy-31(ok296) (A), gcy-33(ok232) (B), gcy-35(ok769) (C) or gcy-36(db66) (D) mutants treated with daf-2(RNAi), daf-16(RNAi) or control RNAi (Con). All experiments in (A–D) were performed at the same time, and the same groups of WT (Con), daf-2(RNAi), and daf-16(RNAi) were compared to individual gcy mutants. (E–H) Survival curves of N2 WT strain versus gcy-31(ok296) (E), gcy-33(ok232) (F), gcy-35(ok769) (G), or gcy-36(db66) (H) mutants treated with daf-12(RNAi) or control RNAi (Con). All experiments in (E–H) were performed at the same time, and the same groups of WT (Con) and daf-12(RNAi) were compared to individual gcy mutants. Statistics for curve comparisons are shown in the figure, n=24–45 per group (A–D), n=25–43 per group (E–H). Mean lifespan and 75th percentile lifespan of (A–D) are shown in Supplementary Figure S3, and mean lifespan and 75th percentile lifespan of (E–H) are shown in Supplementary Figure S4.
Figure 6.
GCYs modulate lifespan independently of germline signalling or sensory perception. (A–D) Survival curves of N2 wildtype (WT) strain versus gcy-31(ok296) (A), gcy-33(ok232) (B), gcy-35(ok769) (C), or gcy-36(db66) (D) mutants treated with glp-1(RNAi) or control RNAi (Con). All experiments in (A–D) were performed at the same time, and the same groups of WT (Con) and glp-1(RNAi) were compared to individual gcy mutants. (E–H) Survival curves of N2 WT strain versus gcy-31(ok296) (E), gcy-33(ok232) (F), gcy-35(ok769) (G), or gcy-36(db66) (H) mutants treated with osm-3(RNAi), odr-7(RNAi), or control RNAi (Con). All experiments in (E–H) were performed at the same time, and the same groups of WT (Con), osm-3(RNAi), and odr-7(RNAi) were compared to individual gcy mutants. Statistics for curve comparisons are shown in the figure, n=34–48 per group (A–D), n=24–46 per group (E–H). Mean lifespan and 75th percentile lifespan of (A–D) are shown in Supplementary Figure S5, and mean lifespan and 75th percentile lifespan of (E–H) are shown in Supplementary Figure S6.
Figure 7.
GCYs modulate lifespan independently of dietary restriction or egl-9 knockdown. (A–D) Survival curves of N2 wildtype (WT) strain versus gcy-31(ok296) (A), gcy-33(ok232) (B), gcy-35(ok769) (C), or gcy-36(db66) (D) mutants subjected to ad libitum feeding or dietary restriction (DR). All experiments in (A–D) were performed at the same time, and the same groups of WT (ad libitum) and WT (DR) were compared to individual gcy mutants. (E–H) Survival curves of N2 WT strain versus gcy-31(ok296) (E), gcy-33(ok232) (F), gcy-35(ok769) (G), or gcy-36(db66) (H) mutants treated with egl-9(RNAi) or control RNAi (Con). All experiments in (E–H) were performed at the same time, and the same groups of WT (Con) and egl-9(RNAi) were compared to individual gcy mutants. Statistics for curve comparisons are shown in the figure, n=24–38 per group (A–D), n=27–46 per group (E–H). Mean lifespan and 75th percentile lifespan of (A–D) are shown in Supplementary Figure S7, and mean lifespan and 75th percentile lifespan of (E–H) are shown in Supplementary Figure S8.
Environmental O2 levels modulate lifespan in C. elegans
A relevant question of this study was whether environmental O2 levels are important for GCYs to affect lifespan. To answer this question, we first systematically profiled the lifespan of C. elegans under different levels of environmental O2. To do so, we cultured N2 strain C. elegans on solid nematode growth media (NGM) in air-tight chambers containing different concentrations of O2 balanced with gaseous nitrogen. We tested a range of moderately low O2 levels including 4, 8, and 12%, normoxic 21%, and hyperoxic 40%. Of note, it has been documented that N2 strain prefers to live under 5–12% O2 (Gray et al, 2004; Zimmer et al, 2009). We did not attempt to include extremely low O2 levels, as they have limited physiological relevance for aerobic species, although 0.5% O2 (Mehta et al, 2009) or 1% O2 (Honda et al, 1993) has been shown to increase lifespan. As shown in Supplementary Figure S9, we observed that lifespan of C. elegans increased as environmental O2 level decreased, though survival curves and mean lifespan under 8 and 12% O2 were similar. This similarity between 8 and 12% O2 agrees with the understanding that these levels represent preferred O2 conditions by C. elegans. Using mean lifespan under 8–12% O2 as a reference, we found that mean lifespan of animals increased by ∼7% under 4% O2 but decreased by ∼5% under 21% O2 and by ∼10% under 40% O2. Lifespan under 40% O2 also decreased compared to 21% O2, agreeing with a previously observed lifespan-shortening effect of 60% O2 (Honda et al, 1993). We noted that Honda et al only showed an induction of longevity under 1% O2, and comparatively, our measurement was more sensitive that might be related to a few differences in our study: (1) We did lifespan assay at 25°C, whereas Honda et al used 20°C. Environmental temperature is a lifespan regulator (Lee and Kenyon, 2009), and has been shown to determine the effect of a lifespan regulator (Leiser et al, 2011). (2) Honda et al added 5-fluoro-2′-deoxyuridine (FUdR) to C. elegans media to inhibit progeny development in the lifespan assay. However, FUdR significantly increases lifespan of nematodes (Aitlhadj and Sturzenbaum, 2010; Van Raamsdonk and Hekimi, 2011), thus we did not use FUdR, but instead, we transferred worms to fresh plates every 2 days in their reproductively active stage, as described in the recent literature (Greer et al, 2007). (3) We subjected worms to varied O2 levels since the very beginning of life (the timed-egg laying (TEL) stage). By contrast, Honda et al exposed worms to varied O2 levels since 4 days after hatching, which skipped the embryonic stage and early adulthood (C. elegans develop into adulthood in ∼45 h after hatching at 20°C). Accumulated evidence has indicated that early-life events have profound effects on lifespan and ageing (Gavrilov and Gavrilova, 2004; Vaiserman, 2008; Waterland, 2009). Regardless, our data have shown that moderate decrease or increase in environmental O2 levels near a probable range for wild soil habitat (e.g., 8–21%) can have opposing effects on lifespan in C. elegans.
Gas-induced lifespan effects are independent of and counteracted by GCYs
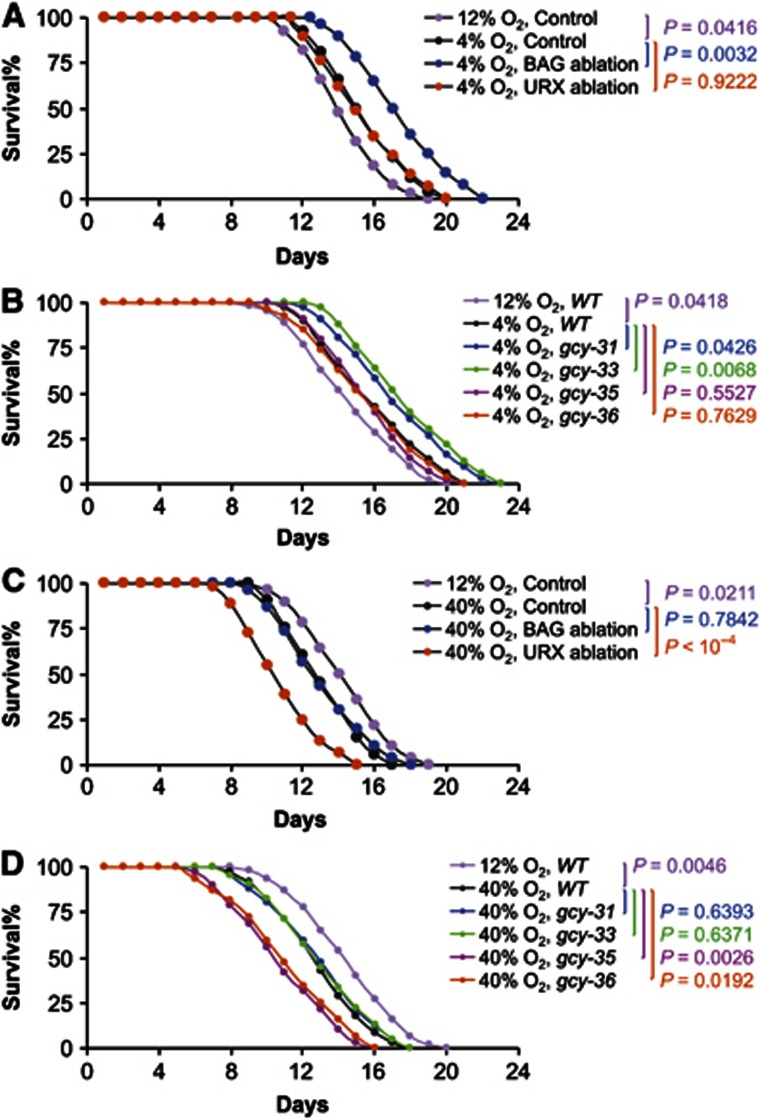
We then studied whether BAG and URX neurons are involved in the lifespan-promoting effect of low O2 environment. Laser ablation was used to kill BAG or URX neurons in wildtype animals, which were maintained under 4% O2 condition versus 12% O2 as control. Ablation controls lived longer at 4% O2 compared to 12% O2 (Figure 8A; Supplementary Figure S10A). Loss of URX neurons did not significantly alter lifespan compared to ablation controls under 4% O2 (Figure 8A; Supplementary Figure S10A), indicating that URX neurons do not exert lifespan modulation at 4% O2, which is consistent with the literature showing that URX neurons are inactive at low O2 environment (Busch et al, 2012). By contrast, loss of BAG neurons extended lifespan compared to ablation controls at 4% O2 (Figure 8A; Supplementary Figure S10A). Thus, BAG neurons can counteract the pro-longevity of low O2. Since GCY-31/33 in BAG neurons is important for lifespan control, we examined if these GCYs were involved in the lifespan effect of low O2, and found that gcy-31(ok296) and gcy-33(ok232) mutants had further extended lifespan compared to wildtype at 4% O2 (Figure 8B; Supplementary Figure S10B). By contrast, gcy-35(ok769) and gcy-36(db66) mutants showed similar lifespan to wildtype under 4% O2 (Figure 8B; Supplementary Figure S10B). Thus, genetic mutant and cell ablation studies consistently demonstrated that the pro-longevity effect of low O2 is independent of and counteracted by actions of BAG neurons and comprised GCY-31/33, while URX neurons and comprised GCY-35/36 are not involved in this process. In parallel with studying low O2, we used laser ablation to kill neurons in animals under 40% O2 versus 12% O2 as control. Ablation controls showed significantly shorter lifespan at 40% O2 compared to 12% O2 (Figure 8C; Supplementary Figure S10C). Ablation of URX neurons at 40% O2 further shortened lifespan compared to ablation controls (Figure 8C; Supplementary Figure S10C). In contrast to URX neurons, ablation of BAG neurons at 40% O2 did not significantly change lifespan (Figure 8C; Supplementary Figure S10C). Since GCY-35 and GCY-36 are important for URX neurons to affect lifespan, we determined if these GCYs were involved in the lifespan-modulating effect of high O2. We found that lifespan-shortening effect of 40% O2 was further enhanced in gcy-35(ok769) and gcy-36(db66) mutants (Figure 8D; Supplementary Figure S10D). Also consistent with absent effect from ablating BAG neurons (Figure 8C; Supplementary Figure S10C), gcy-31(ok296) or gcy-33(ok232) mutants showed similar lifespan compared to wildtype control at 40% O2 (Figure 8D; Supplementary Figure S10D). In sum, the anti-longevity effect of high O2 is independent of and counteracted by actions of URX neurons and comprised GCY-35/36, without involving BAG neurons or comprised GCY-31/33. We appreciate that BAG and URX neurons can mediate aerotaxis behaviours in response to O2 changes, for example, low O2-induced BAG activation drives animals to navigate away from low O2 environment, and high O2-induced URX activation drives animals to navigate away from high O2 environment (Cheung et al, 2005; Bretscher et al, 2008; Hallem and Sternberg, 2008; Zimmer et al, 2009). However, since our laboratory C. elegans were cultivated under stable O2 levels in dishes with UV-killed bacteria, apparently aerotaxis is not critically involved in the lifespan phenotypes revealed in these models.
Figure 8.
GCYs modulate lifespan independently of and counteractively with gas conditions. (A) Survival curves of C. elegans following laser ablation of BAG neurons or URX neurons under 4% O2 versus ablation controls under 4% O2 and 12% O2, respectively (n=26–38 per group). (B) Survival curves of gcy-31 mutants, gcy-33 mutants, gcy-35 mutants, and gcy-36 mutants under 4% O2 versus wildtype controls under 4% O2 and 12% O2, respectively (n=27–43 per group). (C) Survival curves of C. elegans following laser ablation of BAG neurons or URX neurons under 40% O2 versus ablation controls under 40% O2 and 12% O2, respectively (n=28–44 per group). (D) Survival curves of gcy-31 mutants, gcy-33 mutants, gcy-35 mutants, and gcy-36 mutants under 40% O2 versus wildtype controls under 40% O2 and 12% O2, respectively (n=32–45 per group). Statistics for curve comparisons are shown in the figure. Mean lifespan and 75th percentile lifespan of (A–D) are shown in Supplementary Figure S10.
Pro-longevity action of moderately high CO2 and its counteraction by GCY-9
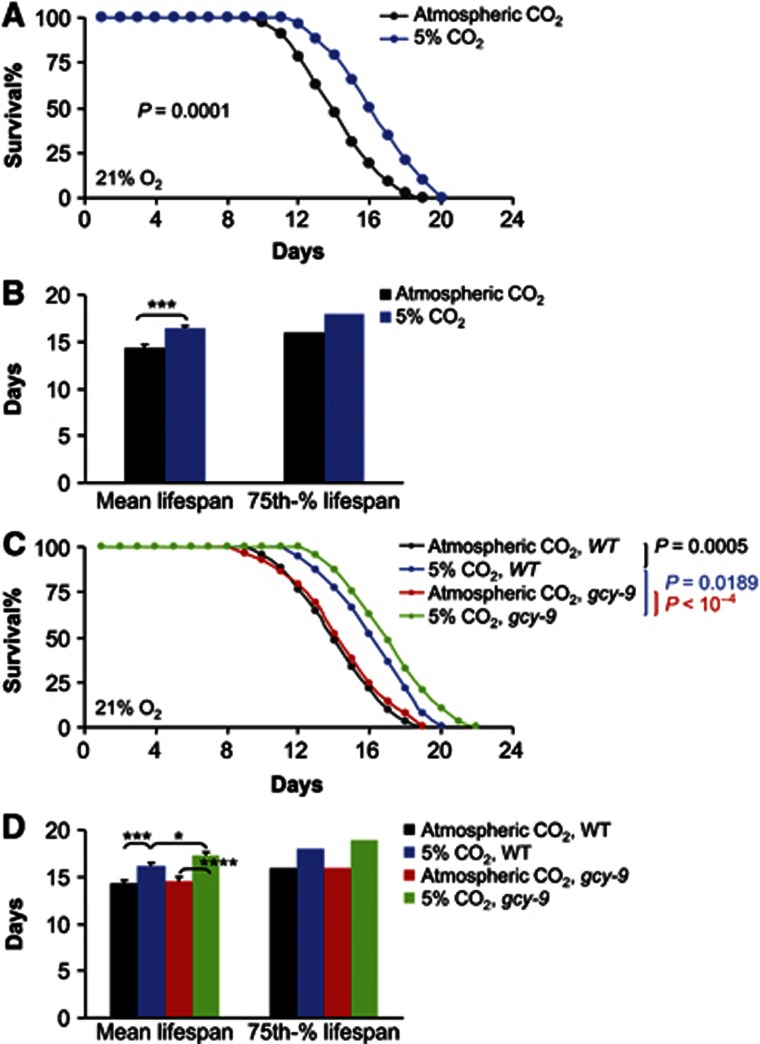
In addition to O2 sensing, recent research showed the existence of CO2 sensing in C. elegans (Bretscher et al, 2008; Hallem and Sternberg, 2008; Hallem et al, 2011). Of interest, BAG neurons can sense CO2 in addition to low O2 (Hallem and Sternberg, 2008), and GCY-9 was shown to specifically mediate CO2 sensing of BAG neurons (Hallem et al, 2011). We hence tested whether environmental CO2 levels may affect lifespan, and if so, how BAG neurons and GCY-9 are involved. Our test targeted 5% CO2, because in well-fed C. elegans, both behavioural and neuronal responses to CO2 linearly increase from 0% to 5%, and approximately hit a plateau beyond 5% (Bretscher et al, 2008; Hallem and Sternberg, 2008; Hallem et al, 2011), indicating that 5% CO2 represents a condition without causing obvious toxic effects. In our experiments, C. elegans were cultured in air chambers containing 5% CO2 and 21% O2, and control animals under atmospheric 21% O2 and 0.039% CO2. We found that 5% CO2 led to a significant increase in lifespan (Figure 9A), and the mean lifespan increased by 14.4% (Figure 9B). According to the literature, CO2 sensing is induced by a neural circuit that involves BAG neurons (Hallem and Sternberg, 2008; Hallem et al, 2011), and GCY-9 was identified as a CO2 sensor specifically expressed in BAG neurons (Hallem et al, 2011). Thus, we employed gcy-9 mutants to test if this CO2 sensor has an effect on lifespan. Our results revealed that, in the presence of 5% CO2, gcy-9 mutants lived longer than wildtype animals (Figure 9C) and mean lifespan increased by 7.1% (Figure 9D). Thus, GCY-9 in BAG neurons counter-regulates lifespan modulation by environmental CO2, a phenomenon that resembles the counteractive effect of GCY-31/33 in BAG neurons against the lifespan effect of low O2. Under atmospheric air condition that contains only trace amount of CO2 (0.039%), gcy-9 mutants exhibited similar lifespan compared to wildtype (Figure 9C and D), indicating that the counteractive action of GCY-9 against CO2 relies on increased levels of environmental CO2. In summary, different GCYs in BAG neurons can synergize to counter-regulate the pro-longevity actions of CO2 and low O2 environment.
Figure 9.
BAG neurons and GCY-9 counteract CO2-induced lifespan extension. (A, B) Survival curves (A) and mean lifespan and 75th percentile lifespan (B) of wildtype C. elegans under atmospheric CO2 (n=32) versus 5% CO2 (n=52). (C, D) Survival curves (C) and mean lifespan and 75th percentile lifespan (D) of wildtype controls and gcy-9(tm2816) mutants under atmospheric CO2 versus 5% CO2 (n=29–45 per group). *P<0.05, ***P<0.001, ****P<0.0001, statistics for curve comparisons are shown in the figure. Error bars represent mean±s.e.m.
Discussion
BAG and URX neurons antagonize each other to control lifespan balance
C. elegans can detect environmental and internal factors via sensory nervous system to direct behavioural and physiological responses. Of note, some forms of sensory perception have been negatively associated with lifespan of C. elegans, and the underlying basis involves a few types of sensory neurons (Finch and Ruvkun, 2001; Antebi, 2007; Bishop and Guarente, 2007a; Mair and Dillin, 2008; Kenyon, 2010). For example, inhibition of gustatory neurons ASI and ASG or olfactory neurons AWA and AWC were shown to increase lifespan (Apfeld and Kenyon, 1999; Alcedo and Kenyon, 2004). At signalling level, neurons were also demonstrated to be responsible for lifespan regulation by germline or DAF-16 signalling (Wolkow et al, 2000; Lin et al, 2001; Boulias and Horvitz, 2012). In this work, through targeting an antagonistic neural pathway consisting of BAG and URX neurons, we found that BAG neurons have an inhibitory role while URX neurons have a stimulatory role on the lifespan of C. elegans. We appreciate that the literature has related BAG and URX neurons to O2 sensing-dependent behavioural effects such as aggregation (Cheung et al, 2004; Rogers et al, 2006), social activities (Coates and de Bono, 2002; Gray et al, 2004; Cheung et al, 2005; Bretscher et al, 2008; Hallem and Sternberg, 2008), and immune response (Styer et al, 2008; Reddy et al, 2009). While it remains to be studied if these behavioural and physiological effects are integrated with lifespan regulation, we found that ablation of BAG or URX neurons or loss-of-function of GCYs did not significantly affect the time on food of C. elegans, and indeed, the lifespan phenotypes of these models are independent of DR. Overall, the antagonistic actions of BAG and URX neurons in lifespan control that we discovered seem to provide a set point for lifespan balance, which represents a previously unappreciated paradigm of lifespan regulation. Presumably, an ecological advantage of having this two-way neural regulation is to buffer an organism’s lifespan from being either unhealthily decreased or excessively increased, and therefore maintain the homeostasis of lifespan. If so, then identification of this regulatory system can lend a pronounced support to a notion that lifespan, in addition to being genetically defined, is also safeguarded by the nervous system.
BAG and URX neurons control lifespan independently of canonical pathways
In this study, we demonstrated that BAG and URX neurons and their comprised GCYs do not mediate the lifespan-modulating effects from canonical pathways such as DAF-2/DAF-16, DAF-12, or germline signalling, or the pathways involved in sensory perception or DR. Thus, BAG and URX neurons act to control lifespan independently of these canonical pathways. Through our cross-pathway analyses, it is also clear that lifespan upregulation or downregulation by manipulating these canonical pathways is significantly buffered through the actions of BAG-URX neural system. For example, BAG neurons counteract against the pro-longevity effects of inhibiting DAF-2, germline signalling, sensory perception, or food intake, while URX neurons counteract against the lifespan-shortening effects of DAF-16 or DAF-12 inhibition. These observations can be further supportive of the unique action of the BAG-URX network to control lifespan balance. In the context of diverse environmental changes (such as food and sex context) that affect lifespan, this neural network employs the antagonistic actions of BAG versus URX neurons to exert two-way counter influences on the lifespan modulations induced by these environmental changes. While these environmentally driven physical and physiological activities constitute important contents of life, it has been noticeable that a tradeoff exists between lifespan and life activities such as metabolism and reproduction. We speculate that the BAG-URX neural network may work to balance life activities and lifespan in order to achieve optimal survival. In the scenario of food and sex, for example, food limitation and reproductive restriction have been known to promote longevity but clearly also compromise these important life activities; however, the existence of counter-regulatory BAG neurons can offset these negative corollaries. Reciprocally, excess of food or reproductive activities enriches the life content but at the expense of shortened lifespan, whereas URX neurons can act to attenuate such lifespan loss. Therefore, the BAG-URX counter-regulatory system may safeguard overall biological homeostasis of an organism balanced with lifespan. And to this end, it is predicted that BAG-URX system may work above the levels of the lifespan-modulating pathways that respond to specific events, and undoubtedly further research is much needed to evaluate the potential role of this network in homeostatic integration of lifespan control and life activities.
Independent controls of lifespan by BAG-URX versus O2/CO2 are counteractive
In the literature, BAG and URX neurons have been studied for gas sensing, and it was shown that BAG neurons can sense low-level environmental O2, while URX neurons sense high-level environmental O2 (Coates and de Bono, 2002; Cheung et al, 2004; Gray et al, 2004; Chang et al, 2006; Rogers et al, 2006; Zimmer et al, 2009). Prior to our study, the literature have limitedly shown lifespan increase in C. elegans under extremely low O2 (0.5–1%) and lifespan shortening under extremely high O2 (60%) (Honda et al, 1993; Mehta et al, 2009). In this work, we also aimed to understand if gas sensing is important for BAG and URX neurons to regulate lifespan. To do that, we first systematically profiled the lifespan of C. elegans under different gas concentrations, and revealed that lifespan exhibits an inverse correlation with environmental O2 levels including a range of 4–12% O2 that probably exists in natural soil environment where wild worms live. Also, for the first time, we revealed that moderately high CO2 (5%) leads to a striking increase in lifespan in C. elegans. However, the most intriguing observation is that the pro-longevity or anti-longevity phenotypes of changing environmental O2 or CO2 are not only independent of but also counteracted by different GCYs in the BAG-URX network. Specifically, BAG neurons work via GCY-31/33 or GCY-9 to counteract the pro-longevity effect of low O2 or moderately high CO2, whereas URX neurons work via GCY-35/36 to counteract the lifespan shortening from high O2. Altogether, our findings indicate that there are GCY-independent parallel pathways that mediate the lifespan effects of O2 and CO2, but these parallel pathways are counterbalanced by GCYs to maintain lifespan balance. Indeed, neuronal targets of O2 and CO2 are not limited to BAG and URX neurons (Chang et al, 2006; Rogers et al, 2006; Hallem and Sternberg, 2008), and parallel pathways that mediate the pro-longevity effect of low O2 probably include HIF-1, since activation of HIF-1 has been shown to increase lifespan (Mehta et al, 2009; Muller et al, 2009; Leiser et al, 2011). Also, parallel pathways that mediate the anti-longevity effect of high O2 probably include insulin/IGF-1 signalling, since hyperoxia has been reported to activate insulin/IGF-1 signalling (Honda et al, 2008). Regarding the lifespan-regulating actions of GCYs in BAG-URX neurons, question still remains regarding if these actions are independent of gas sensing, or whether some logical relationship may exist between the biochemical pathway that mediates sensory transduction and the downstream pathways that regulate lifespan. While these outstanding questions call for future research, the findings in this work can lead to the conclusion that BAG and URX neurons work via different GCYs to control lifespan homeostasis.
Materials and methods
Nematode strains
All strains were maintained under standard conditions (Brenner, 1974) and fed with UV-killed E. coli OP50. Wildtype animals are C. elegans Bristol strain N2. Other strains used in this study are OH4841 [otIs92[Pflp-10::gfp]]; CZ3714 [gcy-31(ok296) X]; CX9552 [gcy-31(ok296) X; kyEx2027[WRM0630cF03, Podr-1::DsRed2]]; CZ3715 [gcy-33(ok232) V]; DSC1194 [gcy-33(ok232) V; aeEx1024[F57F5, Podr-1::DsRed2]]; AX1295 [gcy-35(ok769) I]; CX9560 [gcy-35(ok769) I; kyEx2033[Pgcy-35::gcy-35, Podr-1::DsRed2]]; AX1297 [gcy-36(db66) X]; CX9621 [gcy-36(db66) X; kyEx2087[Pgcy-36::gcy-36, Podr-1::DsRed2]]; CX9620 [gcy-33(ok232) V; kyEx2086[Pgcy-31::gcy-33, Podr-1::DsRed2]]; DSC1233 [gcy-33(ok232) V; aeEx1043[Pgcy-36::gcy-33, Podr-1::DsRed2]]; DSC1213 [gcy-35(ok769) I; aeEx1033[Pgcy-36::gcy-35, Podr-1::DsRed2]]; DSC1101 [gcy-33(ok232) V; gcy-31(ok296) X]; CX10131 [gcy-35(ok769) I; gcy-31(ok296) X]; CX7102 [lin-15(n765); qaIs2241[Pgcy-36::egl-1, Pgcy-35::gfp, lin-15(+)]]; XA2262 [gcy-33(ok232) V; gcy-31(ok296) X; qaIs2241[Pgcy-36::egl-1, Pgcy-35::gfp, lin-15(+)]]; EAH2 [gcy-9(tm2816) X].
Lifespan analysis
Freshly streaked E. coli OP50 bacteria were inoculated into liquid LB with and grown at 37°C to mid-log phase to seed standard NGM plates at 150 μl/plate. FUdR was not used in lifespan assay plates to prevent lifespan interference (Aitlhadj and Sturzenbaum, 2010; Van Raamsdonk and Hekimi, 2011). Seeded plates were left at room temperature for ∼12 h to let bacteria suspension precipitate and grow into a confluent lawn. The next day, bacteria were killed by exposing lawn surface to UV radiation of 0.9999, joules/cm2 for 10 min using Stratagene UV Statalinker 2400. Lifespan measurement was performed as previously described (Apfeld and Kenyon, 1999). Strains were first cultured under standard growth conditions for two generations to minimize non-genetic effects on lifespan. Then, 10–20 third-generation adult hermaphrodites were allowed 6–8 h for timed egg laying (TEL), and progenies sired during TEL continued to grow into late L4 larvae before being transferred to new plates at 30 animals per plate. Animals were passed to fresh plates every day during the reproductively active stage, and then passed every 5–7 days until the end of experiment. Each plate was scored every day for live animals, dead animals that no longer responded to body touch, and censored animals that ruptured, bagged, or crawled off plates. Lifespan was counted from Day 1 of adulthood. All lifespan assays were done at 25°C and in atmospheric environment unless otherwise mentioned.
O2 and CO2 treatment. Worms were exposed to specific O2 and CO2 from the beginning of life to death. To ensure gas exposure throughout a worm’s lifespan, both initial TEL plates and subsequent culture plates were maintained in air-tight chambers containing specific gas ingredients. Worms were briefly exposed to atmospheric environment only during scoring survival and passing plates. Gas chambers were refilled with customized pre-mixed gases (Tech Air) every day right after they were opened. Different concentrations of O2 (4, 8, 12, or 40%) or O2 (21%) plus CO2 (5%) were balanced with gaseous nitrogen.
Laser ablation
Synchronized eggs were produced through 2-h TEL, and left at 20°C for 12–14 h to hatch and grow into young L1 larvae. L1 larvae were mounted on 2% agarose pads containing 3 mM sodium azide for anaesthesia. Ablations of BAG and URX neurons were performed in OH4841 strain, which carries integrated transgenic marker Pflp-10::gfp to label flp-10 expressing neurons including BAG and URX (Kim and Li, 2004). Ablation was performed with MicroPoint Laser System (Photonic Instruments, Inc.) connected to a multi-channel epifluorescence Zeiss Axioplan 2 compound microscopy. Target neurons were identified through combined Nomarski and epifluorescence imaging with × 400 magnification, and ablated with a laser microbeam adjusted to minimal sufficient energy strength as previously described (Bargmann and Avery, 1995). Operated animals were recovered in S-basal droplets for 20 min and grown at 20°C until late L4 stage for lifespan assay. Ablation controls were randomly obtained from the same pool of L1 larvae, and handled in parallel with experimental animals at each step except that they did not undergo laser treatment.
Transgenic rescue
Experiment of gcy-31(ok296) rescue was based on using wildtype gcy-31 genomic DNA in strain CX9552, which expresses extrachromosomal arrays of C. elegans fosmid WRM0630cF03 in gcy-31(ok296) mutant background. Experiment of gcy-33(ok232) BAG-specific rescue was based on strain CX9620, which expresses extrachromosomal arrays of gcy-33 cDNA from BAG-specific gcy-31 promoter in gcy-33(ok232) mutant background. Experiment of gcy-35(ok769) whole-worm rescue was based on strain CX9560, which expresses extrachromosomal arrays of gcy-35 cDNA from gcy-35 promoter in gcy-35(ok769) mutant background. Experiment of gcy-36(db66) rescue was based on strain CX9621, which expresses extrachromosomal arrays of gcy-36 cDNA from gcy-36 promoter in gcy-36(db66) mutant background. All these strains were previously established and kindly provided by C Bargmann (Zimmer et al, 2009). Experiment of gcy-33(ok232) whole-worm rescue was performed through injection of C. elegans cosmid F57F2, which contains gcy-33 genomic DNA at 0.5 ng/μl in gcy-33(ok232) mutants, and four independent transgenic lines were analysed. Experiment of gcy-33(ok232) O2-sensing neuron rescue was performed through injection of gcy-33 cDNA plasmid directed by gcy-36 promoter at 20 ng/μl in gcy-33(ok232) mutants, and six independent transgenic lines were analysed. The expression plasmid was generated by subcloning gcy-33 cDNA PCR product into pSM vector containing 1-kb gcy-36 promoter (a kind gift from C Bargmann) using MscI and Bsu36I. The PCR product was obtained using primers 5′-gttggccaatgtacggattggtcattg-3′ and 5′-cgcccttaggctacataatactgcaaactttagttttc-3′. Experiment of gcy-35(ok769) O2-sensing neuron rescue was performed through injection of gcy-35 cDNA plasmid directed by gcy-36 promoter at 20 ng/μl in gcy-33(ok232) mutants, and three independent transgenic lines were analysed. The expression plasmid was generated by subcloning gcy-35 cDNA PCR product into pSM vector containing 1-kb gcy-36 promoter using NheI and EcoRV. The PCR product was obtained using primers 5′-ttgcgaagagtagaatattgg-3′ and 5′-agcacagggagaaagagcat-3′.
RNAi
C. elegans were fed bacterial strain HT115 transformed with L4440 vector expressing daf-16, daf-12, glp-1, egl-9, osm-3 or odr-7 RNAi (Fraser et al, 2000), HT115 transformed with pAD48-daf-2 RNAi construct (Dillin et al, 2002), or HT115 transformed with empty vector L4440 or pBluescript. Bacteria were grown in LB media containing 100 μg/ml ampicillin at 37°C overnight with vigorous shaking, and 3-ml bacterial culture was concentrated into 300 μl to seed one RNAi agar plate containing 100 μg/ml ampicillin and 6 mM isopropylthiogalactoside. RNAi treatments were generally provided to eggs collected from 3-day-old hermaphrodites lysed with bleaching solution (equal volumes of bleach and 1 M NaOH). In all, 30–40 eggs per plate density were used and continuously cultured at 25°C till hatching and adulthood. daf-2 RNAi and daf-16 RNAi were provided to Day 1 hermaphrodites. Worms were passed to new RNAi plates every day during their reproductively active stage and then every 5–7 days until the end of lifespan assay.
Feeding analysis and intervention
Time spent on food. Assay for the amount of time animals spend on bacterial lawn was adapted from the literature-established food leaving assay methods (Shtonda and Avery, 2006; Bendesky et al, 2011). Assay plates were prepared the same as lifespan analysis plates. Hermaphrodites in Day 2 adulthood were transferred to assay plates (30 worms per plate), and allowed to feed for 15 min to adapt to the plates before observation. Worms in plates were continuously observed for 15 min, and the amount of time that worms spent on bacteria lawn was recorded. Numbers of worms that stayed versus left bacterial lawn during each minute of 15-min duration were also recorded and analysed. Three independent assays were done per treatment group.
Dietary restriction. DR plates were prepared with the literature-established modified solid DR method (Chen et al, 2009). Freshly grown OP50 bacteria were pelleted and resuspended in S Medium to 5 × 109 cfu per ml, and 150 μl of this bacteria suspension was used to seed one DR agar plate, which was modified from standard NGM plate by removing peptone, increasing agar concentration to 2%, and adding 100 μg/ml ampicillin. Seeded DR plates were left at room temperature overnight to dry out excessive liquid in bacteria suspension, and then exposed to UV radiation of 0.9999, joules/cm2 for 10 min to kill bacteria. Synchronized C. elegans in their Day 1 adulthood were transferred to DR plates and maintained under DR treatment until end of life.
Statistical analysis
ANOVA and appropriate post hoc analysis were used for comparisons involving more than two groups. Two-tailed Student’s t-tests were used for comparisons involving only two groups. Lifespan analyses were performed using Kaplan–Meier survival analysis. Comparisons of survival curves used log-rank (Mantel-Cox) test to calculate P-values. All data were presented as mean±s.e.m. P<0.05 was considered as statistically significant.
Supplementary Material
Acknowledgments
We sincerely thank C Bargmann for providing gcy-31, gcy-33, gcy-35, and gcy-36 transgenic rescue lines, rescue DNA plasmids and gcy-35 gcy-31 double mutants; E Hallem for gcy-9 mutant strain; J-Y Sze for RNAi strains; D Hall for assistance with C. elegans neuroanatomy; and labs of S Emmons, H Buelow, and C Rubin for general assistance. This work was supported by Einstein startup funds and partially by NIH R01 grants AG031774 and DK078750 (all to DC). DC is a recipient of Irma T Hirschl Scholarship.
Author contributions: DC conceived the project and designed the study. TL participated in experimental design and performed experiments. TL and DC did data analysis and discussion. DC and TL wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aitlhadj L, Sturzenbaum SR (2010) The use of FUdR can cause prolonged longevity in mutant nematodes. Mech Ageing Dev 131: 364–365 [DOI] [PubMed] [Google Scholar]
- Alcedo J, Kenyon C (2004) Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron 41: 45–55 [DOI] [PubMed] [Google Scholar]
- Altun ZF, Herndon LA, Crocker C, Lints R, Hall DH (2012) Wormatlas, A database of behavioral and structural anatomy of Caenorhabditis elegans. The C. elegans Research Community 4-16-2012. Ref Type: Internet Communication [Google Scholar]
- Antebi A (2007) Genetics of aging in Caenorhabditis elegans. PLoS Genet 3: 1565–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C (1999) Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402: 804–809 [DOI] [PubMed] [Google Scholar]
- Arantes-Oliveira N, Apfeld J, Dillin A, Kenyon C (2002) Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295: 502–505 [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Avery L (1995) Laser killing of cells in Caenorhabditis elegans. In Epstein HF, Shakes DC (eds.) Caenorhabditis elegans: Modern Biological Analysis of an Organism San Diego: Academic Press, , pp 225–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendesky A, Tsunozaki M, Rockman MV, Kruglyak L, Bargmann CI (2011) Catecholamine receptor polymorphisms affect decision-making in C. elegans. Nature 472: 313–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop NA, Guarente L (2007a) Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet 8: 835–844 [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L (2007b) Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447: 545–549 [DOI] [PubMed] [Google Scholar]
- Boulias K, Horvitz HR (2012) The C. elegans microRNA mir-71 acts in neurons to promote germline-mediated longevity through regulation of DAF-16/FOXO. Cell Metab 15: 439–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher AJ, Busch KE, de Bono M (2008) A carbon dioxide avoidance behavior is integrated with responses to ambient oxygen and food in Caenorhabditis elegans. Proc Natl Acad Sci USA 105: 8044–8049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broue F, Liere P, Kenyon C, Baulieu EE (2007) A steroid hormone that extends the lifespan of Caenorhabditis elegans. Aging Cell 6: 87–94 [DOI] [PubMed] [Google Scholar]
- Busch KE, Laurent P, Soltesz Z, Murphy RJ, Faivre O, Hedwig B, Thomas M, Smith HL, de Bono M (2012) Tonic signaling from O(2) sensors sets neural circuit activity and behavioral state. Nat Neurosci 15: 581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI (2006) A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biol 4: e274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Thomas EL, Kapahi P (2009) HIF-1 modulates dietary restriction-mediated lifespan extension via IRE-1 in Caenorhabditis elegans. PLoS Genet 5: e1000486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung BH, Arellano-Carbajal F, Rybicki I, de Bono M (2004) Soluble guanylate cyclases act in neurons exposed to the body fluid to promote C. elegans aggregation behavior. Curr Biol 14: 1105–1111 [DOI] [PubMed] [Google Scholar]
- Cheung BH, Cohen M, Rogers C, Albayram O, de Bono M (2005) Experience-dependent modulation of C. elegans behavior by ambient oxygen. Curr Biol 15: 905–917 [DOI] [PubMed] [Google Scholar]
- Coates JC, de Bono M (2002) Antagonistic pathways in neurons exposed to body fluid regulate social feeding in Caenorhabditis elegans. Nature 419: 925–929 [DOI] [PubMed] [Google Scholar]
- Dillin A, Crawford DK, Kenyon C (2002) Timing requirements for insulin/IGF-1 signaling in C. elegans. Science 298: 830–834 [DOI] [PubMed] [Google Scholar]
- Finch CE, Ruvkun G (2001) The genetics of aging. Annu Rev Genomics Hum Genet 2: 435–462 [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J (2000) Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature 408: 325–330 [DOI] [PubMed] [Google Scholar]
- Gavrilov LA, Gavrilova NS (2004) Early-life programming of aging and longevity: the idea of high initial damage load (the HIDL hypothesis). Ann NY Acad Sci 1019: 496–501 [DOI] [PubMed] [Google Scholar]
- Gerisch B, Rottiers V, Li D, Motola DL, Cummins CL, Lehrach H, Mangelsdorf DJ, Antebi A (2007) A bile acid-like steroid modulates Caenorhabditis elegans lifespan through nuclear receptor signaling. Proc Natl Acad Sci USA 104: 5014–5019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerisch B, Weitzel C, Kober-Eisermann C, Rottiers V, Antebi A (2001) A hormonal signaling pathway influencing C. elegans metabolism, reproductive development, and life span. Dev Cell 1: 841–851 [DOI] [PubMed] [Google Scholar]
- Gray JM, Karow DS, Lu H, Chang AJ, Chang JS, Ellis RE, Marletta MA, Bargmann CI (2004) Oxygen sensation and social feeding mediated by a C. elegans guanylate cyclase homologue. Nature 430: 317–322 [DOI] [PubMed] [Google Scholar]
- Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A (2007) An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol 17: 1646–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Spencer WC, McWhirter RD, Zeller G, Henz SR, Ratsch G, Miller DM III, Horvitz HR, Sternberg PW, Ringstad N (2011) Receptor-type guanylate cyclase is required for carbon dioxide sensation by Caenorhabditis elegans. Proc Natl Acad Sci USA 108: 254–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem EA, Sternberg PW (2008) Acute carbon dioxide avoidance in Caenorhabditis elegans. Proc Natl Acad Sci USA 105: 8038–8043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda S, Ishii N, Suzuki K, Matsuo M (1993) Oxygen-dependent perturbation of life span and aging rate in the nematode. J Gerontol 48: B57–B61 [DOI] [PubMed] [Google Scholar]
- Honda Y, Tanaka M, Honda S (2008) Modulation of longevity and diapause by redox regulation mechanisms under the insulin-like signaling control in Caenorhabditis elegans. Exp Gerontol 43: 520–529 [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C (1999) Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362–366 [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366: 461–464 [DOI] [PubMed] [Google Scholar]
- Kenyon CJ (2010) The genetics of ageing. Nature 464: 504–512 [DOI] [PubMed] [Google Scholar]
- Kim K, Li C (2004) Expression and regulation of an FMRFamide-related neuropeptide gene family in Caenorhabditis elegans. J Comp Neurol 475: 540–550 [DOI] [PubMed] [Google Scholar]
- Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G (1997) daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942–946 [DOI] [PubMed] [Google Scholar]
- Lans H, Jansen G (2007) Multiple sensory G proteins in the olfactory, gustatory and nociceptive neurons modulate longevity in Caenorhabditis elegans. Dev Biol 303: 474–482 [DOI] [PubMed] [Google Scholar]
- Lee SJ, Kenyon C (2009) Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr Biol 19: 715–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiser SF, Begun A, Kaeberlein M (2011) HIF-1 modulates longevity and healthspan in a temperature-dependent manner. Aging Cell 10: 318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C (2001) Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet 28: 139–145 [DOI] [PubMed] [Google Scholar]
- Mair W, Dillin A (2008) Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem 77: 727–754 [DOI] [PubMed] [Google Scholar]
- Mehta R, Steinkraus KA, Sutphin GL, Ramos FJ, Shamieh LS, Huh A, Davis C, Chandler-Brown D, Kaeberlein M (2009) Proteasomal regulation of the hypoxic response modulates aging in C. elegans. Science 324: 1196–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DB, Hudson ML, Waters E, O'Shea M (1999) Soluble guanylyl cyclases in Caenorhabditis elegans: NO is not the answer. Curr Biol 9: R546–R547 [DOI] [PubMed] [Google Scholar]
- Muller RU, Fabretti F, Zank S, Burst V, Benzing T, Schermer B (2009) The von Hippel Lindau tumor suppressor limits longevity. J Am Soc Nephrol 20: 2513–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G (1997) The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389: 994–999 [DOI] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, Kim DH (2009) A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323: 382–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Persson A, Cheung B, de Bono M (2006) Behavioral motifs and neural pathways coordinating O2 responses and aggregation in C. elegans. Curr Biol 16: 649–659 [DOI] [PubMed] [Google Scholar]
- Sengupta P, Colbert HA, Bargmann CI (1994) The C. elegans gene odr-7 encodes an olfactory-specific member of the nuclear receptor superfamily. Cell 79: 971–980 [DOI] [PubMed] [Google Scholar]
- Shtonda BB, Avery L (2006) Dietary choice behavior in Caenorhabditis elegans. J Exp Biol 209: 89–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styer KL, Singh V, Macosko E, Steele SE, Bargmann CI, Aballay A (2008) Innate immunity in Caenorhabditis elegans is regulated by neurons expressing NPR-1/GPCR. Science 322: 460–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabish M, Siddiqui ZK, Nishikawa K, Siddiqui SS (1995) Exclusive expression of C. elegans osm-3 kinesin gene in chemosensory neurons open to the external environment. J Mol Biol 247: 377–389 [DOI] [PubMed] [Google Scholar]
- Vaiserman AM (2008) Epigenetic engineering and its possible role in anti-aging intervention. Rejuvenation Res 11: 39–42 [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk JM, Hekimi S (2011) FUdR causes a twofold increase in the lifespan of the mitochondrial mutant gas-1. Mech Ageing Dev 132: 519–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland RA (2009) Is epigenetics an important link between early life events and adult disease? Horm Res 71(Suppl 1): 13–16 [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S (1986) The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci 314: 1–340 [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G (2000) Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science 290: 147–150 [DOI] [PubMed] [Google Scholar]
- Yu S, Avery L, Baude E, Garbers DL (1997) Guanylyl cyclase expression in specific sensory neurons: a new family of chemosensory receptors. Proc Natl Acad Sci USA 94: 3384–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shao Z, Zhai Z, Shen C, Powell-Coffman JA (2009) The HIF-1 hypoxia-inducible factor modulates lifespan in C. elegans. PLoS ONE 4: e6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer M, Gray JM, Pokala N, Chang AJ, Karow DS, Marletta MA, Hudson ML, Morton DB, Chronis N, Bargmann CI (2009) Neurons detect increases and decreases in oxygen levels using distinct guanylate cyclases. Neuron 61: 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.