Abstract
The numerous functions of the important class of molecular chaperones, heat shock proteins 70 (Hsp70), rely on cycles of intricate conformational changes driven by ATP-hydrolysis and regulated by cochaperones and substrates. Here, we used Förster resonance energy transfer to study the conformational dynamics of individual molecules of Ssc1, a mitochondrial Hsp70, in real time. The intrinsic dynamics of the substrate-binding domain of Ssc1 was observed to be uncoupled from the dynamic interactions between substrate- and nucleotide-binding domains. Analysis of the fluctuations in the interdomain separation revealed frequent transitions to a nucleotide-free state. The nucleotide-exchange factor Mge1 did not induce ADP release, as expected, but rather facilitated binding of ATP. These results indicate that the conformational cycle of Ssc1 is more elaborate than previously thought and provide insight into how the Hsp70s can perform a wide variety of functions.
Keywords: chaperones, Hsp70, single molecule, single pair Förster resonance energy transfer
Introduction
Heat shock proteins 70 (Hsp70) are molecular chaperones ubiquitously present in prokaryotes and in almost all subcellular compartments of eukaryotic cells (Bukau et al, 2006; Kampinga and Craig, 2010; Mayer, 2010; Richter et al, 2010; Hartl et al, 2011). They perform a variety of different functions including folding of newly synthesised proteins, prevention of aggregation of unfolded proteins, remodelling of protein complexes and translocation of proteins across cellular membranes. All these different functions depend on the ability of Hsp70 to bind to hydrophobic segments in proteins in an ATP-dependent manner. The binding affinity to the substrate is regulated by the N-terminal nucleotide-binding domain (NBD). The NBD is connected to the substrate-binding domain (SBD) by a flexible linker. The SBD consists of the binding pocket for the substrate that can be covered by an alpha-helical lid. Binding of ATP to the NBD induces conformational changes in the SBD resulting in opening of the lid and thereby reducing the affinity to the substrate. The intrinsic ATP hydrolysis rate is slow but is stimulated by the presence of substrate and J-proteins. ATP hydrolysis results in trapping of the substrate in the binding pocket of the SBD. The conformational cycle is completed by the release of ADP and subsequent ATP rebinding aided by nucleotide-exchange factors.
The major Hsp70 in the mitochondrial matrix of yeast is Ssc1 (Craig et al, 1987; Neupert and Brunner, 2002; Voos and Rottgers, 2002). It is essential for cell viability and serves an important role in a number of different processes. Protein translocation into the mitochondrial matrix is performed by the TOM and TIM23 complexes in the outer and inner mitochondrial membranes, respectively (Koehler, 2004; Chacinska et al, 2009; Mokranjac and Neupert, 2010; Endo et al, 2011). Ssc1 functions as a molecular motor in the TIM23 complex driving the translocation of unfolded proteins across mitochondrial membranes. The membrane-associated protein Tim44 recruits Ssc1 from the mitochondrial matrix to the translocation channel where its function is regulated by the J-protein, Tim14 and the J-like protein, Tim16 (Mokranjac et al, 2006). In the mitochondrial matrix, Ssc1 assists folding and prevents aggregation of imported proteins in conjunction with the J-protein Mdj1 and the nucleotide-exchange factor Mge1 (Kang et al, 1990; Rowley et al, 1994; Horst et al, 1997; Craig et al, 2006). Ssc1 itself requires a specialized chaperone Hep1 for folding and maintenance of the structure and function (Sichting et al, 2005; Blamowska et al, 2012). In higher eukaryotes, changed expression patterns and/or mutations of mitochondrial Hsp70 have recently been implicated in neurodegenerative disorders and cancerogenesis (Deocaris et al, 2009).
We recently developed FRET sensors to investigate the conformational dynamics of Ssc1 on the single molecule level (Mapa et al, 2010). Our data demonstrate that the ATP state of Ssc1 is well defined with two domains docked together and an open SBD. In contrast, the ADP state appears rather heterogeneous, both in respect to domain–domain interaction and the degree of opening of the SBD. We also showed that substrates have an active role in determining the conformation of Ssc1 and are required for full undocking of the two domains and for closure of the SBD. Furthermore, comparison with the major bacterial Hsp70, DnaK, which shares over 50% sequence identity with Ssc1, revealed important differences with respect to the conformations of the ADP-bound state of the two chaperones possibly explaining the specialisation of Ssc1.
These previous experiments were performed on Ssc1 in solution using a spFRET burst analysis with pulsed interleaved excitation (Müller et al, 2005) and multi-parameter detection (Eggeling et al, 2001; Widengren et al, 2006; Kudryavtsev et al, 2012) enabling extraction of quantitative distance distributions from the FRET data on the single molecule level. However, burst analysis experiments only allow a snap shot of the protein dynamics, as the time scale that is measured for each molecule is limited by the passage time of the molecule through the observation volume, which is in the order of 1 ms. This prevents the detection and quantification of dynamic conformational changes with slower timescales. For Ssc1, such dynamics are possibly hidden in the broad distance distributions in the presence of ADP. Indeed, an indication of the presence of conformational dynamics of Ssc1 in the ADP state on the ms timescale or slower came from the analysis of the FRET efficiencies as a function of donor fluorescence lifetime for individual bursts (Mapa et al, 2010). The nature of this dynamics, however, remained unclear.
In the present study, we analysed the conformational dynamics of individual Ssc1 molecules in real time over minutes. To this end, we encapsulated Ssc1 in lipid vesicles, immobilised the vesicles on the surface and monitored the conformational dynamics of Ssc1 by spFRET using total internal reflection fluorescence (TIRF) microscopy. With the single molecule TIRF setup, the kinetics of the dynamic fluctuations can be directly measured on individual molecules. To extract the kinetic information, the single molecule FRET traces were analysed using a hidden Markov modelling approach (McKinney et al, 2006). Data presented here show that the heterogeneities observed previously in the ADP-bound state are due to dynamic fluctuations between different conformations. Intriguingly, the dynamics of the SBD are uncoupled from the dynamics observed between the SBD and NBD. The dynamics of the SBD could be described by a linear three-well model where transitions between the two end states can only occur via the intermediate state. Analysis of the dynamics observed between the domains revealed the presence of a nucleotide-free state. Surprisingly, measurements of the dynamics of Ssc1 in the presence of Mge1 showed that Mge1 did not stimulate release of ADP but rather binding of ATP. We also monitored the cycling of Ssc1 and found that peptide binding occurs to the nucleotide-bound conformation of Ssc1.
Results
Heterogeneity of Ssc1 is due to dynamic conformational changes
We recently developed two FRET-based sensors to follow the conformational dynamics of Ssc1 (Mapa et al, 2010) (Supplementary Figure 1A). The first sensor, named Ssc1(448,590) or lid sensor, reflects the degree of opening of the SBD. One fluorophore is attached to the base of the SBD while a second is bound to the lid. The second sensor, named Ssc1(341,448) or domain sensor, has labels on the SBD and NBD and is sensitive to the interaction between the domains. In this study, we have monitored the FRET efficiency of the same sensors over timescales of 30 ms to minutes to study the conformational dynamics of individual molecules of Ssc1 in real time (Figure 1). The sensors were encapsulated in lipid vesicles (Boukobza et al, 2001; Okumus et al, 2004) to avoid effects from direct immobilisation of the protein (Supplementary Figure 1B). The vesicles, which contained a small fraction of biotinylated lipids, were immobilised using a biotin–avidin–biotin linkage and visualised using TIRF microscopy (Supplementary Figure 1C). Depending on the nucleotide present and the absence or presence of substrate peptide in the vesicles, we observed either static FRET signals over the course of the measurement or dynamic switching between different FRET efficiencies in both FRET sensors. Representative single molecule data are shown in Figure 1.
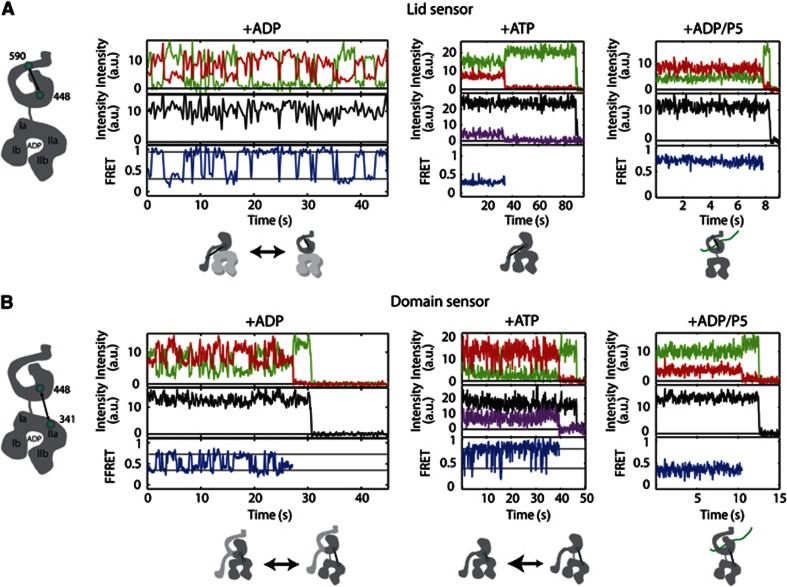
Figure 1.
Representative data from spFRET TIRF experiments. Time traces of the donor and acceptor intensity (upper graphs), total intensity (middle graphs) and FRET efficiency (lower graphs) of the lid sensor (Ssc1(448, 590)) (A) or domain sensor (Ssc1(341, 448)) (B) in the presence of 1 mM ADP (left panels), collected at 250 ms/frame from the lid sensor and 30 ms/frame for the domain sensor, 1 mM ATP (middle panels) at a time resolution of 100 ms/frame, and 1 mM ADP and 5 μM P5 (right panels) at a time resolution of 30 ms/frame. The magenta lines display the total acceptor intensity after direct acceptor excitation (at 633 nm) measured using msALEX (Kapanidis et al, 2005). With the exception of the left panel in (A), all traces show photobleaching of the acceptor molecule, where the acceptor intensity drops to zero along with the concomitant rise in the donor signal, followed by photobleaching of the donor molecule at the end of the trace. Schematic depictions of the labelling positions are shown to the left of each panel and schematic representations of the conformational changes being observed in each spFRET trace are shown below the respective trace.
The lid sensor, Ssc1(448,590), in the presence of ADP showed dynamic switching between a closed conformation (very high FRET) and an open conformation (intermediate FRET) (Figure 1A, left panel). The anti-correlation between the donor (green) and acceptor (red) signals (upper graph) with a constant overall fluorescence intensity (black, middle graph) is a clear signature for FRET.
In the presence of ATP, no dynamics of the lid sensor, Ssc1(448,590), were observable (Figure 1A, middle panel) and the lid was in a wide-open conformation (low FRET). Ssc1 is known to bind substrate peptides with high affinity in the ADP state. Consequently, when we measured Ssc1(448,590) with ADP in the presence of the model substrate peptide P5, we observed static FRET traces with a closed lid (high FRET) (Figure 1A, right panel).
Similar experiments were performed with the Ssc1(341,448) sensor of Ssc1, referred to as the domain sensor. The left panel in Figure 1B shows a typical trace for a Ssc1(341,448) molecule in the presence of ADP collected with 30 ms time resolution. The FRET signal displays switching between an intermediate FRET state (partially undocked) and a high FRET state, referred to as the docked conformation (Figure 1B, left panel). The two distinct conformations interchange on a timescale of ∼1 s. The FRET changes of the lid sensor in the presence of ADP occur on clearly slower timescale suggesting that the SBD and interdomain dynamics are not directly coupled.
For the domain sensor, Ssc1(341,448), in the presence of ATP, we observed cycling of the interdomain distance between a tightly docked conformation (high FRET) and a partially undocked state (intermediate FRET) (Figure 1B, middle panel). The dwell time of the partially undocked conformation is short and transitions to the docked state occurred within a few frames. To exclude photophysical effects of the acceptor as the cause for these fast fluctuations, we performed experiments using millisecond-alternating laser excitation (ALEX) (Kapanidis et al, 2005; Margeat et al, 2006) to directly probe the photophysics of the acceptor molecule. No dynamics in the acceptor signal were observed until the acceptor photobleached, excluding photophysics of the acceptor as cause of the FRET fluctuations. In the presence of substrate, the FRET signal of the domain sensor, Ssc1(341,448), was static over the course of the measurement with a low FRET efficiency indicating that the domains are completely undocked (Figure 1B, right panel).
From inspection of the traces in Figure 1, significant differences in the dynamics of the two sensors are visible in the presence of different nucleotides and with peptide. When the peptide is bound, no dynamics were observed on the timescale analysed for either sensor. In the presence of ATP, the SBD showed no dynamics on the measured timescale while slow fluctuations were clearly observed in the presence of ADP. The domain sensor shows a significantly faster dynamics than the lid sensor in the presence of ADP and even faster transitions in the presence of ATP.
Conformational dynamics of the SBD
To quantitatively analyse the dynamics observable in the spFRET traces, we applied a hidden Markov model (HMM) (Baum and Petrie, 1966; McKinney et al, 2006) approach. From the HMM analysis, the most likely kinetic parameters and FRET efficiencies were determined for a two-state model. There was no improvement in the description of the data when models with more states were applied. From the HMM results, the most likely description of the state of the system at each time point in each trace can be determined, yielding the underlying FRET efficiency of the states, the kinetic rates between them and the so-called Viterbi path (Supplementary Figure 2A). The results of the HMM analysis for the lid sensor, Ssc1(448,590), in the presence of ADP are visualised in a transition density plot showing the number of transitions to a final state depending on the initial configuration of the system (Supplementary Figure 2B). The transitions are clustered into two regions of the plot suggesting the existence of two conformations with distinct FRET efficiencies. The cluster in the upper left corner of the transition probability plot represents transitions from a conformation with an open lid (E≈35%) to a conformation with a closed lid (E≈95%). The other cluster represents transitions in the opposite direction.
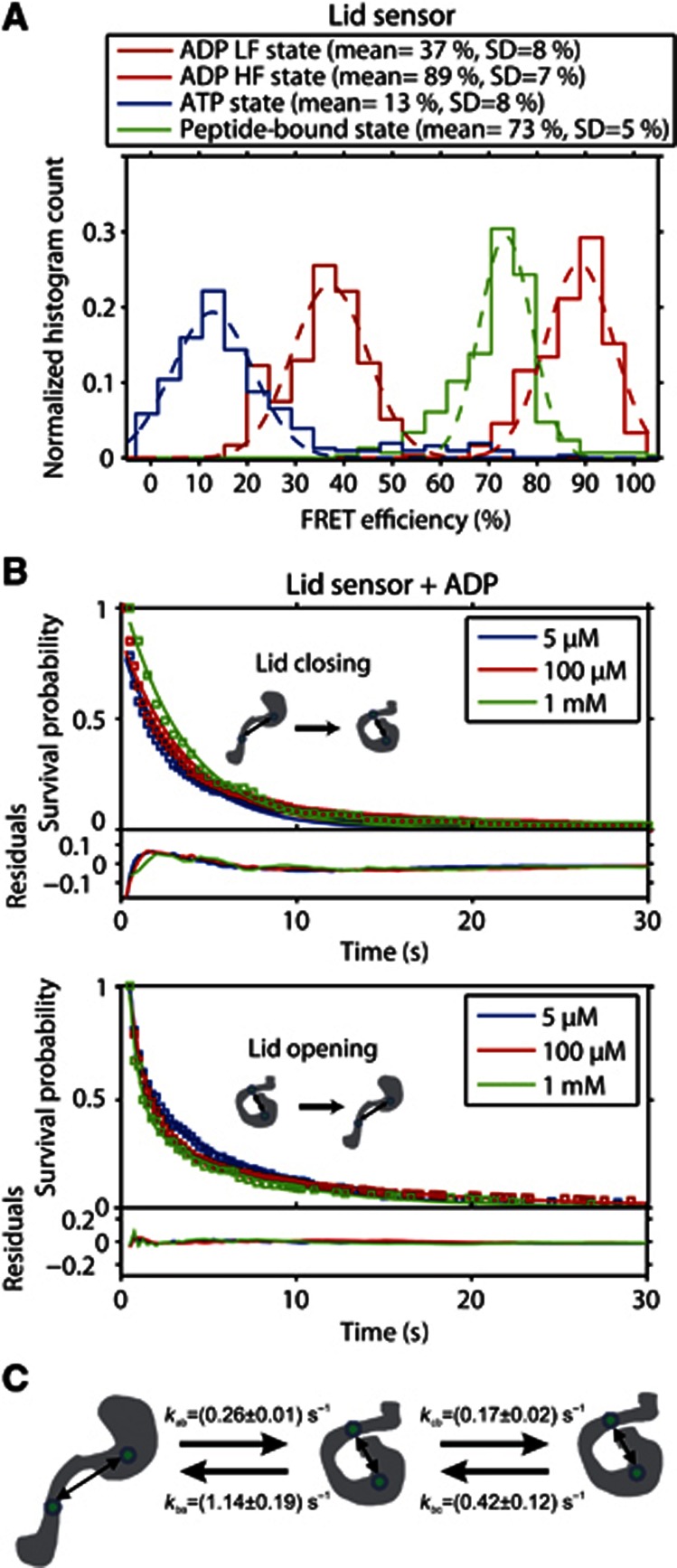
Histograms of the FRET efficiency for the states found by the HMM analysis of the lid sensor in the presence of ADP are shown in Figure 2A together with molecule-wise histograms of the static traces observed in the lid sensor in the presence of ATP and ADP/P5. In the presence of ADP, the SBD cycles between an open conformation with intermediate FRET efficiency (E=37%, d=6.7 nm) and a tightly closed state (E=89%, d=4.3 nm). These conformations are distinct from the conformations observed with bound substrate or in the presence of ATP (Figure 2A). In the presence of ATP, the lid is more widely open (E=13%, d=8.4 nm) and when the substrate is bound, the lid is closed (E=73%, d=5.1 nm). These values are in excellent agreement with the FRET efficiencies we determined previously (Mapa et al, 2010) (Table I). Although the conformation of the lid is closed in the presence of substrate, it does not close as tightly as the lid-closed state when only ADP is present. Obviously, the bound substrate prevents complete closure of the lid. In the experiments with bound substrate, we observed a subpopulation (19% of the molecules) showing identical dynamic behaviour as in the presence of ADP alone (data not shown). We attributed this subpopulation to complexes that did not have a substrate bound. Although the bulk concentration of substrate peptide was high enough to ensure the presence of several substrates in each vesicle, the local concentration inside the vesicles may have been lower due to interaction of the hydrophobic substrate with the lipid bilayer. This subpopulation was excluded from further analysis.
Figure 2.
Dynamics of the substrate-binding domain. (A) FRET efficiency histograms of the two SBD conformations identified by the HMM analysis in the presence of ADP and molecule-wise FRET histograms of the SBD conformation in the presence of ATP (blue) and in the presence of ADP and P5 (green). Single Gaussian fits to the data are shown as dashed lines. The average FRET value and the standard deviation are given in the figure. HF, High FRET; LF, Low FRET. (B) Upper panel. Cumulative dwell-time histogram (open squares) and single-exponential fits (solid lines) for the lid closing transitions. Lower panel. Cumulative dwell-time histogram (open squares) and double-exponential fits (solid lines) for the lid opening transitions. The cumulative dwell-time histograms were calculated from the HMM analysis for the lid sensor, Ssc1(448, 590), in the presence of 1 mM ADP (green), 100 μM ADP (red), and 5 μM ADP (blue). The residuals of the fits are shown in the lower panels of the figures. (C) A linear 3-well model of the dynamics of the SBD of Ssc1 in the ADP state derived from a double-exponential fit of the lid-opening transition and a single-exponential fit of the lid closing transition. The rates are given as average values with the standard error for the measurements at different ADP concentration.
Table 1. FRET efficiencies and distances in Ssc1(341,448) and Ssc1(448,590).
Abbreviations: HF, high FRET state; LF, low FRET state.
The HMM analysis does not only allow the quantification of the FRET efficiencies of the states observed in a single molecule FRET trace but also the determination of the dwell-time distributions for each state. We quantified the rate of the conformational changes as a function of ADP concentration (Figure 2). The dwell-time distributions are shown as cumulative distributions, i.e., the number of molecules remaining in the respective state over time, to avoid reducing the available information by binning the dwell times (Gebhardt et al, 2006).
From the transition density plot in Supplementary Figure 2B, it is clear that the SBD of Ssc1 cycles between at least two different conformations in the presence of ADP. We have used the HMM analysis to construct cumulative dwell-time histograms for the closed lid and open lid states (Figure 2B). In the presence of 1 mM ADP, the average dwell time of the lid closing transition was 4.22 s (Figure 2B, upper graph), and the average dwell time of the lid opening transition was 3.80 s (Figure 2B, lower graph). Both transition rates do not change significantly with ADP concentration. An interesting feature of the observed SBD dynamics is that the dwell-time distribution of the closed lid state does not fit to a single-exponential decay. This suggests that there is more than one closed lid state present in the SBD. The lid sensor may not be sensitive to the transitions between these two closed states due to the labelling position of the FRET pair. Different models were used to fit the data (Supplementary Figure 3). Although a stretched exponential describes the data well and could account for an ensemble of states with a closed lid, the decay time distribution resulting from this fit is extremely broad, ranging over ∼3 orders of magnitude. Alternatively, the data can be fit well to a biexponential model (Figure 2B, lower panel and Supplementary Figure 3). Hence, we assumed a linear model with three states, two of which are indistinguishable by the FRET efficiency of the lid sensor, consistent with the presence of three different conformations proposed for the SBD of DnaK in the presence of ADP (Schlecht et al, 2011). After lid closure, the SBD can either switch back to an open lid conformation, giving rise to the fast component of the dwell-time decay, or fluctuate to the other closed lid conformation. In the latter case, it will take longer to return to the open lid conformation, giving rise to the slower component of the kinetics. The double-exponential fit was used to extract the rates kba, kbc, and kcb in the model in Figure 2C. The rate from the open to the closed lid state kab was determined from a mono-exponential fit to the dwell-time histograms in Figure 2B (upper panel). The rates indicate that ∼83% of the proteins are in the open lid state, and the closed lid states are populated by ∼10% for the intermediate state in Figure 2C and ∼8% for the right state in Figure 2C. This agrees well with previous spFRET experiments in solution where 90% of the molecules were observed in an open lid conformation (Mapa et al, 2010).
Dynamics of the domain–domain interactions
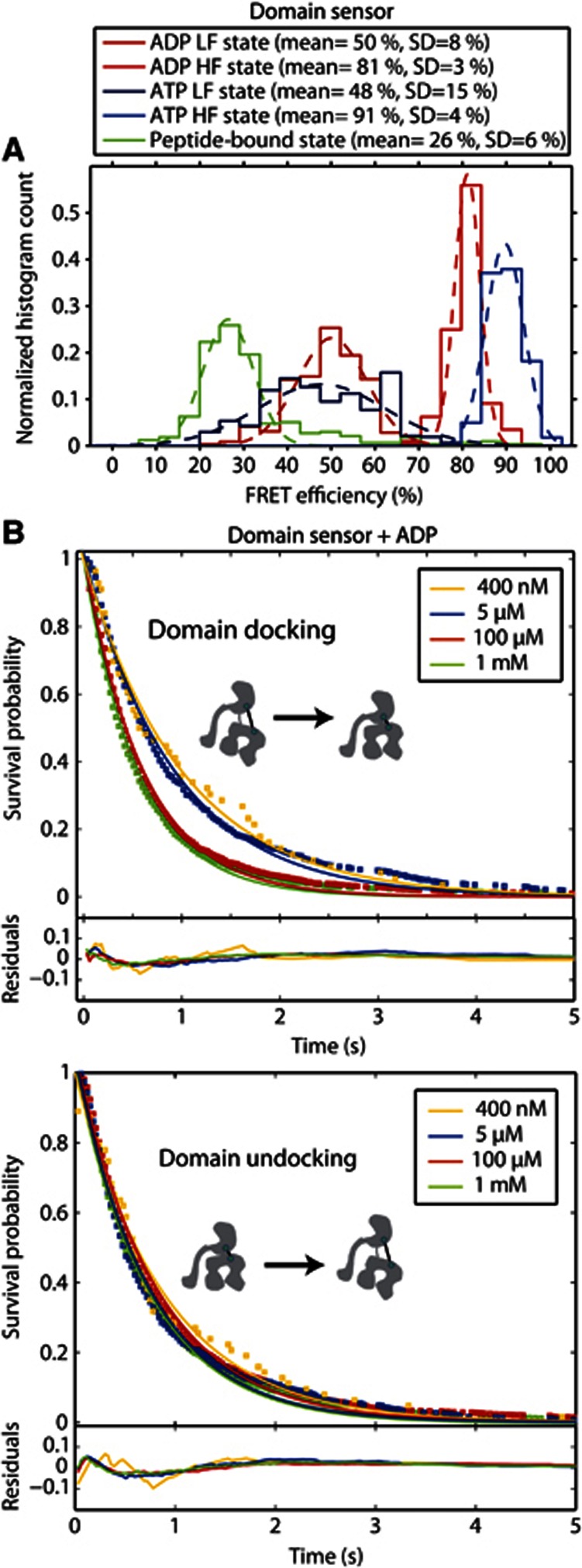
The HMM analysis of the domain sensor in the presence of ADP or ATP also showed the existence of two distinct states (Supplementary Figure 2C-D), a docked conformation with high FRET efficiency (E=81%, d=4.8 nm for ADP and E=91%, d=4.2 nm for ATP) and an intermediate conformation (E=50%, d=6.1 nm for ADP and E=48%, d=6.2 nm for ATP). The docked conformation in the presence of ADP is not as tightly docked as in the presence of ATP. The distribution of FRET efficiencies in the different conformations of the domain sensor in the presence of ADP and ATP calculated by the HMM analysis are compared to the molecule-wise FRET efficiencies of substrate-bound Ssc1 in Figure 3A. From single molecule burst analysis with Ssc1 in the presence of substrate, we have shown the Ssc1 adapts a well-defined undocked conformation with a narrow distance distribution (Mapa et al, 2010). The spFRET TIRF assay using the encapsulated Ssc1 domain sensor also showed an undocked conformation when substrate was bound (E=27%, d=7.2 nm, Figure 3A). The excellent correspondence between the results determined using burst analysis and from TIRF experiments (Table I) strongly indicate there are no artifacts introduced by possible interactions of Ssc1 with the lipids or other encapsulation-related effects.
Figure 3.
Dynamics of the interdomain separation. (A) FRET efficiency histograms of the two interdomain conformations identified by the HMM analysis in the presence of ADP (dark/light red) and ATP (dark/light blue), together with the molecule-wise FRET histogram of the domain sensor in the presence of ADP and P5 (green). Single Gaussian fits to the data are shown as dashed lines. The average FRET value and the standard deviation of the fits are given in the figure legend. HF, High FRET; LF, Low FRET. (B) Cumulative dwell-time histograms (open squares) and mono-exponential fits (solid lines) of the domain-docking transitions (upper panel) and the domain-undocking transitions (lower panel) calculated from the HMM analysis for domain sensor, Ssc1(341, 448), in the presence of 1 mM ADP (green), 100 μM ADP (red), 5 μM ADP (blue) and 400 nM ADP (gold). The residuals of the fits with respective color-coding are shown in the lower graphs.
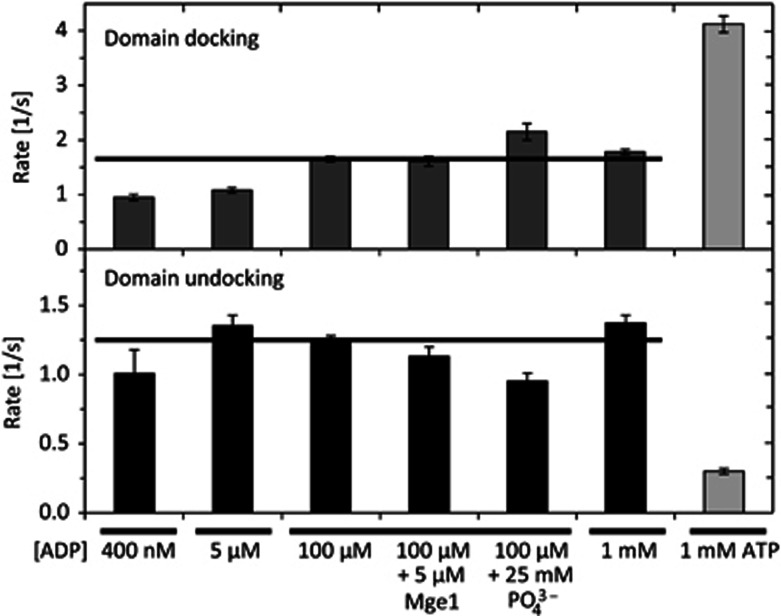
For the domain sensor, the two conformations observed in the presence of ADP as well as the two conformations observed in the presence of ATP are clearly different from the substrate-bound conformation. While the respective high FRET populations have clearly different mean FRET efficiencies, the intermediate FRET subpopulations observed in the presence of ATP and ADP have surprisingly similar average FRET values. The greater width of the intermediate-FRET state observed in the presence of ATP can be attributed to the increased statistical uncertainty of the analysis because of the low number of frames the molecule spends in this conformation. This suggests that the intermediate FRET state observed both in the presence of ADP and ATP may be the same conformation, e.g., a nucleotide-free conformation, and that Ssc1 switches between this conformation and a nucleotide-dependent conformation. To test this hypothesis, the kinetics of the domain sensor were measured as a function of ADP concentration. The cumulative dwell-time distributions and corresponding fits are displayed in Figure 3B. The kinetic rates were a factor of four faster for the interdomain dynamics compared to the intra-SBD dynamics, as qualitatively observed above. In addition, the rate of domain docking was dependent on the ADP concentration, increasing by almost a factor of 2 from 0.95 s−1 at 400 nM ADP to 1.78 s−1 at 1 mM ADP (Figure 4, upper panel). At the same time, the rate of domain undocking was 1.35 s−1 in the presence of 1 mM ADP and was independent of ADP concentrations down to 400 nM (Figure 4, lower panel). These data strongly support the interpretation of the intermediate FRET state as a nucleotide-free conformation and the conformational change is induced by binding and release of nucleotide.
Figure 4.
Rates of domain docking and undocking. Rates of the domain-docking and undocking transitions determined from the HMM analysis of the domain sensor under various conditions. The error bars are the standard error of the mean from single-exponential fits of three randomly chosen subsets of the transitions with the exception of the error bar for 400 nM, which was determined from two independent experiments. The horizontal lines were added as a guide to the eye.
To further test this hypothesis, we investigated the rates of domain docking and undocking in the presence of 25 mM phosphate and 100 μM ADP (Figure 4, Supplementary Figure 4 and Supplementary Table I). The affinity of Hsp70s for ADP has been shown to increase in the presence of phosphate in solution (Russell et al, 1998; Brehmer et al, 2001; Arakawa et al, 2011). We observed a decrease in the rate of domain undocking in the presence of 100 μM ADP with 25 mM phosphate by 24% from 1.25 s−1 to 0.95 s−1. Interestingly, also the rate of domain docking increased by 30% from 1.65 s−1 to 2.15 s−1. Both rate changes agree well with the increased affinity of Ssc1 for ADP assuming that the observed conformational change corresponds to binding and release of ADP.
Influence of Mge1 on the interdomain dynamics
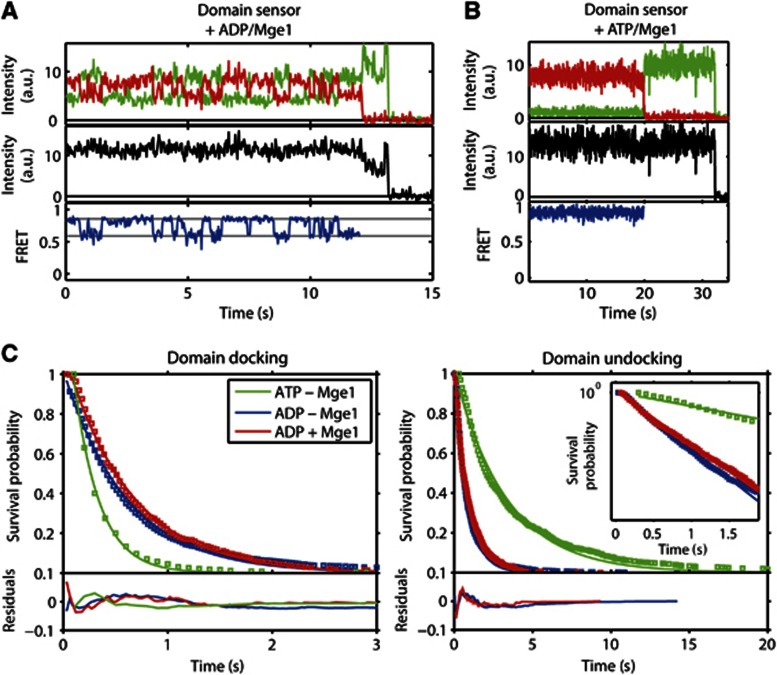
The nucleotide-exchange factor Mge1 was proposed to mediate release of ADP from Ssc1 (Dekker and Pfanner, 1997; Miao et al, 1997) and thus likely stabilise its nucleotide-free conformation. Having identified a conformation that we speculate to be a nucleotide-free state, we used our single-molecule assay to directly observe the influence of Mge1 on the rates of ADP and ATP binding and release. We expected that Mge1 would stimulate the release of ADP and thus shift the equilibrium towards the nucleotide-free conformation of Ssc1. However, in the presence of 100 μM ADP and 5 μM Mge1, transitions between the partially undocked conformation and docked conformation were still visible (Figure 5A). In contrast, a constant FRET signal with high FRET efficiency is observed for the domain sensor in the presence of 5 μM Mge1 and 1 mM ATP (Figure 5B), indicating tightly docked domains and suggesting that Ssc1 is in the nucleotide-bound conformation. This is in sharp contrast to what was measured in the absence of Mge1 where rapid transitions between a partially undocked (putative nucleotide-free) conformation and docked (nucleotide-bound) conformation were observed. This suggests that Mge1 stimulates ATP binding rather than ADP release from Ssc1. Figure 5C shows the cumulative dwell-time histograms for the domain docking and undocking transitions for the domain sensor, Ssc1(341,448), with 100 μM ADP in the presence and absence of Mge1. The transition rates for 1 mM ATP in the absence of Mge1 is shown for comparison. The rates derived from single-exponential fits to the histograms are shown in Figure 4 and given in Supplementary Table I. Mge1 had no effect on the rate of ADP binding (domain-docking transition) and only a very minor decrease of the ADP release rate (domain-undocking transition) was observed.
Figure 5.
Influence of the nucleotide-exchange factor Mge1. (A, B) Representative time traces of the donor (green) and acceptor (red) fluorescence intensity (upper panel), total fluorescence intensity (black, middle panel) and FRET efficiency (blue, lower panel) of the domain sensor, Ssc1(341, 448), in the presence of (A) 1 mM ADP and 5 μM Mge1 and (B) 1 mM ATP and 5 μM Mge1. (C) Cumulative dwell-time histograms (open squares) and single-exponential fits (solid lines) of the domain-docking transitions (left) and the domain-undocking transitions (right) calculated from the HMM analysis for the domain sensor, Ssc1(341, 448), in the presence of 100 μM ADP (blue), 100 μM ADP and 5 μM Mge1 (red), and 1 mM ATP (green). The residuals of the fits are shown in the lower panels of the figures. The inset in the right figure shows an expansion of the first seconds of the survival probability with a logarithmic y-axis.
Cycling of Ssc1 in the presence of ATP and substrate
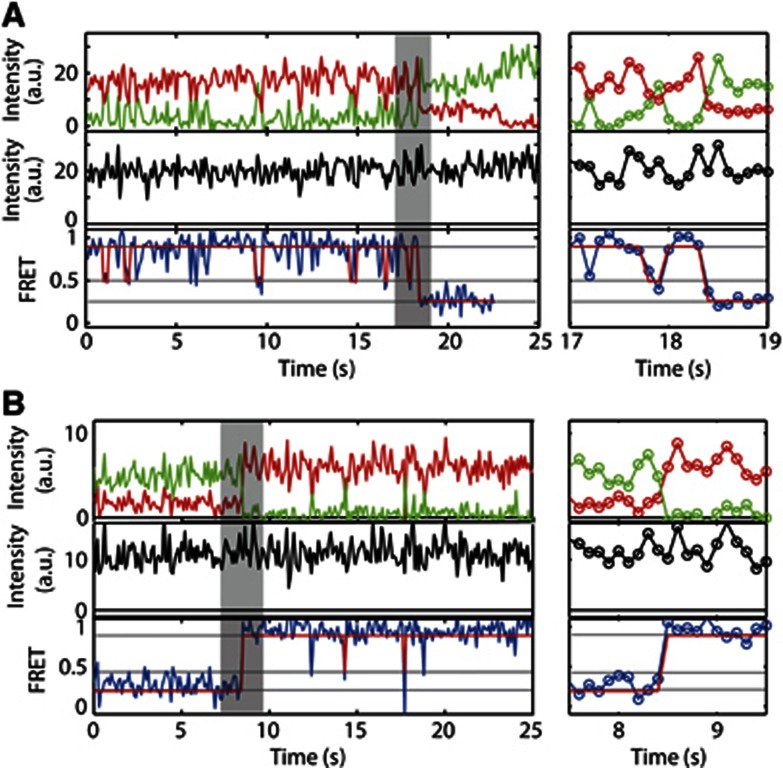
The experiments presented thus far describe the conformational dynamics of Ssc1 in specific phases of its conformational cycle. Having characterised the different conformations, we can use spFRET to investigate the kinetics of the complete chaperone cycle of Ssc1 in the presence of ATP and substrate. The domain sensor, Ssc1(341,448), was encapsulated in vesicles with 5 mM ATP and 0.5 mM P5. We observed dynamics of the interdomain distance switching between the nucleotide-bound conformation (E=91%) and the nucleotide-free conformation (E=50%), similar to what was observed for ATP only (Figure 6). We do not distinguish between the ATP- and ADP-bound conformation, since the FRET efficiencies of the two states are very similar in this construct. In addition to the nucleotide binding and release, transitions to and from the peptide bound state (E≈30%) were observed. The substrate-bound state had a significantly longer dwell time than the putative nucleotide-free state, as expected from the constant FRET signal observed when substrate was bound.
Figure 6.
Cycling of Ssc1(341, 448) in the presence of ATP and substrate. (A, B) Representative traces of the domain sensor, Ssc1(341, 448), in the presence of 5 mM ATP and 0.5 mM P5 peptide substrate where the donor (green) and acceptor (red) fluorescence intensities (upper panel), total fluorescence intensity (black, middle panel) and FRET efficiency (blue, bottom panel) are shown for a transition (A) from a peptide-free state to the peptide-bound state and (B) from the peptide bound-state to the peptide-free state. The Viterbi path from the HMM analysis is shown in red. The graphic on the right is an expanded plot of the transition region highlighted in the left plot in grey.
In all substrate-binding transitions observed, Ssc1 switched directly from a nucleotide-bound conformation to the substrate-bound conformation without populating the putative nucleotide-free state (Figure 6A). One possible explanation could be that, due to the short dwell time of the intermediate FRET state, the transitions through the putative nucleotide-free conformation to the substrate-bound conformation occur too quickly to be detected by the HMM analysis. However, for the measured dwell time of 240 ms in the intermediate FRET state and the number of transitions observed to the substrate-bound conformation (40), we rule out that a transition through the putative nucleotide-free state is necessary for substrate binding. Otherwise, we would have observed it in at least some of the transitions. Similarly, only transitions from the peptide-bound conformation to the nucleotide-bound state were observed (Figure 6B).
Discussion
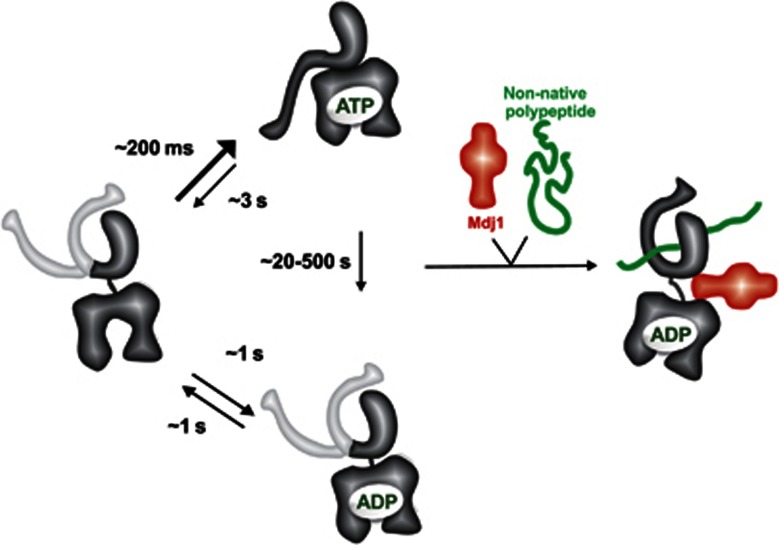
In this study, we explored the conformational dynamics of individual molecules of Ssc1, the mitochondrial member of the Hsp70 family of molecular chaperones, in real time using spFRET measurements on a TIRF microscope. Our data show that the conformational cycle of Ssc1 is even more intriguing than previously thought. The heterogeneities we observed previously in spFRET burst analysis measurements performed in the presence of ADP (Mapa et al, 2010) are now identified to be due to dynamic switching between different conformations shown schematically in Figure 7. In the presence of substrate, both the lid conformation and interaction of the domains were static on the timescale of the measurement (∼1 minute). In the absence of substrate, we observed dynamical switching between different conformational states for both the domain and lid sensors. Interestingly, the dynamics of the SBD of Ssc1 were slower than what was observed for the interdomain interactions. Also, in contrast to the interdomain dynamics, the SBD dynamics were not dependent on ADP concentration. Several studies have found evidence for allosteric communication between the SBD and the NBD in other proteins of the Hsp70 family (Marcinowski et al, 2011; Zhuravleva and Gierasch, 2011). Our data support a similar picture for Ssc1 where the protein undergoes a conformational transition to a static structure (Figure 1) at high ATP concentrations or in the presence of a substrate. Consistent with what has been observed for DnaK (Zhuravleva and Gierasch, 2011), the two domains of Ssc1 behave independently in the presence of ADP and the absence of a substrate.
Figure 7.
Conformational cycle of Ssc1. A schematic diagram focusing on the different conformational states of Ssc1 observed during the conformational cycle of Ssc1. Ssc1 fluctuates between nucleotide-bound and nucleotide-free states. Interactions of Ssc1 with the substrate occur from the nucleotide-bound conformation. When the substrate is bound, no dynamics are observed, neither for the interdomain sensor nor the lid sensor. Upon dissociation of the substrate, Ssc1 returns to the nucleotide-bound conformation and the dynamics resume.
The SBD of Ssc1 adopted a well-defined, non-fluctuating, open conformation in the presence of ATP. In the presence of peptide substrate, the SBD closed to another well-defined and non-fluctuating conformation. Measurements performed in the presence of ADP showed interesting dynamic behaviour, and we identified at least three different conformational states of the SBD. Molecules spent the majority of time (83%) in an open lid conformation (though not as widely open as in the presence of ATP). However, occasionally the lid would close. The SBD dynamics were non-exponential, suggesting that at least three different conformational states of the SBD may coexist in the presence of ADP. This is consistent with recent experiments performed on DnaK where Schlecht et al showed that the SBD can adopt at least three different conformations: closed, open and intermediate open (Schlecht et al, 2011). Similar conformational flexibility has also recently been shown for the SBD of BiP, the Hsp70 chaperone in the endoplasmic reticulum (Marcinowski et al, 2011). The data presented here also show that the helical lid of SBD of Ssc1 closes on peptide substrate but that, in the absence of any substrate, its SBD can adopt an even more closed conformation. The relative distance between the ß-subdomain and the helical lid will thus probably depend on the nature and the conformation of a particular substrate bound to the SBD. We speculate that the lid of the SBD of Hsp70 chaperones closes as far as its bound substrate permits, a mechanism which would allow tight binding of a broad spectrum of substrates ranging from the completely unfolded proteins during transport into organelles to the large protein aggregates during propagation of prions.
Analyses of the interdomain interactions revealed dynamic behaviour both in the presence of ADP as well as in the presence of ATP. Determination of FRET efficiencies by an HMM analysis revealed that a 50% FRET state is observed in the presence of both ADP and ATP. We interpret this 50% FRET state to be a nucleotide-free state of Ssc1. This notion is supported by the following observations: this state was populated in the presence of either ADP or ATP, the rate of domain docking depended on the ADP concentration, the rate of the ADP binding and release changed in the presence of phosphate, and no dynamics were observable in the presence of ATP and the nucleotide-exchange factor Mge1. Interestingly, the putative nucleotide-free state of Ssc1 was rarely populated in the presence of ATP, but became significantly populated in the presence of ADP. This suggests that the affinity of Ssc1 for the two nucleotides is very different. This finding has very interesting implications for the function of Ssc1. We have recently shown that Ssc1 has a tendency to aggregate and speculated that it is the nucleotide-free state of the chaperone that is prone to aggregation (Sichting et al, 2005; Blamowska et al, 2010), though the presence of this state was not directly shown until now. Similar observations were subsequently made with mammalian mitochondrial Hsp70 (Zhai et al, 2008) and Hsp70 from chloroplasts (Willmund et al, 2008). Furthermore, recent molecular dynamics simulations of bovine Hsp70 (Woo et al, 2009) revealed that the nucleotide-free form of its NBD exhibits a large degree of flexibility, far larger than in the presence of either ATP or ADP. In the case of mitochondrial Hsp70s, it is also the NBD that is aggregation-prone (Zhai et al, 2008; Blamowska et al, 2010). Though the reason for this high aggregation propensity of mitochondrial Hsp70s remains unclear, the data presented here suggest that the aggregation-prone conformation of Ssc1 emerges due to the spontaneous release of ADP in the absence of bound substrate. Under normal conditions, the lifetime of the ADP-bound conformation and thereby of the nucleotide-free state of Ssc1 will likely be limited by the presence of nucleotide-exchange factors and the high concentration of ATP in mitochondria. However, the ADP-bound state and thereby the nucleotide-free state could be significantly populated when ATP levels in mitochondria drop. Another possibility for populating the ADP-bound and nucleotide-free states in mitochondria may come from the potential idle cycling of Ssc1 in the absence of a translocating chain at the TIM23 complex (Mokranjac et al, 2003; Mayer, 2004; Pais et al, 2011). Interestingly, we have recently identified a specialized chaperone in mitochondria, Hep1, which helps maintain Ssc1 in a soluble and functional form (Sichting et al, 2005). Intriguingly, all Hep1 proteins analysed so far bind to their respective Hsp70 chaperones only upon depletion of ATP (Sichting et al, 2005; Willmund et al, 2008; Zhai et al, 2008). The mere fact that Hep1 is conserved in all eukaryotic species analysed strongly suggests that, during the evolution of mitochondria from its prokaryotic ancestors, it was easier to develop and maintain a specialized protein to stabilise this apparently very important conformation of mitochondrial Hsp70 rather than to make mitochondrial Hsp70 itself more stable.
We investigated the effect of the nucleotide-exchange factor Mge1 on the dynamics of Ssc1. We expected that Mge1 would stimulate the release of ADP (Dekker and Pfanner, 1997; Miao et al, 1997) and thus likely stabilise the nucleotide-free form of Ssc1. Surprisingly, we observed no effect of Mge1 on the dynamics of Ssc1 in the presence of ADP. Rather, Mge1 abolished the interdomain dynamics in the presence of ATP. These data suggest that Mge1 does not perform nucleotide exchange on Ssc1 by mediating release of ADP but rather by stimulating ATP binding. In principle, the absence of dynamics could also be explained by stabilisation of the ATP-bound state through Mge1, but this possibility can be ruled out by the fact that Mge1 dissociates from Ssc1 within milliseconds upon ATP binding (Mapa et al, 2010). Thus, the dwell times of the nucleotide-free state must be significantly shorter in the presence of Mge1, a least shorter than 4 ms to not be detectable in our experiments. The exact mechanism for how Mge1 facilitates this >60-fold increase in the ATP binding rate is not yet known. It could be speculated that Mge1 increases the rate of ATP binding by inducing a conformational change in the NBD that specifically facilitates binding of ATP but, interestingly, not of ADP. This additionally points to an important role of Mge1 in inducing a conformation conducive to binding of the terminal phosphate group.
The detailed spFRET study of the conformational dynamics of Ssc1 under different conditions allowed us to observe the complete chaperoning cycle of Ssc1 in the presence of both ATP and substrate. We observed that Ssc1 undergoes multiple nucleotide release and re-binding cycles before the stable binding of the substrate occurs (Figure 7). Binding of the substrate was observed to occur directly from a nucleotide-bound state and substrate release to lead directly back to the ATP-bound state. Hence, the conformational cycle of Ssc1 is more elaborate than previously thought. It remains to be determined whether other members of the Hsp70 family of chaperones have similar nucleotide-free conformations that are significantly populated. However, the presence of this additional state, as well as the potential presence of cochaperones that specifically recognise it, offers a possibility for an additional level of regulation of Hsp70 chaperones and thus provides a clue as to the mechanism via which Hsp70 s are capable of performing such a large variety of functions.
Materials and methods
Protein expression, purification and labelling
The FRET-based sensors Ssc1(341, 448) and Ssc1(448, 590) were prepared as previously described (Mapa et al, 2010) and are only mentioned here briefly. The amino acids D341, I448 and D590 were mutated to cysteines using the QuikChange mutagenesis protocol (Stratagene; Cedar Creek, TX) in the plasmid pETDuet1-Hep1-Ssc1 (Sichting et al, 2005). Cysteine mutants of Ssc1 were expressed in BL21(DE3) ΔdnaK::52 cells (a kind gift from Dr M Mayer, University of Heidelberg) and purified on a His6-Mge1 column using a modified protocol for purification of Ssc1 from yeast mitochondria (Weiss et al, 2002). Cysteine variants were labelled stoichiometrically with Atto532 maleimide (donor, Atto-Tech GmbH, Siegen, Germany) and Atto647N maleimide (acceptor, Atto-Tech GmbH) as described in (Mapa et al, 2010).
Vesicle encapsulation
For the observation of single molecules over timescales of 30 ms to minutes by TIRF microscopy, Ssc1 was immobilised at the surface of the sample cell. To avoid artifacts from the immobilisation, we have used vesicle encapsulation of Ssc1 and subsequent immobilisation of the vesicles (Okumus et al, 2004). Lipid films containing 300 μg 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Polar Lipids, Alabaster, USA) and 6 μg biotinylated lipid (1,2-dipalmitodyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl) (sodium salt)) were prepared by mixing dissolved lipid in chloroform, evaporation of the chloroform under a stream of nitrogen and subsequent removal of residual chloroform in vacuum. The lipid films were then hydrated with buffer containing 400 nM Ssc1 together with different concentrations of ADP, ATP and/or substrate peptide P5 (Metabion, Munich, Germany) for 30 min at 4 °C. This procedure results in the formation of multilamellar vesicles (MLVs). The MLVs were then extruded 31 times through polycarbonate membranes with a pore diameter of 200 nm using an extruder (Liposofast Basic, Avestin Europe GmbH, Mannheim, Germany). This procedure breaks up the MLVs into large unilamellar vesicles with a homogenous size distribution of 200 nm diameter. The vesicles were separated from free protein either by size-exclusion FPLC or by an Amicon spin column with a molecular weight cut-off of 100 kDa. After purification, the vesicles were immobilised on a quartz prism covered with a PEG/3% biotin-PEG layer using a biotin–streptavidin–biotin linkage to the biotinylated lipids. The prism was mounted on an inverted microscope (Nikon TE2000-U) for TIRF microscopy.
The number of encapsulated proteins per vesicle is described by a Poisson distribution. The Ssc1 concentration of 400 nM was chosen to maximise the number of vesicles encapsulating exactly one protein. The number of proteins per vesicle was quantified by counting the number of bleaching steps. All intensity traces showing more than one distinct bleaching step were discarded from the analysis.
Single molecule TIRF microscopy
Single molecule TIRF microscopy was performed on a home-build setup described previously (Schluesche et al, 2007). Fluorescence was excited by the evanescent field of a totally reflected laser beam from a 532 nm solid-state laser (GCL-100-L, CrystaLaser, Reno, USA) or a 633 nm HeNe laser (Laser 2000 Wessling, Germany) for ms ALEX measurements. Fluorescence from single molecules was collected by an objective (CFI Plan Apochromat 60x WI, NA 1.2, Nikon, Germany), separated according to the spectral range of donor and acceptor by a dichroic mirror and additional emission filters and imaged on different regions of an EMCCD camera (iXon+, Andor Technology). After mapping of donor and acceptor channels and identification of single molecules, time traces of the intensity of donor and acceptor fluorescence were extracted from the resulting movies using MATLAB software developed in our lab.
As molecules photobleach during single molecule experiments, one needs to choose what timescales to investigate. Initial movies were recorded at different excitation powers and data collection rates to gain insight into whether dynamics are present and, if so, on what timescale they occur. The excitation intensity and data collection rates were then varied to optimally study the dynamics.
Determination of underlying FRET efficiency states and transitions by Hidden Markov Modelling
For the statistical analysis of FRET time traces from many molecules, we have used HMM to recover the state sequence for each molecule. HMM has been developed in the 1960s (Baum and Petrie, 1966) to resolve signals overlaid by noise, and was applied to speech recognition problems for several decades (Rabiner and Juang, 1986). A typical example for a biophysical application of HMM is modelling of a time trace of the current through a single ion channel switching between an open and a closed state (Qin et al, 2000). Analysis of single molecule fluorescence data using HMM has been studied theoretically (Andrec et al, 2003), and Ha and coworkers have applied HMM to extract the number of states, their FRET efficiencies and rates from spFRET time traces (McKinney et al, 2006).
The number of FRET efficiency states present in the sample can either be determined by over-fitting with a high number of states and a cluster analysis (McKinney et al, 2006) or by using Bayesian maximum evidence (Bronson et al, 2009). For the data of Ssc1(341,448) in the presence of ADP or ATP and for Ssc1(448,590) in the presence of ADP, two states were sufficient to model the data and additional states did not significantly improve the likelihood of the model. The end points of all idealised transitions found by the HMM algorithm (Supplementary Figure 2A) are plotted over the transition start points in transition probability plots (Supplementary Figure 2B-D).
HMM was used to extract FRET efficiencies and rates from dynamic traces (McKinney et al, 2006). It was performed using a software developed in our lab employing the HMM MATLAB toolbox developed by Kevin Murphy. Each trace is modelled using an individual parameter set comprised of two FRET efficiencies and two transition rates to account for molecule-to-molecule variations of the conformational states. The transitions detected by the HMM analysis were plotted in transition probability plots and clusters of transitions in these plots were selected for dwell-time analysis. All dwell-time histograms contained at least 500 transitions from at least 100 molecules. Cumulative dwell-time distributions were fitted by least-squares fitting using MATLAB.
Supplementary Material
Acknowledgments
We wish to thank P Robisch and M Malesic for expert technical assistance, Gregor Heiss for his assistance with the HMM analysis, Barbara Treutlein for the advice on vesicle encapsulation and Walter Neupert and Andreas Ladurner for support. We gratefully acknowledge the financial support of the Deutsche Forschungsgemeinschaft (SFB 1035 to DCL, MO 1944/1-1 and SFB594 to DM), the German-Israeli Foundation, the Ludwig-Maximilians-University Munich (LMUInnovativ BioImaging Network), the Center for NanoScience (CeNS) and Nanosystems Initiative Munich (NIM).
Author contributions: MS and LV performed the experiments and analysed the data. KM purified and labelled the protein. DM and DCL guided the project. MS and LV prepared the figures. MS, DM and DCL designed the experiments, discussed the results and wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Andrec M, Levy RM, Talaga DS (2003) Direct determination of kinetic rates from single-molecule photon arrival trajectories using hidden Markov models. J Phys Chem A 107: 7454–7464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa A, Handa N, Shirouzu M, Yokoyama S (2011) Biochemical and structural studies on the high affinity of Hsp70 for ADP. Protein Sci 20: 1367–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum LE, Petrie T (1966) Statistical inference for probabilistic functions of finite state Markov chains. Ann Math Stat 37: 1554–1563 [Google Scholar]
- Blamowska M, Neupert W, Hell K (2012) Biogenesis of the mitochondrial Hsp70 chaperone. J Cell Biol 199: 125–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blamowska M, Sichting M, Mapa K, Mokranjac D, Neupert W, Hell K (2010) ATPase domain and interdomain linker play a key role in aggregation of mitochondrial Hsp70 chaperone Ssc1. J Biol Chem 285: 4423–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukobza E, Sonnenfeld A, Haran G (2001) Immobilization in surface-tethered lipid vesicles as a new tool for single biomolecule spectroscopy. J Phys Chem B 105: 12165–12170 [Google Scholar]
- Brehmer D, Rüdiger S, Gässler CS, Klostermeier D, Packschies L, Reinstein J, Mayer MP, Bukau B (2001) Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat Struct Biol 8: 427–432 [DOI] [PubMed] [Google Scholar]
- Bronson JE, Fei J, Hofman JM, Gonzalez RL Jr, Wiggins CH (2009) Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys J 97: 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, Weissman J, Horwich A (2006) Molecular chaperones and protein quality control. Cell 125: 443–451 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N (2009) Importing mitochondrial proteins: machineries and mechanisms. Cell 138: 628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig E, Huang P, Aron R, Andrew A (2006) The diverse roles of J-proteins, the obligate Hsp70 co-chaperone. Rev Physiol, Biochem Pharmacol 156: 1–21 [DOI] [PubMed] [Google Scholar]
- Craig EA, Kramer J, Kosic-Smithers J (1987) SSC1, a member of the 70-kDa heat shock protein multigene family of Saccharomyces cerevisiae, is essential for growth. Proc Natl Acad Sci USA 84: 4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker PJ, Pfanner N (1997) Role of mitochondrial GrpE and phosphate in the ATPase cycle of matrix Hsp70. J Mol Biol 270: 321–327 [DOI] [PubMed] [Google Scholar]
- Deocaris CC, Kaul SC, Wadhwa R (2009) The versatile stress protein mortalin as a chaperone therapeutic agent. Protein Pept Lett 16: 517–529 [DOI] [PubMed] [Google Scholar]
- Eggeling C, Berger S, Brand L, Fries JR, Schaffer J, Volkmer A, Seidel CA (2001) Data registration and selective single-molecule analysis using multi-parameter fluorescence detection. J Biotechnol 86: 163–180 [DOI] [PubMed] [Google Scholar]
- Endo T, Yamano K, Kawano S (2011) Structural insight into the mitochondrial protein import system. Biochim Biophys Acta Biomembr 1808: 955–970 [DOI] [PubMed] [Google Scholar]
- Gebhardt JCM, Clemen AEM, Jaud J, Rief M (2006) Myosin-V is a mechanical ratchet. Proc Natl Acad Sci USA 103: 8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475: 324–332 [DOI] [PubMed] [Google Scholar]
- Horst M, Oppliger W, Rospert S, Schonfeld HJ, Schatz G, Azem A (1997) Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J 16: 1842–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Bio 11: 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang PJ, Ostermann J, Shilling J, Neupert W, Craig EA, Pfanner N (1990) Requirement for hsp70 in the mitochondrial matrix for translocation and folding of precursor proteins. Nature 348: 137–143 [DOI] [PubMed] [Google Scholar]
- Kapanidis AN, Laurence TA, Lee NK, Margeat E, Kong X, Weiss S (2005) Alternating laser excitation of single molecules. Acc Chem Res 38: 523–533 [DOI] [PubMed] [Google Scholar]
- Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20: 309–335 [DOI] [PubMed] [Google Scholar]
- Kudryavtsev V, Sikor M, Kalinin S, Mokranjac D, Seidel CA, Lamb DC (2012) Combining MFD and PIE for accurate single-pair forster resonance energy transfer measurements. Chemphyschem 13: 1060–1078 [DOI] [PubMed] [Google Scholar]
- Mapa K, Sikor M, Kudryavtsev V, Waegemann K, Kalinin S, Seidel CAM, Neupert W, Lamb DC, Mokranjac D (2010) The conformational dynamics of the mitochondrial hsp70 chaperone. Mol Cell 38: 89–100 [DOI] [PubMed] [Google Scholar]
- Marcinowski M, Höller M, Feige MJ, Baerend D, Lamb DC, Buchner J (2011) Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat Struct Mol Biol 18: 150–158 [DOI] [PubMed] [Google Scholar]
- Margeat E, Kapanidis AN, Tinnefeld P, Wang Y, Mukhopadhyay J, Ebright RH, Weiss S (2006) Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys J 90: 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP (2004) Timing the catch. Nat Struct Mol Biol 11: 6–8 [DOI] [PubMed] [Google Scholar]
- Mayer MP (2010) Gymnastics of molecular chaperones. Mol Cell 39: 321–331 [DOI] [PubMed] [Google Scholar]
- McKinney SA, Joo C, Ha T (2006) Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J 91: 1941–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Davis JE, Craig EA (1997) Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae1. J Mol Biol 265: 541–552 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M (2006) Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J 25: 4675–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokranjac D, Neupert W (2010) The many faces of the mitochondrial TIM23 complex. Biochim Biophys Acta 1797: 1045–1054 [DOI] [PubMed] [Google Scholar]
- Mokranjac D, Sichting M, Neupert W, Hell K (2003) Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J 22: 4945–4956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller BK, Zaychikov E, Bräuchle C, Lamb DC (2005) Pulsed interleaved excitation. Biophys J 89: 3508–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W, Brunner M (2002) The protein import motor of mitochondria. Nat Rev: Mol Cell Biol 3: 555–565 [DOI] [PubMed] [Google Scholar]
- Okumus B, Wilson TJ, Lilley DMJ, Ha T (2004) Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J 87: 2798–2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pais JE, Schilke B, Craig EA (2011) Reevaluation of the role of the Pam18:Pam16 interaction in translocation of proteins by the mitochondrial Hsp70-based import motor. Mol Biol Cell 22: 4740–4749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Auerbach A, Sachs F (2000) A direct optimization approach to hidden Markov modeling for single channel kinetics. Biophys J 79: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabiner L, Juang B (1986) An introduction to hidden Markov models. ASSP Mag IEEE 3: 4–16 [Google Scholar]
- Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40: 253–266 [DOI] [PubMed] [Google Scholar]
- Rowley N, Prip-Buus C, Westermann B, Brown C, Schwarz E, Barrell B, Neupert W (1994) Mdj1p, a novel chaperone of the DnaJ family, is involved in mitochondrial biogenesis and protein folding. Cell 77: 249–259 [DOI] [PubMed] [Google Scholar]
- Russell R, Jordan R, McMacken R (1998) Kinetic characterization of the ATPase cycle of the DnaK molecular chaperone. Biochemistry 37: 596–607 [DOI] [PubMed] [Google Scholar]
- Schlecht R, Erbse AH, Bukau B, Mayer MP (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol 18: 345–351 [DOI] [PubMed] [Google Scholar]
- Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M (2007) NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol 14: 1196–1201 [DOI] [PubMed] [Google Scholar]
- Sichting M, Mokranjac D, Azem A, Neupert W, Hell K (2005) Maintenance of structure and function of mitochondrial Hsp70 chaperones requires the chaperone Hep1. EMBO J 24: 1046–1056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Rottgers K (2002) Molecular chaperones as essential mediators of mitochondrial biogenesis. Biochim Biophys Acta 1592: 51–62 [DOI] [PubMed] [Google Scholar]
- Weiss C, Niv A, Azem A (2002) Two-step purification of mitochondrial Hsp70, Ssc1p, using Mge1 (His) 6 immobilized on Ni-agarose. Protein Express Purif 24: 268–273 [DOI] [PubMed] [Google Scholar]
- Widengren J, Kudryavtsev V, Antonik M, Berger S, Gerken M, Seidel CAM (2006) Single-molecule detection and identification of multiple species by multiparameter fluorescence detection. Anal Chem 78: 2039–2050 [DOI] [PubMed] [Google Scholar]
- Willmund F, Hinnenberger M, Nick S, Schulz-Raffelt M, Muhlhaus T, Schroda M (2008) Assistance for a chaperone: Chlamydomonas HEP2 activates plastidic HSP70B for cochaperone binding. J Biol Chem 283: 16363–16373 [DOI] [PubMed] [Google Scholar]
- Woo HJ, Jiang JW, Lafer EM, Sousa R (2009) ATP-induced conformational changes in Hsp70: molecular dynamics and experimental validation of an in silico predicted conformation. Biochemistry 48: 11470–11477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai P, Stanworth C, Liu S, Silberg JJ (2008) The human escort protein Hep binds to the ATPase domain of mitochondrial hsp70 and regulates ATP hydrolysis. J Biol Chem 283: 26098–26106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravleva A, Gierasch LM (2011) Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad SciUSA 108: 6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.